1. Introduction and Motivation
Medical Information Retrieval (IR) is crucial for the development of Clinical Practice Guidelines (CPGs), which are essential tools used daily by medical professionals to support clinical decision-making [
1]. Developing these guidelines requires medical experts to process vast amounts of unstructured, domain-specific data [
2]. In this context, Named Entity Recognition (NER) has emerged as a vital tool for enhancing the accessibility and extraction of relevant information from such data [
3]. By identifying Named Entities (NEs) within unstructured, natural language texts, domain experts can more efficiently extract pertinent information for the development of CPGs [
2]. Modern NER techniques, tailored to specific knowledge domains like medicine, necessitate the training of Machine Learning (ML) models with extensive datasets to optimize performance [
4,
5].
Cloud technologies, such as
Microsoft Azure, offer dynamic and scalable infrastructures that facilitate the provisioning and processing of large datasets, essential for training NER models and enabling advanced information management and extraction [
6]. Building on the introduction of NER, ML, and Cloud as foundational techniques for effective IR in the medical field, this research is motivated by various projects.
RecomRatio [
7] is a European research and development project designed to assist medical professionals in decision making by presenting treatment options along with their respective advantages and disadvantages. It leverages the Content and Knowledge Management Ecosystem Portal (KM-EP) Knowledge Management System (KMS) to facilitate evidence-based patient decisions derived from the medical literature. KM-EP was developed in collaboration between the Fernuniversität Hagen’s Faculty of Mathematics and Computer Science, Chair of Multimedia and Internet Applications [
8], and the FTK e.V. Research Institute for Telecommunications and Cooperation [
9]. The system streamlines the acquisition of new knowledge within medical documents by identifying emerging NEs [
10]. The Stanford Named Entity Recognition and Classification (SNERC) [
11] project concentrates on developing a NER system within the realm of Applied Gaming. It democratizes access to ML, enabling non-experts to identify NEs and classify texts using a rule-based expert system. However, SNERC is limited to relying on Stanford CoreNLP as the underlying foundation for NER. Within the scope of the H2020 project proposal Artificial Intelligence for Hospitals, Healthcare & Humanity (AI4H3), a hub-based architecture was proposed [
12]. This architecture is designed not only to process medical records and patient data but also to act as an assistant in explaining medical decisions. It includes a central Artificial Intelligence (AI) hub and a Semantic Fusion Layer to store data and AI models. AI4H3 employs KlinSH-EP, which is based on the KM-EP system [
13]. The Cloud-based Information Extraction (CIE) [
6] project was started in the context of AI4H3 to support a cloud-based NER pipeline, enabling the processing of large datasets. The CIE architecture aims to allow various users to customize cloud resources and train NER models within the cloud environment. Embedded within CIE, the Framework-Independent Toolkit for Named Entity Recognition (FIT4NER) [
14] project seeks to assist medical experts in medical text analysis using different NER frameworks. This approach enables experimentation with multiple frameworks, comparison of results, and selection of the best method for the specific task at hand. Within the context of FIT4NER and CIE, the Cloud Resource Management for Named Entity Recognition (CRM4NER) system [
15] was developed to enable NER model training using cloud resources, with optimization through automated parameter search and context-dependent fine-tuning using a Large Language Model (LLM). However, CRM4NER is currently limited to the AWS Cloud. Additionally, the need to annotate training data in various formats is crucial for enhancing the compatibility of models developed with tools like CRM4NER [
16]. Therefore, this study focuses on streamlining the NER model development process within another widely used cloud environment, Azure, while supporting diverse data formats for NER model training in the medical domain. This leads to the following Research Questions (RQs), explored in both a bachelor’s thesis [
17] and a master’s thesis inspired by CIE and FIT4NER [
18]:
RQ1: How can a system be developed to support medical experts in creating training data and in training NER models within the Azure Cloud environment?
RQ2: How can medical experts be empowered to manage Azure Cloud resources for NER model training?
This work applies the Nunamaker methodology [
19], a well-established systematic approach to developing information systems. This methodology defines specific Research Objectives (ROs) across the phases of
Observation,
Theory Building,
System Development, and
Experimentation. Building on and extending the FIT4NER framework, this study applies these phases to develop and evaluate a cloud-based NER system for medical experts using Microsoft Azure.
Section 2 addresses the observation objectives, offering a critical review of the current state of research and identifying any gaps relevant to this study.
Section 3 focuses on theory building, creating fundamental models that enable medical experts to develop NER models in the Azure Cloud.
Section 4 details the system development objectives, including the creation of a prototype to assist in NER model development within Azure.
Section 5 covers the experimentation phase, discussing the execution and analysis of both quantitative and qualitative evaluations of the system. The concluding
Section 6 summarizes the findings and insights from this research.
2. State of the Art in Science and Technology
This section concentrates on the observation phase and RO1, which involves a “critical review of the current state of research”. The objective is to identify and discuss Remaining Challenges (RCs) in the areas covered by this article.
As part of the FIT4NER project, a process model was developed to assist medical professionals in extracting knowledge for the creation of CPGs using AI [
2]. This extraction relies on NER, which today often employs ML methods [
20]. The phases described in the model, “Data Management & Curation” and “Analytics”, include the development of training data, the evaluation and selection of appropriate NER frameworks, and the training of ML-supported NER models [
2]. Given that such models require large amounts of data, FIT4NER also leverages the CIE project, which provides use cases for cloud-based resources to train the NER models [
6]. However, medical experts face certain challenges when utilizing ML-supported NER: Firstly, generating high-quality training data is a significant challenge [
20]. Robust NER models require high-quality annotated training data [
16,
20]. Medical texts can exist in various formats such as PDF, DOCX, HTML, or plain text [
21,
22]. In addition, there are specialized formats designed for the exchange of medical information, including Digital Imaging and Communications in Medicine (DICOM) [
23], which is used primarily for medical images but also contains text in its metadata. Other common formats include Fast Healthcare Interoperability Resources (FHIR) [
24] and Consolidated Clinical Document Architecture (CCDA) [
25], which both provide standardized structures for organizing and sharing clinical data. These formats support efficient processing and analysis of medical information. However, to be used effectively in NER systems, these data must first be converted, annotated, and adapted to match the format requirements of the selected NER framework. Two widely used formats for this purpose are CoNLL-2003 [
26] and BioC [
27]. The CoNLL-2003 format, developed for a language technology competition, is used for structured annotated linguistic data, while BioC is an XML-based format specifically designed for representing and exchanging biomedical texts along with their annotations. Secondly, configuring different NER frameworks can be a hurdle for medical professionals due to their varying functions and interfaces [
14]. Thirdly, efficiently managing cloud resources for training ML-supported NER models is another significant challenge [
6,
28]. Cloud configuration in various environments such as Microsoft Azure, Amazon Web Services (AWS), or Google Cloud Platform (GCP) is complex and requires specialized knowledge [
28,
29]. Additionally, cost-effective resource management in the cloud is essential to avoid unnecessary expenses [
30]. Fourthly, medical data often reside in knowledge management systems like Clinical Decision Support Systems (CDSSs) [
31]. Therefore, integrating ML training functions for NER models into these systems would be beneficial for efficient use by medical professionals. These challenges are discussed in detail in the following sections.
