From Pixels to Diagnosis: Implementing and Evaluating a CNN Model for Tomato Leaf Disease Detection
Abstract
1. Introduction
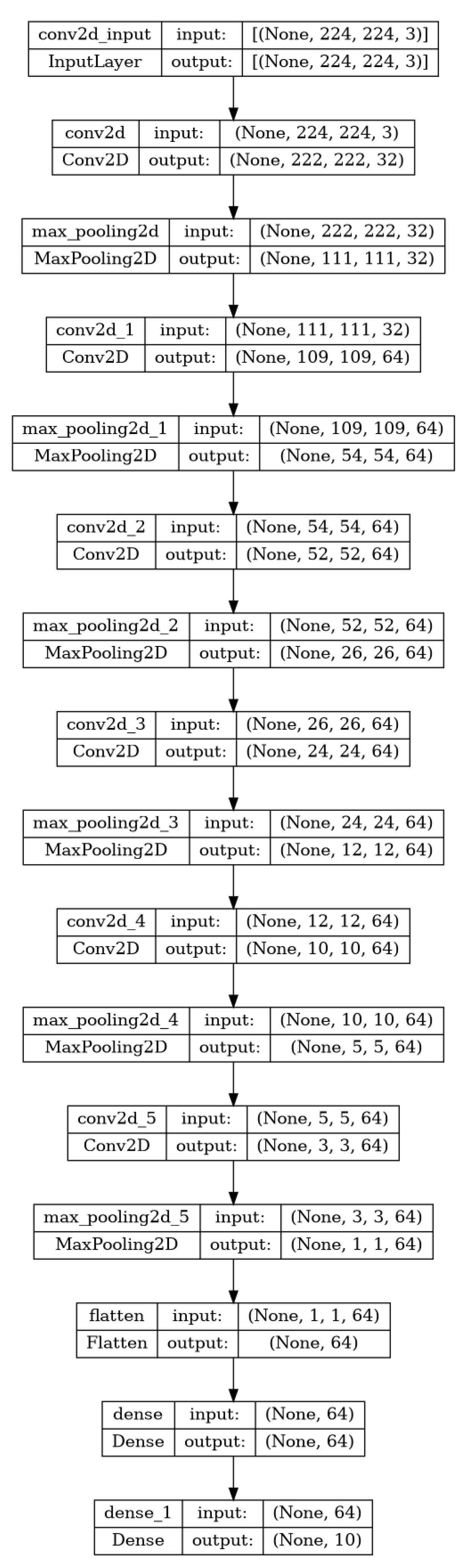
- Develop a Custom CNN: CNNs are a class of deep learning models particularly effective for image identification tasks [2]. They use convolutional layers to extract features from images, followed by fully connected layers to make predictions. Our custom CNN model was trained on a large dataset of tomato leaf images to effectively distinguish healthy leaves and those affected by disease.
- Fine-tune pre-trained models: VGG16 and VGG19 are pre-trained models used in image classification tasks. They are based on deep convolutional architectures trained on the ImageNet dataset. By fine-tuning these models on our specific dataset, we aimed to exploit their pre-learned features and adjust them for the task to identify diseases in tomato plants.
- Performance evaluation: Evaluate the performance of the proposed CNN, VGG16, and VGG19 models and compare their performance metrics.
2. State of the Art
2.1. Machine Learning in Tomato Leaf Disease Detection
2.2. Technologies and Dataset Used
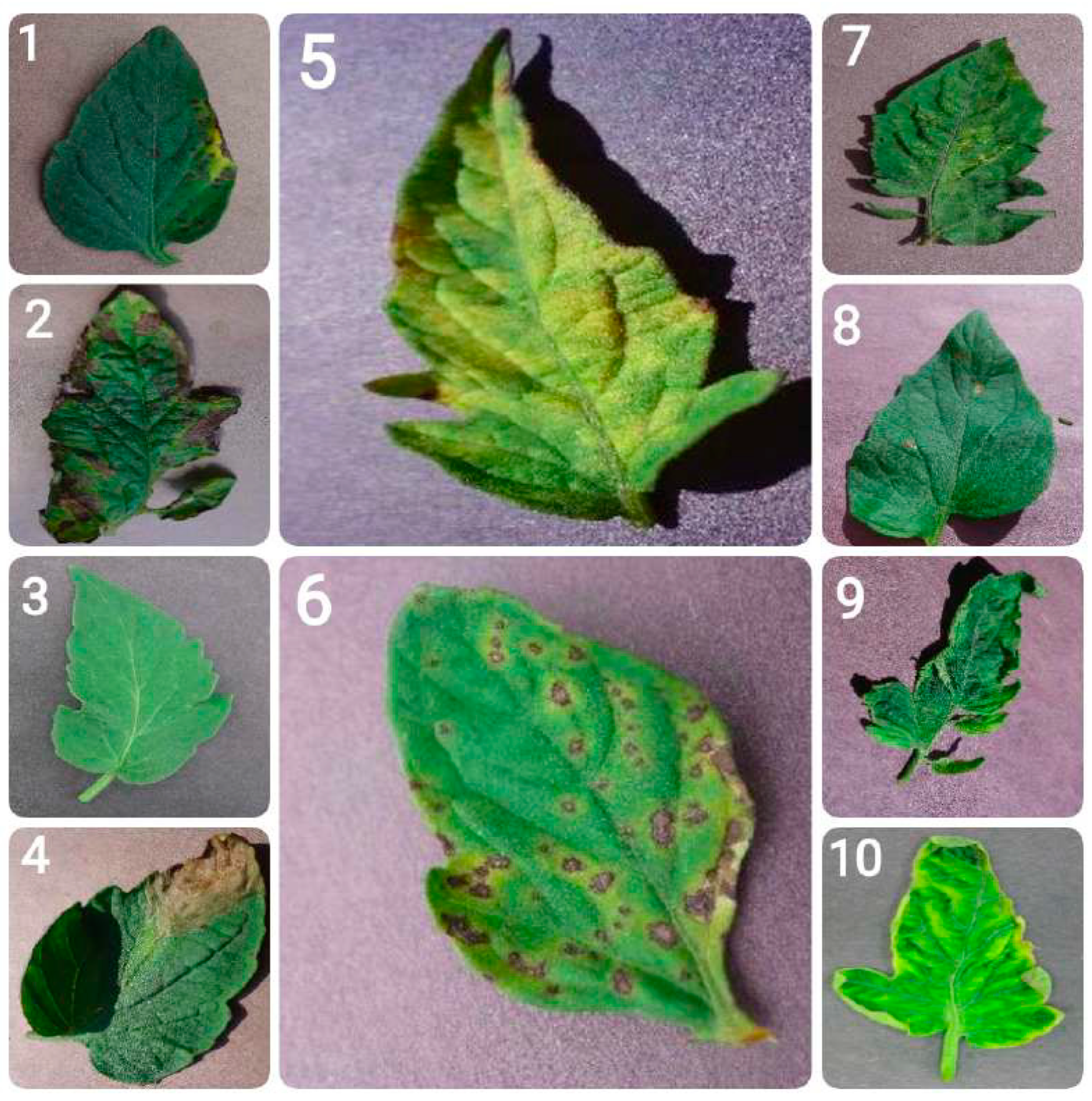
- Bacterial spot: Bacterial spot is caused by the bacterium Xanthomonas campestris pv. vesicatoria, and appears as small, water-soaked spots on leaves. Over time, these spots turn brown and necrotic. In severe cases, the infection can result in leaf drop and reduced fruit quality.
- Early blight: Early blight, caused by the fungus Alternaria solani, is marked by the appearance of concentric rings resembling a ”bullseye” pattern on older leaves. It typically begins as small, dark spots and can result in extensive defoliation, impairing the plant’s ability to carry out photosynthesis.
- Healthy leaves: Tomato leaves in good health exhibit a vibrant green color with a smooth and uniform texture. They show no signs of spots, lesions, discoloration, or pest presence. They are free from wilting or deformities, reflecting proper plant care and maintenance.
- Late blight: This destructive disease, caused by the oomycete Phytophthora infestans, can affect all parts of the tomato plant. It appears as water-soaked lesions that rapidly turn brown and under conditions favorable to the pathogen can lead to the plant’s total collapse.
- Leaf mold: Caused by the fungus Passalora fulva, leaf mold is characterized by yellow spots on the upper surface of leaves and a velvety, olive-green mold on the underside. It thrives in high humidity conditions and can significantly hinder the plant’s ability to photosynthesize.