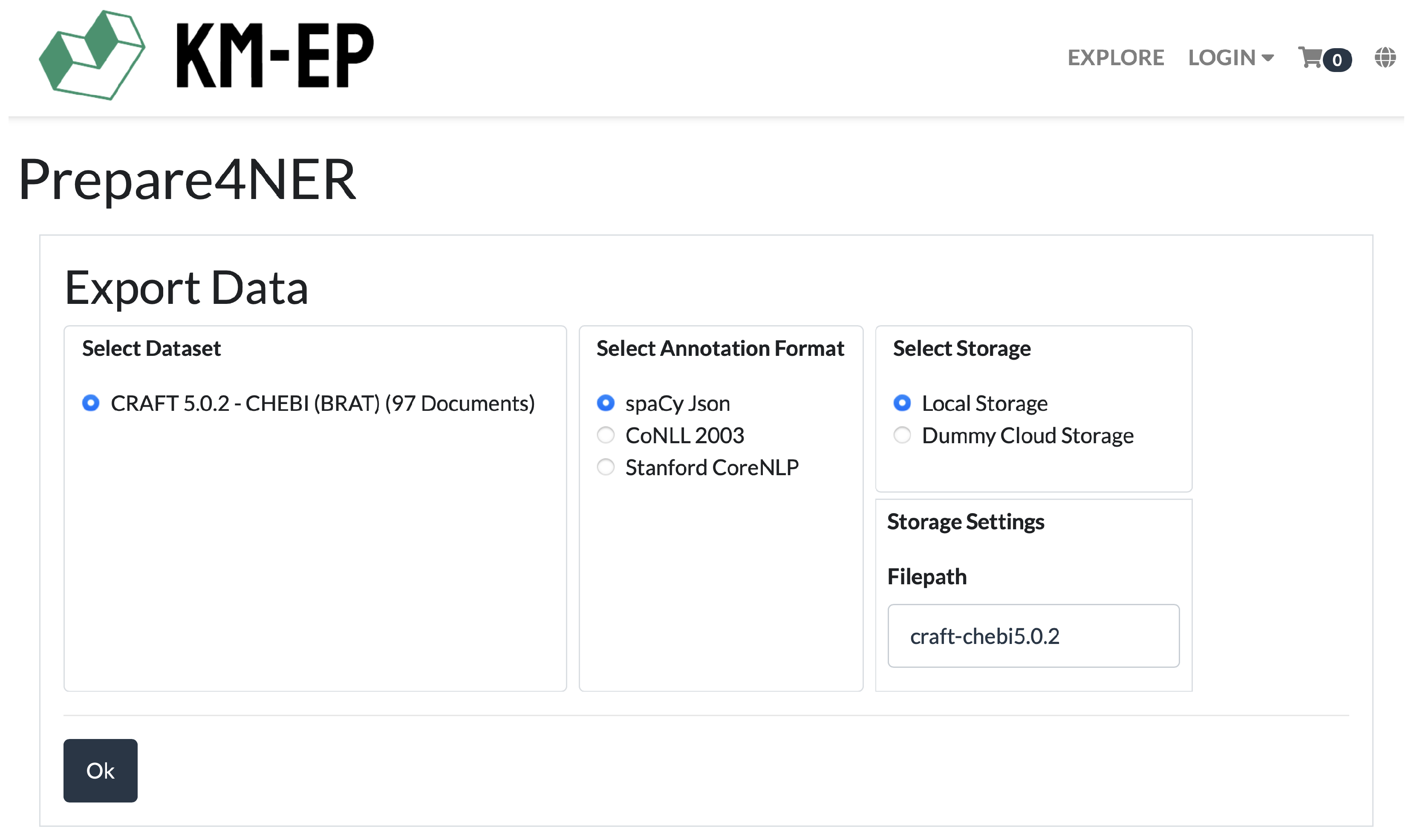
To support medical experts in generating high-quality training data, the
Prepare4NER system was developed [
32]. Prepare4NER assists medical professionals in creating and maintaining extensive training and test datasets across various text formats and NER frameworks [
32] and allows for the easy integration of new NER frameworks through customizable converters. For these reasons, Prepare4NER was selected for creating training data in this work. The first remaining challenge (
RC1) is to integrate Prepare4NER within the Azure Cloud environment. To apply NER for the development of CPGs, various frameworks can be employed [
2]. Recent advancements in transformer models and LLMs like GPT-4 have significantly enhanced NER, document classification, and IR by leveraging large datasets and sophisticated neural architectures [
33]. Platforms like HuggingFace offer pre-trained language models, including those for NER, available for free download [
34]. spaCy is an NER framework that supports these advancements, delivering state-of-the-art performance while being open-source and freely available [
20,
35]. It also allows for the use and fine-tuning of pre-trained models from HuggingFace for NER. Therefore, this work utilizes spaCy and HuggingFace models. The second remaining challenge (
RC2) is to facilitate the use of spaCy with HuggingFace transformer models for medical experts. Several cloud providers offer services for training ML-based NER models [
6], with AWS, GCP, and Microsoft Azure being the most widely used [
29]. These platforms support the full spectrum of services needed for ML projects, including data storage, processing, and AI model development. Microsoft Azure [
36], in particular, is well-suited for integrating cloud computing into NER applications, as it supports the entire machine learning lifecycle. Since AWS was already used for ML-based NER training in the CIE project [
15], this study focuses on Azure to diversify platform usage and leverage its integrated tools and enterprise-level support. An additional emphasis is placed on the accessibility of Azure’s services for medical professionals, ensuring user-friendly tools and seamless access. While the platform offers cost transparency, cost optimization is not a primary concern in this study. The third remaining challenge (
RC3) is to enable medical experts to manage Azure cloud resources and their associated costs. As part of AI4H3 [
37], KlinSH-EP, based on KM-EP, was proposed as a knowledge management system for the medical field [
13]. KM-EP has been used in various research projects, making it well-suited for managing medical information [
7,
11,
13,
38]. It offers a unified user interface and supports essential functions like user and rights management. Integrating cloud-based training features for ML-based NER models into KM-EP would thus be beneficial for medical experts, representing the fourth remaining challenge (
RC4).
To evaluate the prototypes developed in this work, a suitable dataset is required. MIMIC-III [
39] is a comprehensive and freely available database containing anonymized health-related data from more than 40,000 patients. It includes various types of information, such as laboratory results, procedures, medications, and caregiver notes. However, since the corpus is not available in an annotated form for NER, it is not suitable for the experiments in this study. A suitable annotated dataset in the medical domain is the BioNLP Shared Task 2011 Supporting Task-Entity Relation (REL) dataset [
40], which was created as part of the BioNLP Shared Task 2011 and contains annotated texts for various biomedical entities and relationships. However, the dataset is not provided in a standardized format, making its use for NER experiments more complicated compared to other datasets. The Colorado Richly Annotated Full Text (CRAFT) corpus [
41] is available online in standardized formats and comprises 97 medical articles from PubMed with over 760,000 tokens, each meticulously annotated to highlight entities such as genes, proteins, cells, cellular components, biological sequences, and organisms [
41]. Version 5 of the CRAFT corpus includes semantic annotations derived from Open Biomedical Ontologies (OBOs) and is divided into various modules, with this work focusing on the Chemical Entities of Biological Interest (CHEBI) module [
41]. Furrer et al. trained an NER model using the CRAFT-CHEBI corpus and applied various methods [
42], evaluating the models with the OGER system and conducting a comparative analysis of different BiLSTM and Transformer configurations, including BERT [
43] transformers. Their research provides a valuable benchmark for comparison in this work, with the results of NER training on the CRAFT-CHEBI corpus presented in
Table 1. Numerous models from HuggingFace Transformers excel in NER tasks. The first is BERT [
43], which generates deep bidirectional representations from unlabeled text by considering both left and right contexts. This pre-training enables straightforward fine-tuning with an additional output layer, leading to high performance on various tasks, including NER, question answering, and language inference [
43]. RoBERTa [
44] builds on BERT with key differences, such as longer training durations, larger batch sizes, and the removal of the next-sentence prediction objective, allowing for longer text sequences [
44]. GPT-2 [
45] is pre-trained on a large dataset of English text in a self-supervised manner, predicting the next word in a sentence based on continuous text sequences. Finally, ClinicalBERT [
46] is a variant of BERT tailored for clinical text, leveraging its bidirectional transformer architecture and trained on a large corpus of clinical notes [
46]. All these established Transformer models are easily accessible, well documented, and have been utilized in comparable studies, such as those by Furrer et al. [
42]. For these reasons, they were selected for the evaluation experiments in this work.
In this chapter, the current state of the art relevant to this work was critically reviewed. Four remaining challenges were identified. To assist medical experts in creating high-quality training data for ML-based NER models, the Prepare4NER system will be integrated (RC1). Subsequently, medical professionals will be enabled to configure spaCy and HuggingFace transformer models for training with the generated data (RC2). Then, Azure cloud resources will be configured and utilized for the training (RC3). The knowledge management system KM-EP provides a suitable basis for medical data. Therefore, functions for ML-based NER model training by medical experts should be integrated into the system (RC4). The following chapter will describe in detail the creation of suitable models that address all identified remaining challenges (RC1–4).
3. Conceptual Modeling and Design
After conducting a thorough examination of the current state of the art in science and technology, this section focuses on the theory-building phase and aims to address RO2, “The creation of fundamental models for a system that allows medical experts to develop NER models in the Azure Cloud”. This is achieved by taking into account the RCs discussed in
Section 2, reflecting the latest advancements in science and technology. The User-Centered System Design (UCSD) method [
47] is used for design and conceptual modeling, while Unified Modeling Language (UML) [
48] is used as the specification language.