- Septoria leaf spot: This disease, caused by the fungus Septoria lycopersici, is characterized by small, round spots with dark borders and grayish centers. It mainly affects the lower leaves and often leads to premature defoliation.
- Spider mites (two-spotted spider mite): The two-spotted spider mite, Tetranychus urticae, feeds on leaves, causing stippling and a bronzed appearance. In cases of severe infestation, webbing may form, leading to extensive leaf damage and affecting the plant’s strength overall.
- Target spot: Caused by the fungus Corynespora cassiicola, target spot presents as small, water-soaked spots that enlarge into concentric rings with a pale center. It can lead to leaf drop and reduced plant productivity.
- Tomato mosaic virus: This virus causes mottled or mosaic patterns on leaves, along with leaf distortion and reduced fruit quality. It spreads through infected tools, seeds, and human handling.
- Tomato yellow leaf curl virus: This disease, caused by the whitefly Bemisia tabaci, leads to upward curling and yellowing of leaves, stunted plant growth, and reduced fruit production.
- Training set (80%): This is the primary dataset used to train the model. This set should be large enough to allow the model to learn the underlying patterns. It consists of a large number of labeled images that the model uses to learn the features and patterns associated with each class. The model’s parameters are adjusted based on this data to minimize the errors in predictions.
- Validation set (10%): This subset of the dataset is used to evaluate the model’s performance during the training process. It is used to fine-tune the model’s parameters and make decisions about changes in the model architecture. For example, changes are necessary in case of overfitting or underfitting [25]. Overfitting occurs when the model performs well on the training data but poorly on unseen data. It suggests that the model has learned the specific patterns of the training set but not general patterns. Underfitting occurs when the model performs poorly on both training and unseen data. It suggests that the model is too simple to capture the underlying patterns in the data.
- Testing set (10%): This part of the dataset is used for the final evaluation of the model after it has been trained and validated. If the model performs well on the test set, it suggests that the model has learned effectively and can make accurate predictions on new data. Conversely, if there is a significant drop in performance on the test set compared to the training and validation sets, it may indicate that the model has overfitted the training data.
3. Design and Methodology
- We loaded the pre-trained VGG16 and VGG19 models, excluding their top (fully connected) layers.
- The first layers of both models were frozen to retain their feature extraction capabilities while adapting to the new dataset.
- New layers were added on top of each base model, including fully connected layers, a dropout layer to prevent overfitting, and the final output layer to classify the tomato leaves.
4. Results
5. Applying Our Model in a Real-World Tomato Garden
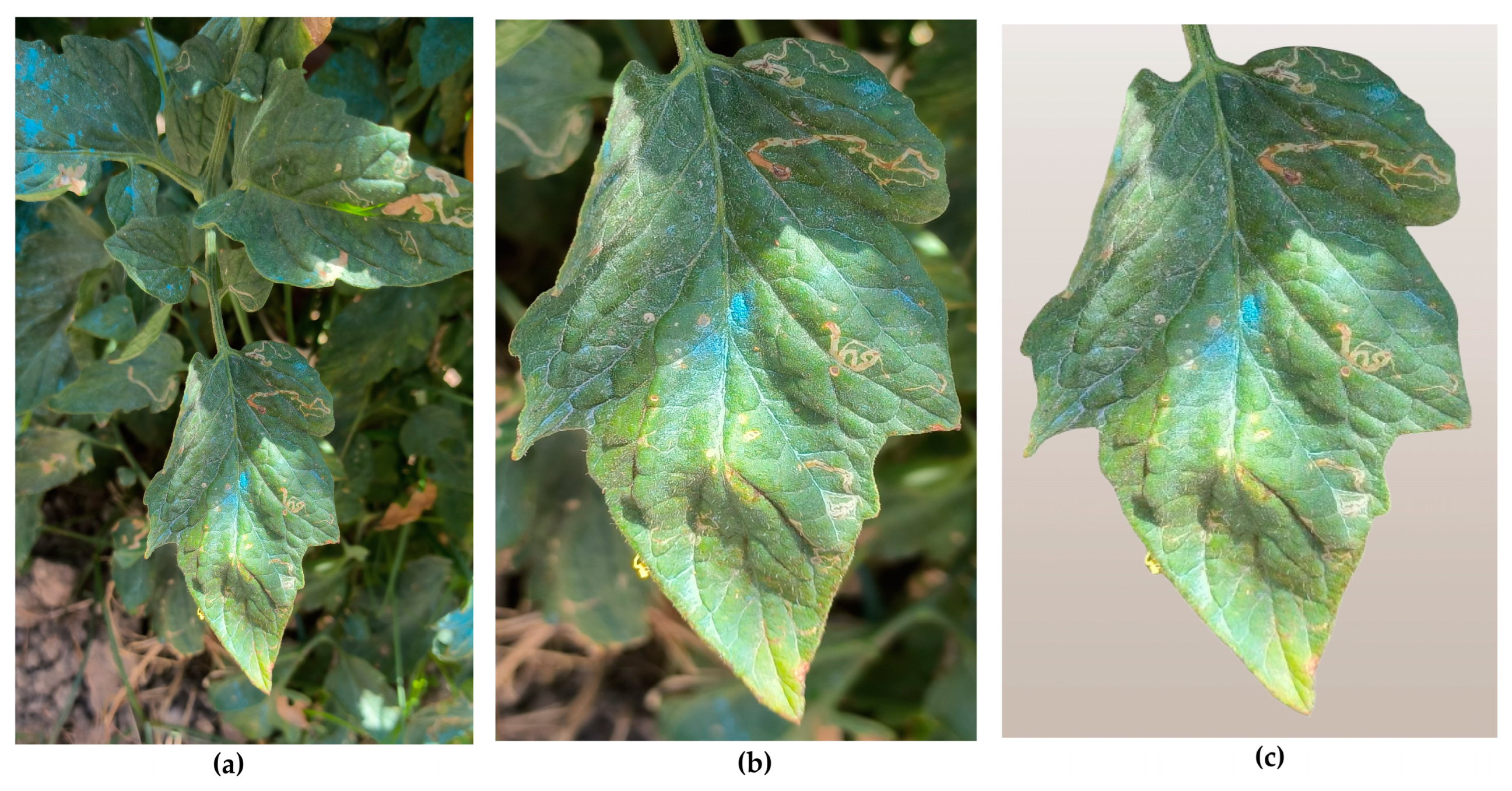
- Original images: this folder contains the raw images as captured by the smartphone, without any modifications.
- Cropped images: this is the same set of images, but with manually cropped photos, in order to focus as much as possible on the specific leaf intended for disease detection. This step was essential to reduce interference from other leaves and background elements present in the original images.
- Background-removed images: this set contains the same images with the previous one, but all images were further processed to remove any remaining background elements, ensuring that only the tomato leaf was present. This step aimed to completely eliminate any distractions and provide the most accurate input for the model. The removed background was replaced by a neutral background. This background was intentionally chosen to be a very pale beige or light brown, in order to prevent any distractions or elements that might interfere with or confuse the model, during the analysis process. By using this neutral background behind the central image of the tomato leaf, the focus was solely on the leaf itself. This approach was consistent with the already defined training, validation, and testing sets, ensuring that the model concentrated on the critical features of the leaf without being influenced by any irrelevant background details.