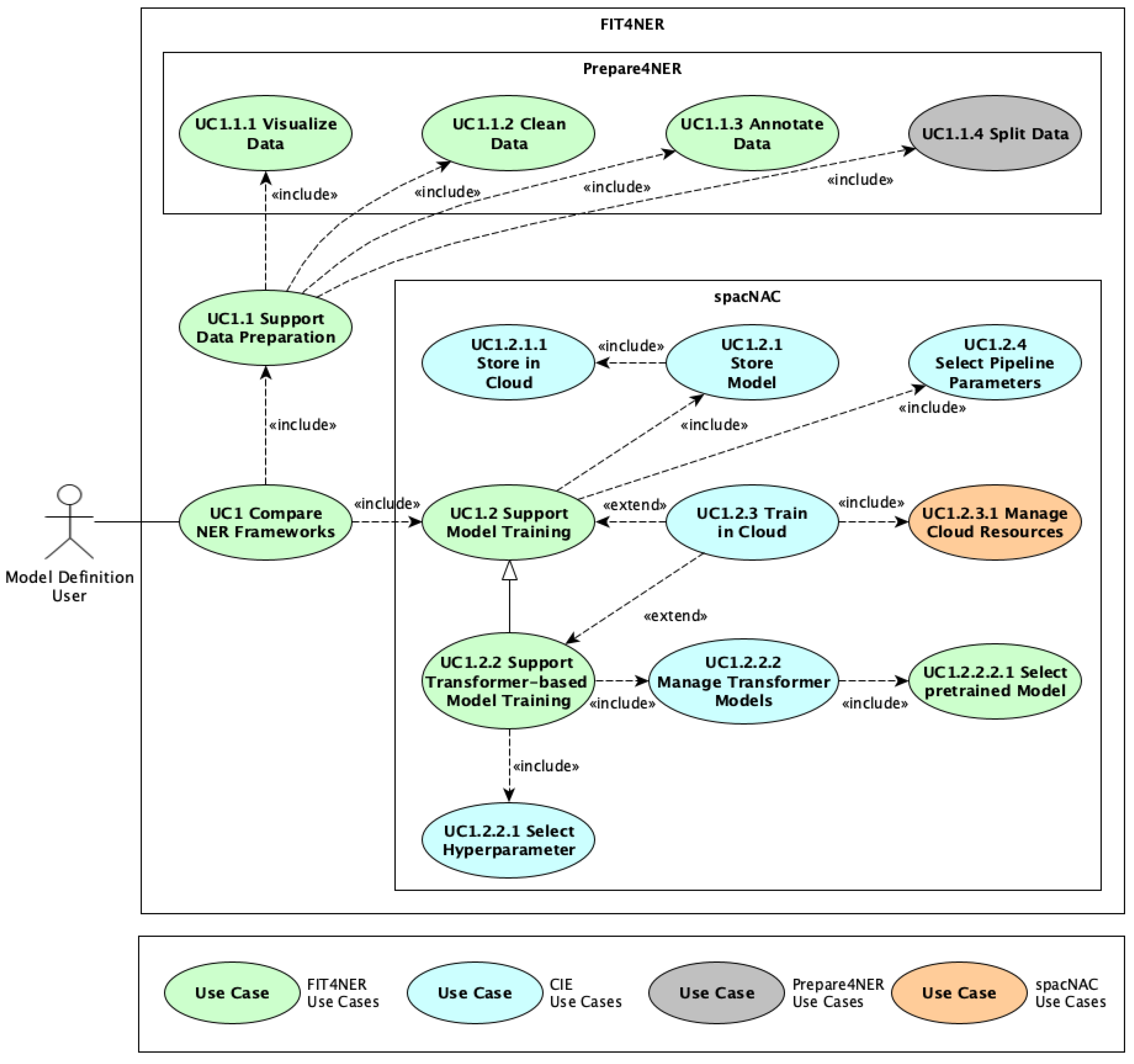
The application context of this work is embedded in the use cases of FIT4NER [
14] and CIE [
6], as illustrated in
Figure 1. FIT4NER use cases are highlighted in green, while CIE use cases are shown in blue. The stereotype defined in FIT4NER, called
Model Definition User, is used, which can be an expert in the relevant knowledge domain but lacks technical knowledge in NER model training [
14]. The initial focus is on the FIT4NER use case “UC1.1 Support Data Preparation”, which leads to the development of a system called “Prepare4NER” [
32].
Figure 1 illustrates the individual use cases of Prepare4NER in gray. This includes the use cases “UC1.1.1 Visualize Data”, “UC1.1.2 Clean Data”, “UC1.1.3 Annotate Data”, and “UC1.1.4 Split Data”. In the use case of “UC1.1 Support Data Preparation”, various document formats are converted to the BioC format [
27]. Data can be visualized using the use case “UC1.1.1 Visualize Data” and cleaned in the “UC1.1.2 Clean Data” use case. In the “UC1.1.3 Annotate Data” use case, domain experts can perform domain-specific annotations or correct existing annotations. The “UC1.1.4 Split Data” use case involves splitting the data into training and testing documents. Additionally, the use case “UC1.2.1.1 Store in Cloud” of CIE [
6] is extended to make the data available for model training in the cloud. Another system that focuses on model training in the Azure Cloud using the spaCy framework is called
“spaCy NER at Azure Cloud (spaCNAC)”. The main use cases in orange in
Figure 1 are “UC1.2.4 Select Pipeline Parameters” and “UC 1.2 Support Model Training”, with the specialization “UC1.2.2 Support Transformer-based Model Training” to allow fine-tuning of transformer models. The use case “UC1.2.2.2.1 Select Pipeline Parameters” supports domain experts in configuring the ML training pipeline. For the training of the models, the “UC1.2.3 Train in Cloud” use case is responsible, which also includes support for domain experts in managing cloud resources in the “UC1.2.3.1 Manage Cloud Resources” use case. The trained models are ultimately stored in the cloud for further use in the respective applications, in the use cases “UC1.2.1 Store Model” and “UC1.2.1.1 Store in Cloud”.
Figure 2 illustrates the primary components of Prepare4NER, divided into a frontend and a backend. For an in-depth explanation of the Prepare4NER theory building, refer to [
32]. The
Data Preparation View (
Figure 2) is the user interface integrated into KM-EP for managing and annotating datasets. The main tasks of the
Data Preparation Subsystem include converting various document formats into the BioC format to establish a uniform data foundation and performing data cleaning to correct inconsistent or erroneous data. Additionally, it facilitates manual annotation by domain experts to label specific NEs for NER model training. To ensure good maintainability, all its components are designed uniformly. They implement an HTTP interface and adhere to a layered architecture. The Data Preparation Controller component delegates requests to two subsystems: the
Document-import Subsystem and the
Corpus-import Subsystem (
Figure 2). The Document-import Subsystem is responsible for parsing and converting documents from various formats into the BioC format, while the Corpus-import Subsystem handles similar tasks for existing corpora. The Controller components,
DocumentImporterController and
CorpusImporterController, employ the Strategy Pattern [
49] (p. 315) in the form of a REST interface to ensure the interchangeability of components as microservices. Thus, extending support for new data formats (such as spaCy JSON, BRAT, etc.) for NER frameworks is limited to the implementation and registration of the corresponding import methods as microservices. Furthermore, the Data Preparation Controller utilizes the Dataset Storage component to store prepared data for further processing. The
Data Provision Subsystem (
Figure 2) manages the conversion of annotated BioC documents into annotation formats compatible with various NER frameworks. It provides the
DataProvisionIF interface, enabling NER model training services such as spaCNAC to consume the annotated documents. As new frameworks emerge, the need to support additional formats may arise. This subsystem mirrors the structure of the Document-import and Corpus-import Subsystems, enabling seamless integration of new strategies through the implementation of converters for these formats. The
Dataset Storage Subsystem consists of the
StorageController, which is responsible for the management of datasets and documents, their BioC representation, as well as the storage and management of NE label collections and their labels (
Figure 2). The
DatasetSplitter Component is responsible for dataset splitting, and the
DataExportController provides the annotated data to the
DataProvision Controller.