5.1. Experiment 1: Background-Removed Images vs. Original Images
- – A total of 21 images showed a rise in confidence, with an average increase of 21.01%.
- – A total of 80 images showed a confidence drop, with an average decrease of 17.80%.
5.2. Experiment 2: Cropped Images vs. Original Images
- – A total of 32 images saw a rise in confidence, with an average increase of 17.85%.
- – A total of 66 images experienced a confidence drop, with an average decrease of 14.76%.
5.3. Experiment 3. Cropped Images vs. Background-Removed Images
- – A total of 35 images experienced a confidence increase, with an average rise of 26.46%.
- – A total of 64 images saw a confidence drop, with an average decrease of 16.47%.
5.4. Findings and Insights
- Magnitude of confidence rise vs. drop: even though more images showed confidence decreases, the confidence rises were more significant in magnitude. A manual inspection confirmed that the confidence drops were relatively small, often not affecting the model’s certainty in a meaningful way. On the other hand, the confidence rises, especially after background removal, were much more substantial, indicating that this preprocessing step provided the model with clearer information on disease features.
- Cropped vs. original performance: the comparison between cropped and original images showed no substantial improvement in model performance, suggesting that while cropping the image helps by focusing the model on the leaf, it does not significantly enhance prediction confidence or accuracy.
- Background removal’s impact: the largest confidence rise occurred in the comparison between background removed and initial images, demonstrating that background removal can greatly help the model focus on the relevant parts of the image. Even though more images showed a drop in confidence, the fact that the rises were larger in magnitude suggests that background removal is an effective preprocessing step, even though it occasionally strips away useful context.
- Cropped vs. background removed: the largest difference in predictions was observed in the comparison between cropped and background removed images. This suggests that cropping alone is not sufficient to maximize model confidence, and that removing the background entirely allows the model to perform more effectively, with greater confidence in key cases. In summary, while background removal sometimes reduces confidence, it generally leads to a more significant rise in confidence where it matters. Cropping alone offers limited improvement, but the combination of careful cropping and background removal appears to provide the most significant boost in model performance.
- Dataset variations and their impact on model performance: another critical observation that emerged during our analysis relates to the dataset used for training compared to the dataset we created through our own image collection and preprocessing. Specifically, we noticed that the model struggled to accurately classify leaves from our custom dataset with high confidence, often failing to correctly identify the leaf category. This occurred despite the preprocessing techniques we applied, as discussed in earlier sections. Upon further investigation into the differences between the two datasets, we identified a key factor contributing to this discrepancy. The issue was not related to image resolution or technical specifications but rather to the intrinsic characteristics of the tomato leaves themselves. In particular, we observed that the tomato leaves in Greece, where our custom dataset was sourced, have distinct morphological differences compared to those in the original dataset. In Greece, the tomato plants commonly cultivated tend to have thinner and narrower leaves, whereas the leaves in the original dataset are typically thicker and broader. This variance in leaf shape, shown in Figure 7, could account for the model’s difficulty in generalizing predictions across both datasets. The model was trained on leaf images with a specific shape and thickness, which limited its ability to recognize the narrower leaf shapes commonly found in our local dataset. This finding underscores the importance of building a more diverse and representative dataset. Although the model achieved respectable confidence levels in certain cases, its performance highlighted the need for greater variability in the training data. As we previously discussed, having a diverse dataset is essential to developing a robust model capable of making accurate predictions across different environmental conditions, leaf variations, and tomato breeds. To improve future performance, it will be crucial to expand the dataset by incorporating images of tomato leaves from a variety of sources, including: (a) different tomato breeds (e.g., cherry tomatoes, heirloom tomatoes, beefsteak tomatoes), (b) various countries and regions, to account for geographical variations in leaf morphology, (c) different stages of growth and health conditions (e.g., diseased vs. healthy leaves, different disease severities), and (d) diverse environmental settings, including different lighting conditions, angles of capture, and weather effects (e.g., dry vs. humid climates).
6. Discussion
7. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| ANOVA | Analysis of variance |
| AR | Average recall |
| CNN | Convolutional neural network |
| CSA | Coordinate space attention |
| CUDA | Compute unified device architecture |
| CuDNN | CUDA deep neural network |
| DCNN | Deep convolutional neural networks |
| FR-CNN | Faster region-based convolutional neural network |
| FPS | Frames per second |
| GLCM | Gray level co-occurrence matrix |
| GPU | Graphics processing unit |
| HCA | Hybrid channel attention |
| LRAM | Location reinforcement attention mechanism |
| mAP | Mean average precision |
| mAP50 | Mean average precision (mAP) at a 50% threshold |
| ML | Machine learning |
| PAC | Perceptual adaptive convolution |
| PFAN | Proximity feature aggregation network |
| RCE | Robust cross-entropy |
| R-CNN | Region-based convolutional neural network |
| ReLU | Rectified linear unit |
| SGD | Stochastic gradient descent |
| SIFT | Scale-invariant feature transform |
| SVM | Support vector machine |
| VGG | Visual geometry group |
References
- Talaei Khoei, T.; Kaabouch, N. Machine Learning: Models, Challenges, and Research Directions. Future Internet 2023, 15, 332. [Google Scholar] [CrossRef]
- De Andrade, A. Best Practices for Convolutional Neural Networks Applied to Object Recognition in Images. arXiv 2019, arXiv:1910.13029. [Google Scholar] [CrossRef]
- Tang, Z.; He, X.; Zhou, G.; Chen, A.; Wang, Y.; Li, L.; Hu, Y. A Precise Image-Based Tomato Leaf Disease Detection Approach Using PLPNet. Plant Phenomics 2023, 5, 42. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.; Nazir, T.; Javed, A.; Masood, M.; Rashid, J.; Kim, J.; Hussain, A. A robust deep learning approach for tomato plant leaf disease localization and classification. Sci Rep. 2022, 12, 18568. [Google Scholar] [CrossRef]
- Zhang, E.; Zhang, N.; Li, F.; Lv, C. A Lightweight Dual-Attention Network for Tomato Leaf Disease Identification. Front. Plant Sci. 2024, 15, 1420584. [Google Scholar] [CrossRef]
- Khan, R.; Ud Din, N.; Zaman, A.; Huang, B. Automated Tomato Leaf Disease Detection Using Image Processing: An SVM-Based Approach with GLCM and SIFT Features. J. Eng. 2024, 2024, 9918296. [Google Scholar] [CrossRef]
- Debnath, A.; Hasan, M.M.; Raihan, M.; Samrat, N.; Alsulami, M.M.; Masud, M.; Bairagi, A.K. A Smartphone-Based Detection System for Tomato Leaf Disease Using EfficientNetV2B2 and Its Explainability with Artificial Intelligence (AI). Sensors 2023, 23, 8685. [Google Scholar] [CrossRef]
- Attallah, O. Tomato Leaf Disease Classification via Compact Convolutional Neural Networks with Transfer Learning and Feature Selection. Horticulturae 2023, 9, 149. [Google Scholar] [CrossRef]
- Pushpa, B.R.; Aiswarya, V.V. Tomato Leaf Disease Detection and Classification Using CNN. Math. Stat. Eng. Appl. 2022, 71, 2921–2930. [Google Scholar]
- Guerrero-Ibañez, A.; Reyes-Muñoz, A. Monitoring Tomato Leaf Disease through Convolutional Neural Networks. Electronics 2023, 12, 229. [Google Scholar] [CrossRef]
- Hossain, M.I.; Jahan, S.; Al Asif, M.R.; Samsuddoha, M.; Ahmed, K. Detecting tomato leaf diseases by image processing through deep convolutional neural networks. Smart Agric. Technol. 2023, 5, 100301. [Google Scholar] [CrossRef]
- Wang, X.; Liu, J. An efficient deep learning model for tomato disease detection. Plant Methods 2024, 20, 61. [Google Scholar] [CrossRef] [PubMed]
- Nagamani, H.S.; Sarojadevi, H. Tomato Leaf Disease Detection using Deep Learning Techniques. Int. J. Adv. Comput. Sci. Appl. 2022, 13, 305–311. [Google Scholar] [CrossRef]
- Trivedi, N.K.; Gautam, V.; Anand, A.; Aljahdali, H.M.; Villar, S.G.; Anand, D.; Goyal, N.; Kadry, S. Early Detection and Classification of Tomato Leaf Disease Using High-Performance Deep Neural Network. Sensors 2021, 21, 7987. [Google Scholar] [CrossRef]
- NVIDIA CUDA Toolkit. Available online: https://developer.nvidia.com/cuda-toolkit (accessed on 4 February 2025).
- OpenCV. Available online: https://opencv.org/ (accessed on 4 February 2025).
- Pandas Library. Available online: https://pandas.pydata.org/ (accessed on 4 February 2025).
- Keras. Available online: https://keras.io/ (accessed on 4 February 2025).
- NymPy. Available online: https://numpy.org/ (accessed on 4 February 2025).
- Scikit-Learn. Available online: https://scikit-learn.org/stable/ (accessed on 4 February 2025).
- Matplotlib. Available online: https://matplotlib.org/ (accessed on 4 February 2025).
- TensorFlow. Available online: https://www.tensorflow.org/ (accessed on 4 February 2025).
- Github Code Repository. Available online: https://github.com/ZamirOsmenaj/tomato-leaf-disease-detection-and-classification (accessed on 4 February 2025).
- Tomato Leaf Disease Detection Dataset. Available online: https://www.kaggle.com/datasets/kaustubhb999/tomatoleaf (accessed on 4 February 2025).