The spaCNAC components (
Figure 3) are divided into two microservices: the frontend and the backend. The
Model Definition Manager View (
Figure 3) serves as a central interface for managing model definitions, which set the configuration for NER model training. The view allows users to create new model definitions, modify existing ones, initiate training, or remove model definitions. When creating a model definition, the view guides the user through a step-by-step process. This process includes the selection of a prepared corpus with the
Corpus UI component, the setup of the model architecture and hyperparameters with the
spaCy Configuration UI component, and the configuration of Azure cloud resources with the
Azure Cloud Resources UI component. The
spaCy Azure NER Service Subsystem (
Figure 3) serves as the backend for the Model Definition Manager View and contains the business logic of the system. The service receives all user interactions through a REST API, processes them with the corresponding manager components (
Corpus Manager,
NER Model Definition Manager,
Cloud Training Config Manager), and returns the results to the view. After the completion of the step-by-step process, the model definition is sent to the
NER Model Deployment Service component, which validates the model definition and transmits the configuration to the
Azure ML Pipeline Subsystem (
Figure 3). There, the
Cloud Deployment Manager component generates the required cloud resources using the
Storage Service and
Compute Service components, as well as an Azure ML pipeline that contains the model definition and configuration for training an NER model. The
Model Manager Controller Subsystem (
Figure 3) uses the
NER Model Deployment Service to retrieve the trained NER model along with the metadata generated during training, such as resource utilization or metrics, and store them in the
Model Registry Subsystem (
Figure 3). Additionally, the Model Manager Controller Subsystem serves as the backend for the
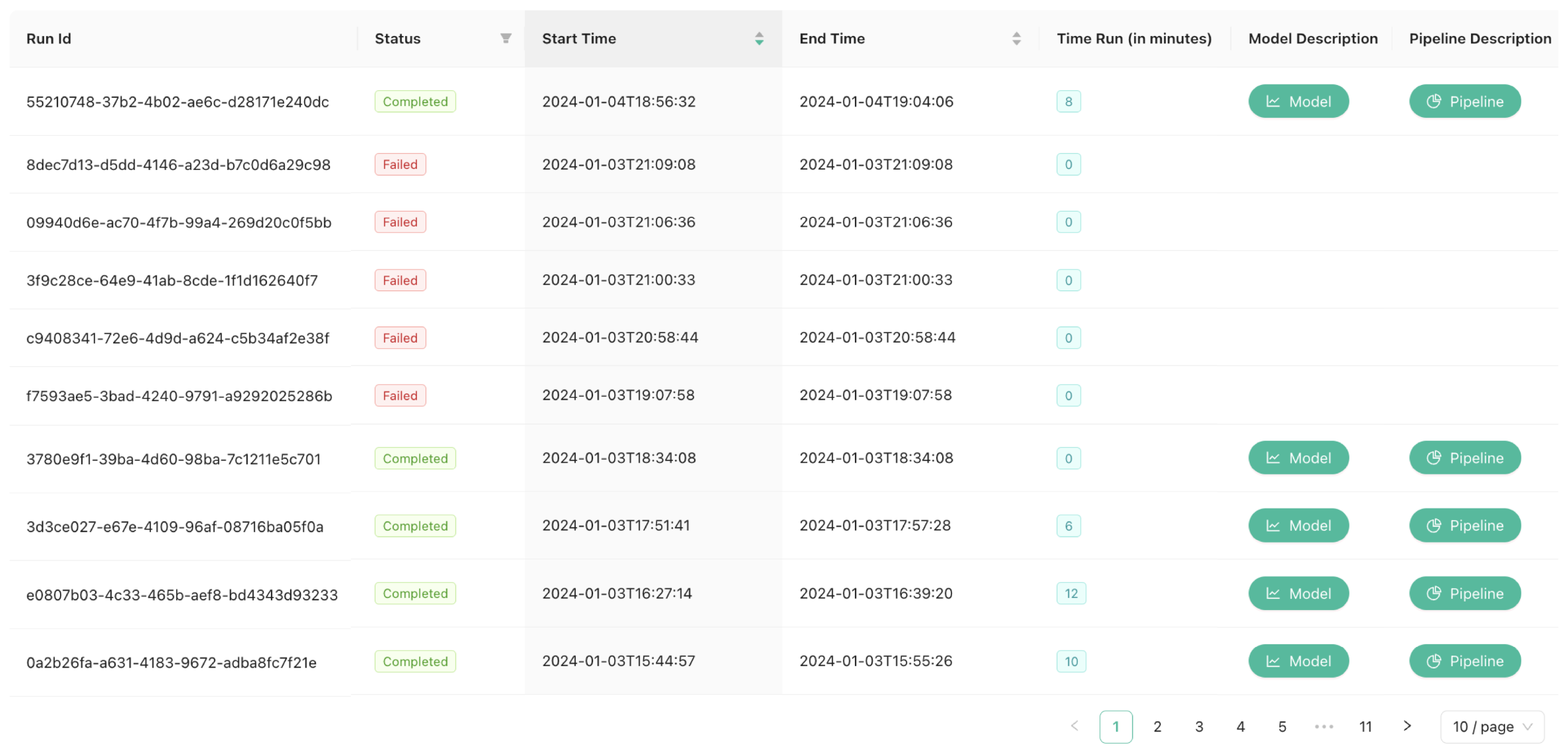
Model Manager View (
Figure 3), which collectively manages all pipelines and NER models. The Model Manager View is responsible for maintaining a comprehensive inventory of all trained NER models and displays all pipelines trained by the NER system. This includes information such as current status, initiation and completion times, total runtime in minutes, model description, and pipeline description.
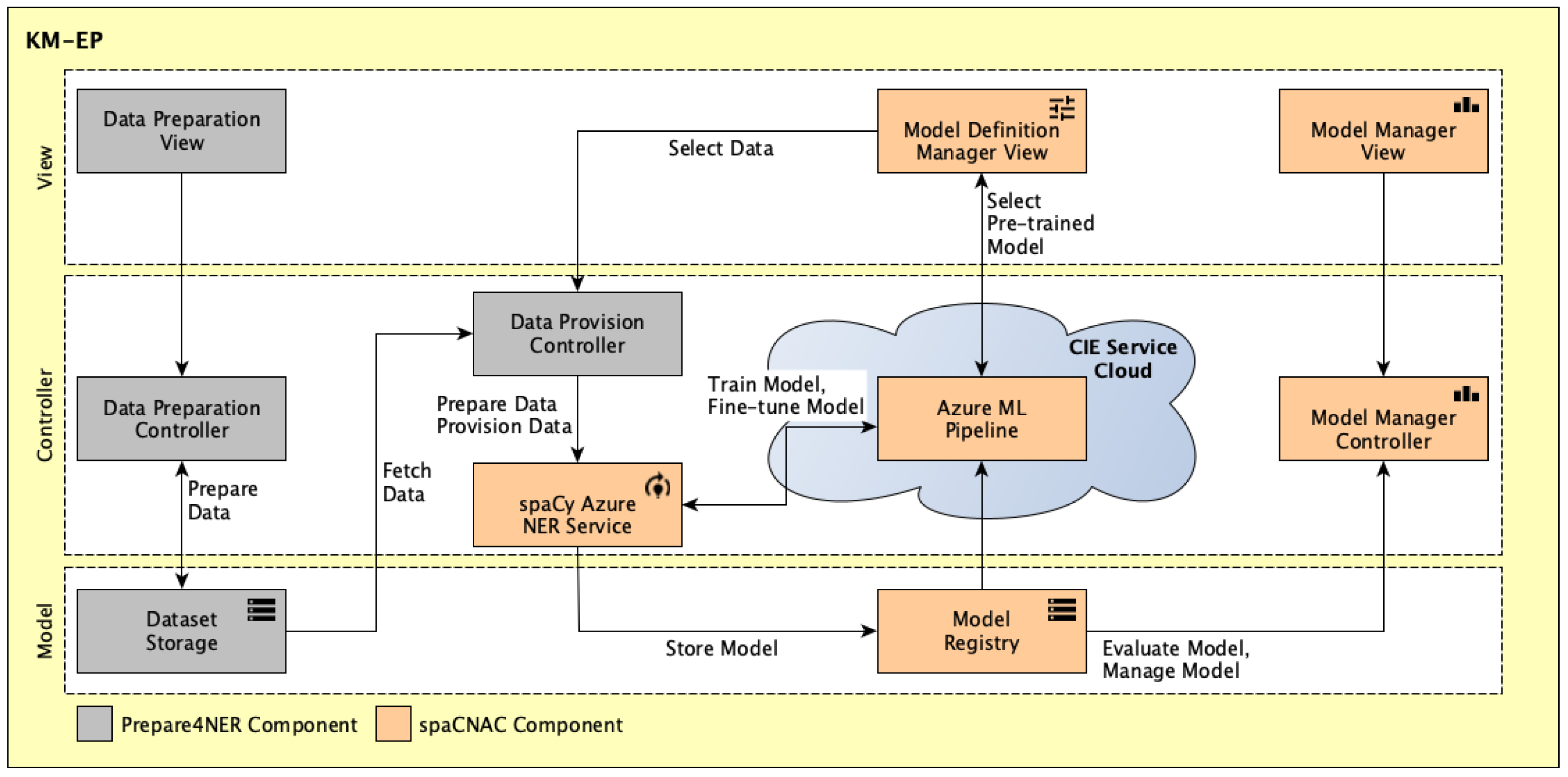
Building upon the CIE [
6] and FIT4NER [
14] general architectures, an integrated architecture is developed that incorporates the Prepare4NER and spaCNAC components. The architecture is depicted in
Figure 4, with the Prepare4NER components shown in gray and the spaCNAC components in orange. The architecture follows a layered structure based on the Model-View-Controller (MVC) pattern [
50], where the user interfaces, business logic, and data storage are separated into distinct components. The
Data Preparation View,
Data Preparation Controller, and
Dataset Storage components implement the “UC1.1 Support Data Preparation” use case and its associated use cases. The
Data Provision Controller component facilitates data provisioning for training using spaCy in the Azure Cloud, which is performed by the
spaCy Azure NER Service component. This is supported by another component called the
Azure ML Pipeline, which runs in the Azure Cloud and performs model training. The component
Model Definition Manager View handles the “UC1.2.4 Select Pipeline Parameter” and “UC1.2.2.1 Select Hyperparameter” use cases. After training, the models are stored using the
Model Registry component and can be retrieved from there for further fine-tuning. The components
Model Manager Controller and
Model Manager View are responsible for managing and displaying the models.