- Filipi, C.; Dos Santos, G.; Papa, J.P. Avoiding Overfitting: A Survey on Regularization Methods for Convolutional Neural Networks. ACM Comput. Surv. 2022, 54, 213. [Google Scholar] [CrossRef]
- Xu, Y.; Goodacre, R. On Splitting Training and Validation Set: A Comparative Study of Cross-Validation, Bootstrap and Systematic Sampling for Estimating the Generalization Performance of Supervised Learning. J. Anal. Test. 2018, 2, 249–262. [Google Scholar] [CrossRef]
- Mishra, A.; Mishra, A.; Tewari, A.K.; Gangrade, J. Deep Transfer Learning for Tomato Leaf Diseases Detection and Classification using Pretrained Models. In Proceedings of the 9th International Conference on Signal Processing and Communication (ICSC), Noida, India, 21–23 December 2023; pp. 290–295. [Google Scholar] [CrossRef]
- Dhivyaa, C.R.; Nithya, K.; Vignesh, T.; Sudhakar, R.; Kumar, K.S.; Janani, T. An Enhanced Deep Learning Model for Tomato Leaf Disease Prediction. In Proceedings of the 8th International Conference on Communication and Electronics Systems (ICCES), Coimbatore, India, 1–3 June 2023; pp. 1322–1331. [Google Scholar] [CrossRef]
- Nguyen, T.-H.; Nguyen, T.-N.; Ngo, B.-V. A VGG-19 Model with Transfer Learning and Image Segmentation for Classification of Tomato Leaf Disease. AgriEngineering 2022, 4, 871–887. [Google Scholar] [CrossRef]