With the description of the theoretical structure in this section, RO2 has been achieved. The following section discusses the prototypical implementation of the developed components and their integration.
4. Implementation
Building on the theoretical framework established in
Section 3, this section delves into the system development of the Prepare4NER and spaCNAC systems. It addresses RO3: The development of a prototype system to assist medical experts in creating NER models in Azure. The chapter begins by describing the implementation of the Prepare4NER components. It delves into the functionality of its components and discusses the technologies, frameworks, and programming languages employed in their development. It also addresses architectural design, dependencies, interfaces, and implementation details, including any specific challenges encountered during the development process. Next, the implementation of the spaCNAC components is explored, following a structure similar to Prepare4NER. The functionality, technologies, implementation details, and challenges specific to each component are described. Subsequently, an outline is given of the integration of the Prepare4NER and spaCNAC systems. The chapter concludes with a comprehensive summary of the effectiveness of the implementation in supporting medical experts and achieving RO3.
Prepare4NER focuses on text annotation and supports different NER frameworks’ data formats. The Prepare4NER components are implemented as separate microservices using Java, Python, Spring Boot, and FastAPI. These microservices feature a uniform design, HTTP interface, and layered architecture to ensure maintainability. The
Data Preparation Controller converts documents to the BioC format, cleans data, and enables expert annotation for NER model training. It provides a
Spring Boot @RestController, which delegates requests to the
DocumentImporter Controller and
CorpusImporter Controller (
Figure 2). These controllers employ the Strategy Pattern [
49] (p. 315), enabling flexible and extensible data processing. For task delegation, the
Data Preparation Controller implements a client class for each addressed controller, using Spring RestTemplate to generate HTTP requests. The endpoints of the subsystems are managed within the configuration of the
Data Preparation Controller. The
StorageController component manages datasets, documents, their BioC representation, and the storage of NE label collections and their labels (
Figure 2). Its
BioCDataset Storage sub-component uses a
LocalStorageProvider class to store BioC collection documents in a configurable folder on a local drive. This architecture supports the implementation of different storage provider classes, enabling the use of alternative storage solutions such as cloud storage. Datasets and documents are managed using Dataset Descriptor and Document Descriptor objects, which are stored in a dedicated database. These descriptor objects contain only metadata about the datasets and documents. The
NELabelStorage sub-component manages the NE label collections used for data annotation. An NE label collection consists of metadata and a list of NE labels. The
DatasetSplitter component, implemented in Python and utilizing the scikit-learn library [
51], is also part of this subsystem. It supports various split strategies, such as Random Split, KFold, and StratifiedKFold [
51], by interacting directly with BioC documents. The
DataProvision Controller (
Figure 2) is responsible for converting annotated BioC documents into the formats required by different NER frameworks. This is completed using its
DataConversion Controller sub-component, which applies the Strategy Pattern [
49] (p. 315). As new NER frameworks become available, the system can easily support them by adding new conversion strategies. Currently, two converters are available: the
CoNLL2003Converter, which transforms data into the CoNLL-2003 format [
26] used by frameworks such as spaCy [
35] and Huggingface Transformers [
34], and the
CoreNLPConverter, which converts data into the Stanford CoreNLP format [
52]. Once converted, the
Dataset Export Controller distributes the data to various storage options, such as local drives or cloud services. To handle large datasets and ensure that the system remains scalable, Prepare4NER should be deployed in the cloud. The Azure Kubernetes Service [
53] offers features such as autoscaling and load balancing, which allow the application to dynamically adjust resources according to demand. However, some challenges may arise when processing very large datasets in real time. To reduce delays, efficient database solutions and caching mechanisms should be used. Additionally, when many users access the system at once (high concurrency), server overloads may occur. In such cases, horizontal scaling and the Azure Load Balancer can help distribute traffic evenly. Future work includes experiments to test scalability under different conditions and to develop strategies to address these challenges. The
DataPreparation UI is integrated into KM-EP and implemented using PHP Symfony controllers and Twig templates. The additional
DocumentEditor UI component, developed independently from KM-EP using the Angular framework in TypeScript, enables domain experts to annotate texts with predefined NE labels. This component also supports nested and overlapping annotations. Originally from a previous project, its integration into KM-EP is loosely achieved through a custom pop-up window. For a detailed description of the Prepare4NER implementation, refer to [