- Wang, Y.; Han, Y.; Wang, C.; Song, S.; Tian, Q.; Huang, G. Computation-Efficient Deep Learning for Computer Vision: A Survey. Cybern. Intell. 2024, 1–24. [Google Scholar]
- Web App. Available online: https://tomato-leaves-prediction-app.streamlit.app/ (accessed on 4 February 2025).
- PlantVillage Dataset. Available online: https://www.kaggle.com/datasets/emmarex/plantdisease (accessed on 4 February 2025).
- Fenu, G.; Malloci, F.M. Evaluating Impacts between Laboratory and Field-Collected Datasets for Plant Disease Classification. Agronomy 2022, 12, 2359. [Google Scholar] [CrossRef]
- Noyan, M.A. Uncovering bias in the PlantVillage dataset. arXiv 2022, arXiv:2206.04374v1. [Google Scholar] [CrossRef]
- Saebi, M.; Nan, B.; Herr, J.E.; Wahlers, J.; Guo, Z.; Zurański, A.M.; Kogej, T.; Norrby, P.-O.; Doyle, A.G.; Chawla, N.V.; et al. On the use of real-world datasets for reaction yield prediction. Chem. Sci. 2023, 14, 4997–5005. [Google Scholar] [CrossRef] [PubMed]
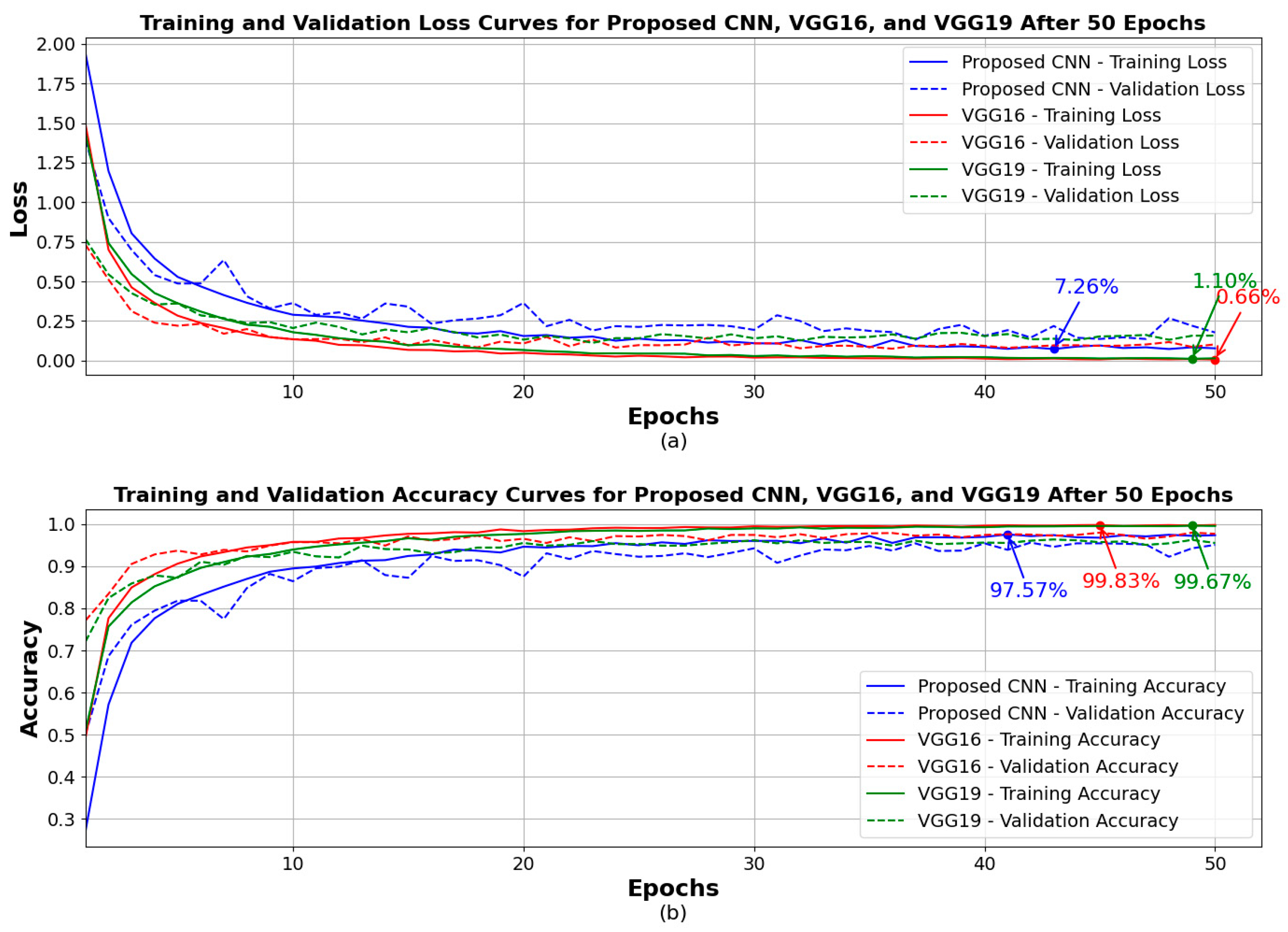
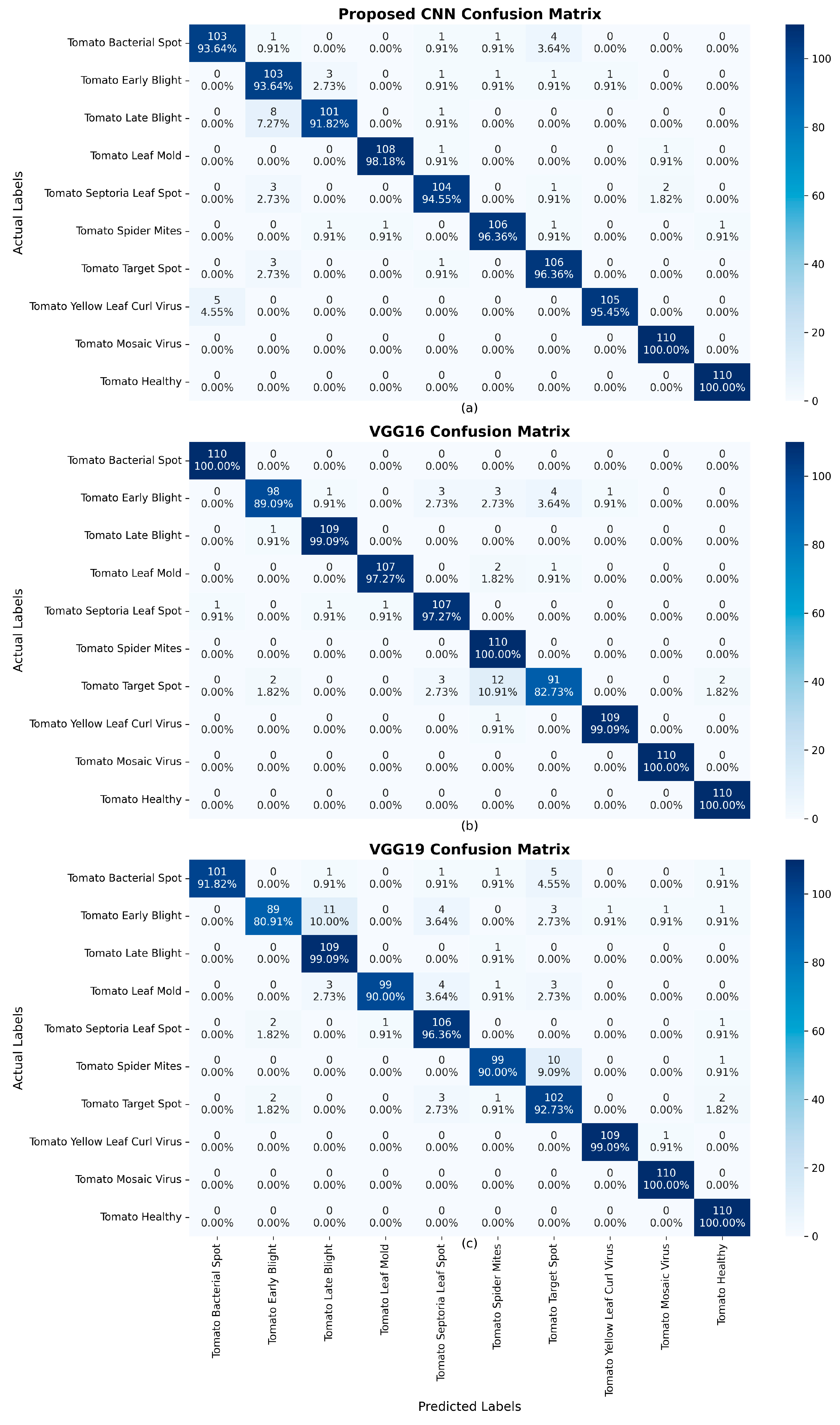
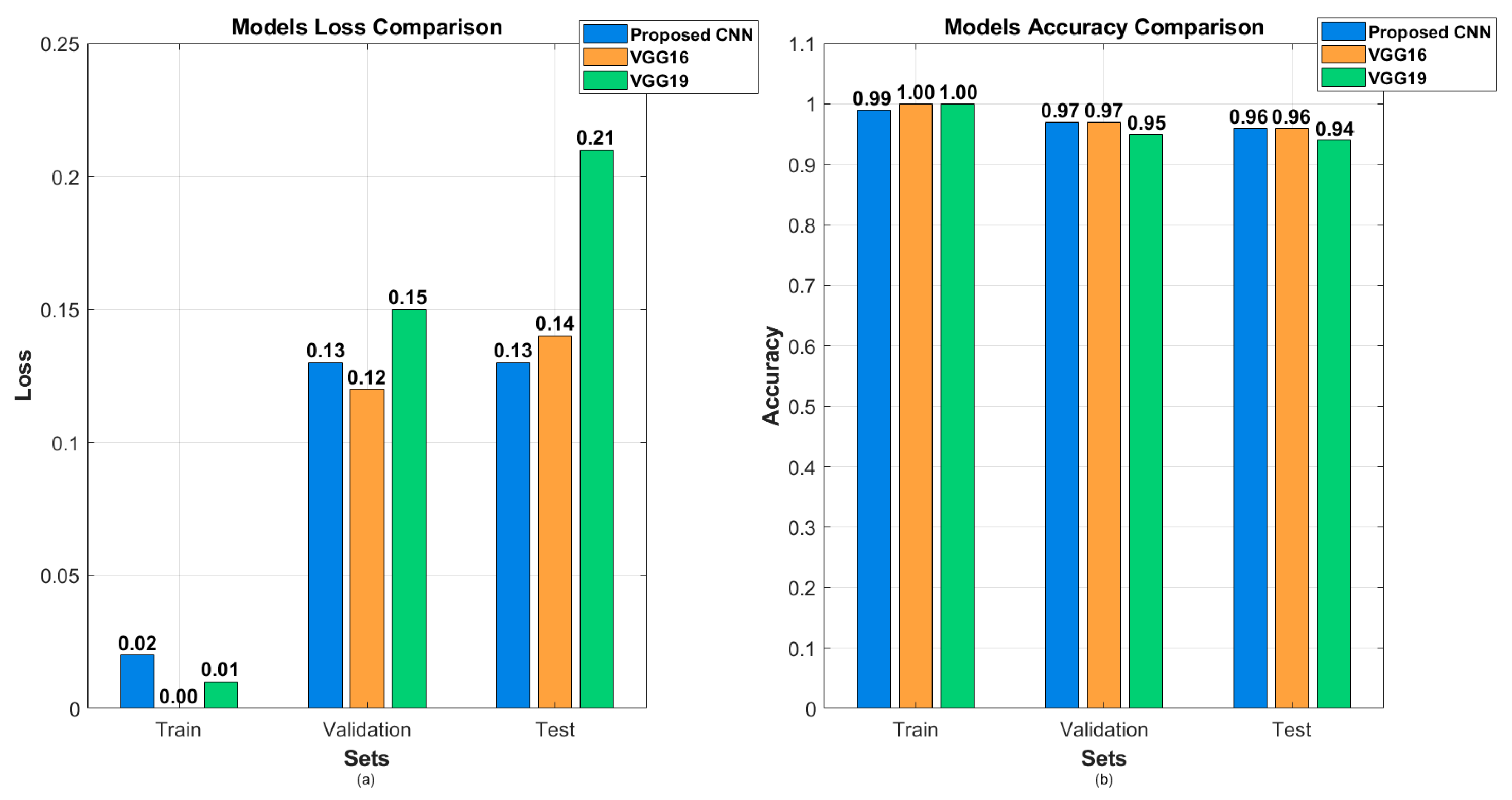

| Model | Accuracy | Loss | Precision | Recall | F1 Score |
|---|---|---|---|---|---|
| Proposed CNN | 96.00% | 12.54% | 96.00% | 96.00% | 96.00% |
| VGG16 (fine-tuned) | 96.45% | 13.55% | 97.00% | 96.00% | 96.00% |
| VGG19 (fine-tuned) | 94.00% | 20.59% | 94.00% | 94.00% | 94.00% |
| Ensemble (three models) | 98.00% | N/A | 98.00% | 98.00% | 98.00% |