32].
spaCNAC empowers domain experts to develop NER models using spaCy in the Azure Cloud, leveraging the data created with Prepare4NER. The frontend is developed using ReactJS, HTML, and TypeScript, while the backend is implemented in Python and FastAPI. The backend interacts with Azure Cloud Services. Both the frontend and the backend are packaged in Docker containers. The Model Definition Manager View (
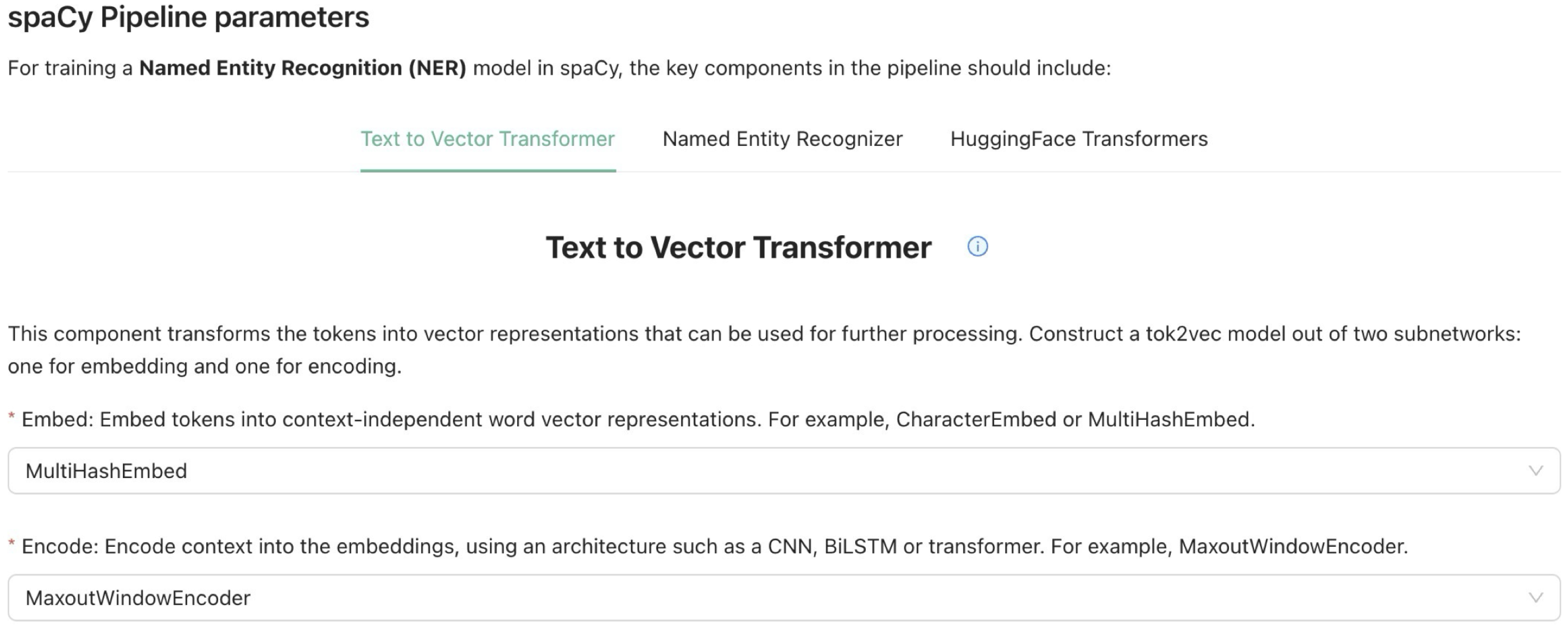
Figure 3) is built on React, integrates data from the spaCy Azure NER Service, and enables navigation between different sub-views for creating or modifying model definitions. The view takes the user through a step-by-step process when creating a model definition. In the first step, the user selects a prepared corpus that has been created, converted into the spaCy format, and divided into training and testing data using Prepare4NER. These data are then uploaded to an Azure Blob container. In the next steps, users set up the spaCy pipeline, choosing from various model types (
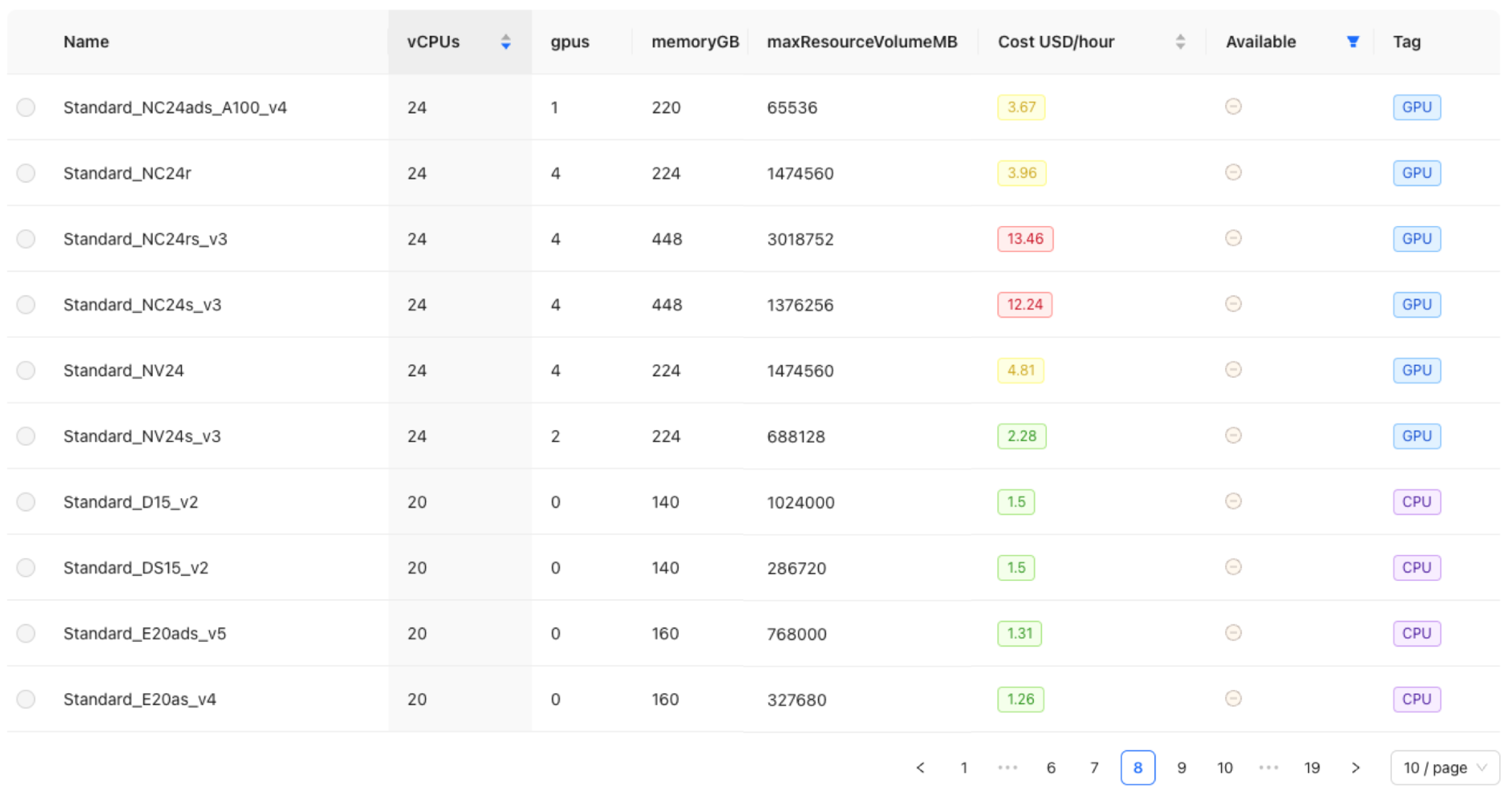
Figure 5) such as “Text to Vector Transformer”, “Named Entity Recognizer”, and “HuggingFace Transformers”. Specific options for embedding and encoder models are available, such as “spacy.MultiHashEmbed.v2” or “spacy.MaxoutWindowEncoder.v2”. Notably, the system provides an overview of available HuggingFace Transformers models to simplify the selection of a pre-trained model for NER training. Users can adjust hyperparameters that significantly influence the model’s performance through a dedicated section. This allows for the modification of six key hyperparameters. For experienced users, there is also an option to make advanced adjustments directly in the spaCy configuration file. After configuration, users select the cloud resources for training, with only compute clusters currently available. A tabular overview facilitates this selection by listing specifications and costs per hour for different clusters, along with sorting and filtering options. The clusters are categorized into three color-coded groups to aid in budget- and requirement-based selection (
Figure 6):
Green: Costing less than $3 per hour, an economical choice
Yellow: Mid-range option between $3 and $10 per hour
Red: High-end selection that costs more than $10 per hour
Once a suitable compute cluster is selected, the user can initiate the training process. The parameters chosen by the user for the training process are saved by the
NER Model Definition Manager in a MySQL database and then passed to the
Cloud Deployment Manager via the
NER Model Deployment Service (
Figure 3). The
Cloud Deployment Manager provisions an Azure ML pipeline and the necessary cloud resources in an Azure workspace for the training process, such as Azure Blob Storage. This requires three key input components: a spaCy configuration file, the training corpus, and the test corpus. After successfully training the NER model, the spaCy models are stored in the
Storage Service (Azure Blob Storage), designated as the output destination within the pipeline. To monitor the model training process, bidirectional communication is established between the Model Definition Manager View and the Azure ML pipeline using WebSockets. This allows for tracking of key performance indicators such as CPU usage, read/write operations, and disk utilization. Upon completion of the training, the trained NER model is stored in the
NER Model Registry.
The
NER Model UI (
Figure 7) maintains an inventory of all trained NER models in the
NER Model Registry and displays pipeline details, including status, initiation and completion times, runtime, and descriptions. Specifically, the model description contains the metrics of the trained NER models, including Precision (P), Recall (R), and F-Score (F1) [
54]. The system allows users to download training results and trained models. Furthermore, it provides detailed descriptions of individual pipelines along with Key Performance Indicators (KPIs) for monitoring resource usage, identifying potential bottlenecks, and optimizing overall system performance. These KPIs include:
CPU utilization: The proportion of total CPU resources consumed by a specific process or collection of processes. Regular monitoring helps identify bottlenecks and optimize resource allocation.
CPU memory utilization: Percentage of total memory resources consumed by one or more processes. High utilization may indicate optimization needs.
Disk read/write operations: Quantifies the speed at which data is read from or written to the disk, which is crucial for identifying potential bottlenecks.
Disk usage: Amount of disk space consumed by a process or group of processes. Regular monitoring is important to ensure adequate availability and to avoid issues related to disk space exhaustion.
By utilizing these performance metrics, users can gain a comprehensive understanding of the cloud resources consumed during the NER model training process. These insights aid in future decision-making regarding model training exercises by improving pipeline efficiency and optimizing resource allocation, thus enabling more effective and efficient resource utilization.