| Comparison | Number of Images with Different Predictions | Average Confidence Rise | Average Confidence Drop |
|---|---|---|---|
| Background removed vs. original | 72 | 21.01% | 17.80% |
| Cropped vs. original | 57 | 17.85% | 14.76% |
| Cropped vs. background removed | 43 | 26.46% | 16.47% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Osmenaj, Z.; Tseliki, E.-M.; Kapellaki, S.H.; Tselikis, G.; Tselikas, N.D. From Pixels to Diagnosis: Implementing and Evaluating a CNN Model for Tomato Leaf Disease Detection. Information 2025, 16, 231. https://doi.org/10.3390/info16030231
Osmenaj Z, Tseliki E-M, Kapellaki SH, Tselikis G, Tselikas ND. From Pixels to Diagnosis: Implementing and Evaluating a CNN Model for Tomato Leaf Disease Detection. Information. 2025; 16(3):231. https://doi.org/10.3390/info16030231
Chicago/Turabian StyleOsmenaj, Zamir, Evgenia-Maria Tseliki, Sofia H. Kapellaki, George Tselikis, and Nikolaos D. Tselikas. 2025. "From Pixels to Diagnosis: Implementing and Evaluating a CNN Model for Tomato Leaf Disease Detection" Information 16, no. 3: 231. https://doi.org/10.3390/info16030231
APA StyleOsmenaj, Z., Tseliki, E.-M., Kapellaki, S. H., Tselikis, G., & Tselikas, N. D. (2025). From Pixels to Diagnosis: Implementing and Evaluating a CNN Model for Tomato Leaf Disease Detection. Information, 16(3), 231. https://doi.org/10.3390/info16030231