Next, the implementation of the integrated overall system, combining the KM-EP [
13], Prepare4NER [
32], and spaCNAC systems, is summarized. KM-EP was developed using Symfony and PHP and provides basic functions such as user management and document management. It allows the integration of additional systems through HTTP and REST interfaces. The microservice architectures of Prepare4NER and spaCNAC allow seamless integration into KM-EP. The components were brought together into a coherent system using Docker Compose, which simplifies configuration and deployment in containers, increasing scalability and portability. The integration of Prepare4NER into KM-EP was carried out using Symfony controllers and Twig templates. The controllers process user actions and manipulate the data model. KM-EP forwards requests to the corresponding microservices. The designed data models are implemented as PHP classes and are passed to templates after subsystem calls, which visualize the data. In KM-EP, Prepare4NER is accessible through the navigation bar, while the DocumentEditorUI is opened in a new browser tab. spaCNAC uses Python and the FastAPI framework for backend operations, and TypeScript with React for a reactive frontend. The integration of spaCNAC into KM-EP is carried out using an iframe to seamlessly embed spaCNAC into KM-EP, as different technologies are used. In KM-EP, spaCNAC is accessible through the KM-EP navigation menu and the entry “Named Entity Recognition spaCy-Azure”, which provides access to the main functions of the system. A distinction is made between model definition and model manager. The “spaCy NER Model Definition Manager” is integrated into the model definition page, and the “spaCy NER Model Manager” is integrated into the model manager page of KM-EP. To transfer data from Prepare4NER to spaCNAC, it was necessary to develop a new converter that transforms data from the BioC format to the spaCy JSON format. This shows that extending the system to accommodate new NER frameworks is straightforward using the Strategy Pattern [
49] (p. 315). The source code for the converter is shown in Listing 1. First, it is checked whether an empty document is present. The BioC document is read, and a spaCy-compatible object is created. The existing annotations are stored in a list of entities within the object after a validity check. The document is then serialized into JSON and returned. The prototype captures, logs, and reports errors that occur, such as those due to invalid or incomplete data, as a
RuntimeException. For production use, additional edge cases should be covered, and the behavior should be verified through unit tests. Very large documents may also cause problems due to high memory usage. This enables the export of the Prepare4NER datasets to spaCNAC, as illustrated in
Figure 8.
| Listing 1. Java Implementation of the SpacyJsonConverter Class |
@Service
public class SpacyJsonConverter {
private static final Logger logger = LoggerFactory.getLogger(SpacyJsonConverter.class);
public byte[] convert(String documentXml) {
if (documentXml == null || documentXml.trim().isEmpty()) {
throw new IllegalArgumentException("Input XML must not be null or empty.");
}
try {
// Parse the input BioC XML into a BioCDocument
BioCDocumentReader bioCDocumentReader = new BioCDocumentReader(new StringReader(documentXml));
BioCDocument bioCDocument = bioCDocumentReader.readDocument();
// Extract plain text and annotation entities from the BioCDocument
String text = extractText(bioCDocument);
List<Annotation> annotations = extractAnnotations(bioCDocument);
// Initialize spaCy-compatible document
SpacyDocument spacyDocument = new SpacyDocument(text);
for (Annotation annotation : annotations) {
int begin = annotation.getBeginIndex();
int end = annotation.getEndIndex();
// Validate annotation offsets
if (begin >= 0 && end <= text.length() && begin < end) {
spacyDocument.entities.add(new Object[]{begin, end, annotation.getLabel()});
} else {
logger.warn("Skipping invalid annotation: {}", annotation);
}
}
// Wrap in a list as spaCy expects a list of documents
List<SpacyDocument> spacyDocuments = new ArrayList<>();
spacyDocuments.add(spacyDocument);
// Convert Java object to JSON string with pretty printing
ObjectMapper mapper = new ObjectMapper();
String spacyJsonString = mapper.writerWithDefaultPrettyPrinter().writeValueAsString(spacyDocuments);
// Return UTF-8 encoded byte array
return spacyJsonString.getBytes(StandardCharsets.UTF_8);
} catch (Exception e) {
logger.error("Failed to convert BioC document", e);
throw new RuntimeException("Error converting BioC XML to spaCy JSON", e);
}
}
} |
In this chapter, the implementation of the Prepare4NER and spaCNAC systems, as well as their integration into KM-EP, has been detailed, successfully completing RO3. The next chapter will now present experiments that will verify whether the created systems meet the set requirements.
5. Evaluation
The previous chapter described the implementation of the two prototypes, Prepare4NER and spaCNAC, as well as their integration. This chapter focuses on the experimental goal of RO4: The execution and analysis of quantitative and qualitative evaluations of the developed system. Qualitative and quantitative evaluation methods were employed. For qualitative experiments, two different approaches were used: the Cognitive Walkthrough (CW) [
55], a method for evaluating user interfaces, and the Software Technical Review (STR) process, an approach to improving software quality that is also established as an IEEE standard (IEEE Std 1028-2008) [
56]. A CW involves simulating and analyzing the cognitive processes of a user interacting with the user interface. Initially, tasks with specific goals and required action sequences are defined. Subsequently, an expert panel evaluates the expected user interaction, considering criteria such as action availability and labeling, the likelihood of correct action selection, and action complexity. In contrast, the STR process is a technique to analyze the technical aspects of a software product. A lead engineer organizes a review meeting and provides structured inputs, including information such as the software product, review objectives, planned functionality, and a description of the review process. During the review meeting, a group of experts examines the software product and documents all findings and anomalies. The quantitative experiments were evaluated using the metrics P, R, and F1 [
57]. Precision measures how accurately users have selected the correct items. Recall expresses completeness, and the F1-Score combines P and R to measure the accuracy of the results. The chapter begins by detailing the experiments conducted with Prepare4NER. Then, it outlines the experiments performed with the spaCNAC prototypes. This is followed by an overview of the experiments that involve the integration of the two systems. The chapter concludes with a summary of the results in relation to achieving RO4.
For the initial CW evaluation of Prepare4NER [
32], several meetings were held, involving a doctor, two Ph.D. students, and a bachelor’s student, all conducting research in the fields of Natural Language Processing (NLP) and NER. Tasks were prepared for these meetings, covering the entire workflow from importing, annotating, and splitting documents in the data preparation phase to exporting datasets for a specific NER framework and storage providers in the data provisioning phase. During the evaluation, usability insights were discussed and documented, resulting in a total of 12 findings. A key insight was that additional instructions for users should be introduced. The participants of the CW sessions recognized the need for instructions or help texts for several tasks to assist users in performing them more effectively. Similarly, the creation of label collections and labels should be simplified. Furthermore, the support for additional annotation features was emphasized, as the current state of the annotation tool does not support discontinuous, nested, or overlapping annotations. Despite these insights, which are intended for future improvements, the system was deemed suitable for the intended tasks. In addition to the initial CW, Prepare4NER was also assessed using a STR in a single review meeting. This evaluation involved five sets of inputs and included a PhD student with expertise in modeling microservice-based architectures and a bachelor’s student in computer science. The outcome of the STR confirmed that Prepare4NER and its architecture are suitable for the intended purpose. In particular, the strategy pattern facilitates the intended extension of the system for future document formats, NER frameworks, and storage solutions. The insights from the STR session mainly suggest improvements such as better documentation of the HTTP interface of the strategy components to make it easier for system developers to implement new components. In addition, there is a proposal for a self-registering approach for new components to avoid the need to change the system configuration when new strategies are added. Another system improvement would be to allow system developers to test new components before integrating them into the system.
spaCNAC was evaluated both quantitatively and qualitatively. First, a quantitative analysis was performed, followed by a qualitative one. Each evaluation description begins with an explanation of the evaluation process and concludes with an in-depth analysis of the results. The CRAFT-CHEBI corpus was selected, as described in
Section 2, because it is already annotated in standardized data formats for NER. Future work can extend quantitative experiments to additional corpora, such as the BioNLP Shared Task 2011 Supporting Task-Entity Relation (REL) dataset [
40]. The quantitative evaluation of spaCNAC was conducted using the CHEBI annotations of the CRAFT corpus and the metrics P, R, and F1. The CRAFT corpus with CHEBI annotations was converted to the BRAT format [
58] using Clojure Boot according to the official documentation [
59] and then imported via the Prepare4NER import function. The split ratio for training and testing data was set in Prepare4NER so that 100% of the data was used for training, as spaCNAC later splits the dataset into training and testing data itself. The dataset was then exported for spaCNAC using the SpacyJsonConverter (see Listing 1,
Figure 8). Since Prepare4NER exports the dataset as individual documents, they had to be consolidated. The JSON documents were combined into a list using
jq and saved in a single file (see Listing 2). Then, this file was imported into spaCNAC. Within spaCNAC, the Azure compute instances shown in
Table 2 were used for training, some of which offer GPU support. Three quantitative experiments were conducted using four different Transformer models from HuggingFace, as selected in
Section 2: BERT [
43], RoBERTa [
44], GPT-2 [
45], and ClinicalBERT [
46]. The models were trained on the CRAFT CHEBI corpus and partially compared with the work of Ferrur et al. [
42], who also used BERT Transformer models for the NER task.
| Listing 2. Consolidation of CRAFT-CHEBI Documents for spaCNAC using jq |
# The command uses jq to consolidate multiple JSON files into a single array.
# The ’-s’ option reads all input files into an array.
# ’[.[][]]’ accesses the contents of each JSON file and combines them into a single output.
jq -s ’[.[][]]’ prepare4ner-localstorage/undefined/* \
> craft-chebi5.0.2-spacy.json
# The resulting consolidated JSON array is saved to ’craft-chebi5.0.2-spacy.json’.
|
In the first experiment, the impact of computational resources on the performance of spaCy NER models was investigated. The models were trained using the CRAFT corpus on three Azure CPU machines with different specifications: Standard_A2_v2, Standard_D11_v2, and Standard_A2m_v2. The results showed that training on the Standard_A2_v2 machine was aborted due to insufficient memory, while the other two machines successfully completed the training. The models trained on the Standard_D11_v2 and Standard_A2m_v2 machines exhibited significant performance differences. The model trained on the Standard_A2m_v2 machine achieved higher Precision, Recall, and F-Score values than the model trained on the Standard_D11_v2 machine, although it took twice as long to train. This suggests that more powerful computational resources can lead to better NER models, albeit requiring more time. The results are shown in
Table 3.
In the second experiment, the HuggingFace Transformers models BERT, GPT-2, ClinicalBERT, and RoBERTa were used, along with the Standard_NC4as_T4_v3 Azure-VM for training. Models 1 and 2 (BERT and GPT-2) achieved a Precision of 0.84, with GPT-2 showing a higher Recall and F-Score. Despite the longest training duration (129 min), ClinicalBERT achieved the highest precision (0.86), although its recall was only slightly better than BERT’s and lower than GPT-2. RoBERTa stood out: despite the shortest training duration (93 min), it achieved a high Precision of 0.85, the highest Recall (0.82), and the highest F-Score (0.83). This suggests that RoBERTa offers a more balanced performance and efficient training time. Comparing the models used in this study (BERT, GPT-2, ClinicalBERT, and RoBERTa) with those from the referenced research [
42] (OGER, BiLSTM, BERT-IDs, BERT-Spans+OGER, and BERT-IDs+Spans+OGER), it is clear that the models in this work are superior. Every model evaluated in this experiment exceeds the performance of the best model from the research study, BERT-Spans+OGER, which integrates IOBES predictions with OGER annotations in a pipeline fashion. In summary, RoBERTa appears to be the most efficient and balanced model based on these metrics, although all models performed relatively well. However, the choice of model still depends on the specific requirements of a given task.
Table 4 presents the results of the model training. After extensive research, no comparable studies were found that address the training of the CRAFT-CHEBI corpus in the cloud. However, the overarching CIE approach has been utilized to train the GENERMED dataset in the AWS and Azure clouds [
6]. Although the results are not directly comparable, it is evident that training times are at a similar level. Since spaCNAC and prepare4NER are embedded in the FIT4NER [
14] and CIE [
6] projects, the underlying architectures also facilitate transfer to other domains of knowledge, such as history or applied games [
11], as well as the use of other cloud providers such as GCP or AWS. This flexibility fundamentally allows for the avoidance of vendor lock-in.
In the third and final experiment, the learning rate hyperparameter used for training with the RoBERTa model from the HuggingFace library was refined. Three learning rates were tested:
The results indicate that the learning rate significantly impacts the Precision, Recall, and F-Score of the NER models with RoBERTa. The highest Precision was achieved with a learning rate of
The best balance between Precision and Recall, as indicated by the F-Score, was achieved with a learning rate of
This learning rate was also the most efficient, measured by task duration. Comparing the results of this experiment with those from the research [
42], the RoBERTa model with a learning rate of
outperforms all other models. The highest F-Score achieved in this experiment, 0.8337, exceeds the best F-Score of 0.7700 from other studies, which was achieved by the BERT IDs+spans+OGER model. Even the RoBERTa model with the lowest F-Score in this experiment (at a learning rate of
) surpasses all the other models in the research. Thus, the RoBERTa model, with its adjustable learning rate, is superior. The results are shown in
Table 5.
Following the presentation of the three quantitative experiments, the results of the qualitative evaluation will be presented next. spaCNAC aims to facilitate the development of NER models by leveraging spaCy, HuggingFace Transformers, and Azure cloud services within KM-EP. The primary objective of this qualitative evaluation is to confirm user-friendliness and performance. This will be achieved through detailed user feedback on the prototype’s features and functionality via a CW [
55]. It is essential to capture users’ perceptions of whether they find it straightforward to train new NER models, apply Azure cloud services, and evaluate the effectiveness of the prototype’s KM-EP integration. It is important to highlight that the system is designed to support diverse user groups, including medical experts and NER specialists.
The CW session was meticulously prepared. First, tasks to be performed during the CW session were defined. These tasks, listed in
Table 6, cover areas such as NER model definition, training, and management. Participants in the walkthrough session represented various user groups, including a Senior Data Scientist and PostDoc, a Data Scientist who is also pursuing a PhD, and two Senior Software Engineers. Despite their backgrounds as data scientists and NLP experts, participants were encouraged to adopt the perspective of naïve or domain users. This approach was chosen to ensure that the feedback received is as objective as possible, providing an overview of the prototype’s usability, feasibility, effectiveness, and efficiency. To ensure that all participants have a clear understanding of the project’s context, the central ideas and concepts are presented at the beginning. Participants were then guided through the application, gaining insights into the system’s functionality and design considerations.
During the evaluation, usability findings were discussed and documented, resulting in a total of seven findings (
Table 7). Initially, participants did not understand the purpose and function of the “Edit NER Model Definitions” page (CWF1). A potential solution would be to integrate a detailed description of model definitions into the user interface. The participants were also uncertain about the structure and use of the datasets provided by the system (CWF2). A detailed description of these datasets, including their format, would help users utilize them more effectively. Regarding the language selection for NER model training, it was suggested to optimize the process by automatically filling the language field in Step 2 based on the corpus selected in Step 1 (CWF3). Additionally, participants found the navigation bar confusing due to overloaded step descriptions (CWF4). Shorter and more precise step descriptions, along with a visual representation of the steps, would improve clarity. The layout of the NER Model Definitions page was considered too cluttered by users (CWF5). They had difficulties with the functionality of the “Save” and “Cancel” options during the editing process and found navigation between steps unclear. Suggested improvements include introducing “Next” and “Back” buttons for seamless navigation, as well as a clear display of the overall saving status during the editing process. Overall, users felt that the amount of information presented was sometimes overwhelming (CWF6). A concise and clear presentation of the necessary information could help balance providing sufficient data and avoiding information overload. Lastly, users desired a notification popup to confirm when the data was successfully saved (CWF7). Based on the feedback received, several adjustments were made to the system. For CWF1, clear descriptions were added for both the Model Manager and Model Definitions in their respective UI areas. The information on the provided datasets was enhanced by specifying the data type and language of each dataset (CWF2). This update also enables automatic filling of the language field based on the selected dataset, as noted in CWF3. Navigation was improved by relocating the step descriptions to their respective pages, as suggested in CWF4. Finally, in response to CWF7, a notification feature was added that activates each time the “Save” button is clicked. The issues identified in CWF5 and CWF6 remain open and are planned for future improvements. Overall, users provided positive feedback on the tasks performed.
A total of three distinct quantitative and qualitative experiments were conducted for the evaluation of Prepare4NER and spaCNAC. The detailed evaluation of Prepare4NER is described in [
32]. The qualitative experiments identified several usability improvement opportunities for the systems, some of which have already been implemented. Overall, the systems were deemed highly effective for assisting medical experts in creating ML-based NER models in the Azure Cloud. The quantitative experiments also demonstrated that Cloud-based training produces NER models with performance at a state-of-the-art level. In summary, across all experiments, RO4 was achieved.