Abstract
As yet, there is no systematic review focusing on benefits and issues of commercial deep learning-based auto-segmentation (DLAS) software for prostate cancer (PCa) radiation therapy (RT) planning despite that NRG Oncology has underscored such necessity. This article’s purpose is to systematically review commercial DLAS software product performances for PCa RT planning and their associated evaluation methodology. A literature search was performed with the use of electronic databases on 7 November 2024. Thirty-two articles were included as per the selection criteria. They evaluated 12 products (Carina Medical LLC INTContour (Lexington, KY, USA), Elekta AB ADMIRE (Stockholm, Sweden), Limbus AI Inc. Contour (Regina, SK, Canada), Manteia Medical Technologies Co. AccuContour (Jian Sheng, China), MIM Software Inc. Contour ProtégéAI (Cleveland, OH, USA), Mirada Medical Ltd. DLCExpert (Oxford, UK), MVision.ai Contour+ (Helsinki, Finland), Radformation Inc. AutoContour (New York, NY, USA), RaySearch Laboratories AB RayStation (Stockholm, Sweden), Siemens Healthineers AG AI-Rad Companion Organs RT, syngo.via RT Image Suite and DirectORGANS (Erlangen, Germany), Therapanacea Annotate (Paris, France), and Varian Medical Systems, Inc. Ethos (Palo Alto, CA, USA)). Their results illustrate that the DLAS products can delineate 12 organs at risk (abdominopelvic cavity, anal canal, bladder, body, cauda equina, left (L) and right (R) femurs, L and R pelvis, L and R proximal femurs, and sacrum) and four clinical target volumes (prostate, lymph nodes, prostate bed, and seminal vesicle bed) with clinically acceptable outcomes, resulting in delineation time reduction, 5.7–81.1%. Although NRG Oncology has recommended each clinical centre to perform its own DLAS product evaluation prior to clinical implementation, such evaluation seems more important for AccuContour and Ethos due to the methodological issues of the respective single studies, e.g., small dataset used, etc.
1. Introduction
Globally, prostate cancer (PCa) is the number four cancer and the second most common cancer type in males [1,2,3]. About 80% of PCa is localised [4]. Common treatments for localised and regional PCa include active surveillance, radical prostatectomy, and radiation therapy (RT) with and without hormone therapy (androgen deprivation therapy) [4,5]. Recent literature reviews and meta-analysis show that both radical prostatectomy and RT are equally effective for treating the localised PCa [4,5,6]. However, the PCa RT treatment effectiveness depends on the accurate segmentation of tumours (clinical target volumes (CTVs)) as well as adjacent healthy tissues (organs at risk (OARs)) such as the rectum on medical images during the treatment planning process, so as to minimise its side effects, e.g., faecal incontinence, blood in stool, etc. [5,7,8,9,10].
Traditionally, the OARs and CTVs segmentation in PCa RT treatment planning is handled by radiation therapists (RTTs) and/or radiation oncologists (ROs) based on established contouring guidelines [11,12]. However, this manual process is time-consuming and subjective, resulting in notable inter- and intra-observer variations and hence affecting the PCa RT treatment outcomes [13,14,15]. To address these issues, auto-segmentation approaches (e.g., intensity analysis, shape modelling, atlas-based, etc.) have emerged in recent decades [10,15,16,17]. Given the recent popularity of deep learning (DL) in medical imaging, commercial companies have shifted their focus to develop DL-based auto-segmentation (DLAS) software for PCa RT planning. Examples of such software include Carina Medical LLC INTContour (Lexington, KY, USA), Limbus AI Inc. Contour (Saskatchewan, Canada), Mirada Medical Ltd. DLCExpert (Oxford, UK), MVision.ai Contour+ (Helsinki, Finland), Radformation Inc. AutoContour (New York, NY, USA), RaySearch Laboratories AB RayStation (Stockholm, Sweden), Siemens Healthineers AG AI-Rad Companion Organs RT (Erlangen, Germany), and Therapanacea Annotate (Paris, France) [17,18,19,20].
Over the past five years, several review papers that cover the DLAS in PCa RT planning have been available. Their review areas include DLAS architectures/approaches (e.g., convolutional neural network (CNN), etc.) [12,14,15,17], model training and testing dataset sources (such as private and public) [12,14], dataset types (e.g., magnetic resonance imaging (MRI), computed tomography (CT), etc.) [12,13,15], dataset sizes such as 69 patients [12,13,14,15,21], structures segmented (e.g., seminal vesicles (SVs), rectum, etc.) [12,14,15,17,21], reference contour sources (such as at least three ROs) [14,21], contouring guidelines (e.g., Radiation Therapy Oncology Group (RTOG), etc.) [15], evaluation metrics (such as Dice similarity coefficient (DSC)), and model performance (e.g., efficiency, geometric accuracy, etc.) [12,14,15,17]. Most of these review articles were published in 2024, highlighting the significance and urgency of this topic [12,15,17,21]. However, the aforementioned articles, except for two, were narrative reviews and covered a range of disease sites such as breast, lung, and prostate [12,13,17,21]. As yet, there is only one DLAS systematic review focused on the PCa RT planning. Nevertheless, it was published in 2020 and no studies on commercial DLAS software were included [14].
According to the latest narrative reviews (published in 2024) on the DLAS for the RT planning including one by the working group of the influential body, NRG Oncology, the importance of reviewing commercial DLAS software products has been underscored. This is because such reviews can enable the clinical community to realise strengths and weaknesses of the commercial products for consideration of clinical implementation [12,21]. As yet, there is no systematic review focusing on the benefits and issues of the commercial DLAS software for the PCa RT planning despite that a previous critical review on the DLAS for RT planning of multiple cancers covered four studies about performance evaluation of three commercial DLAS products for the PCa RT planning (Carina Medical LLC INTContour, Limbus AI Inc. Contour and Siemens Healthineers AG AI-Rad Companion Organs RT) out of 12 included papers. Hence, the findings of that review paper with a mix of studies of in-house and commercial DLAS models are less relevant to the clinical community due to potential barriers to accessing and implementing the in-house models [15]. Without a systematic review focusing on the commercial DLAS products for the PCa RT planning with evaluation of associated methods for generation of evidence, this would hinder clinical adoption of such technology to take advantage of its potential benefits to a greater extent [22]. The purpose of this article is to systematically review original studies to answer the question: “What are the performances of commercial DLAS software products for the PCa RT planning and methods for their performance evaluation?”
2. Materials and Methods
This systematic review was conducted according to the PICO model (patient/population: PCa patients; intervention: PCa RT planning with the use of commercial DLAS software), comparison (DLAS versus standard practice-manual contouring), and outcome (performance of structure segmentation), and preferred reporting items for systematic reviews and meta-analyses (PRISMA) guidelines [22,23,24,25,26]. Four major steps including literature search, article selection, and data extraction and synthesis were involved [22,23,24,25].
2.1. Literature Search
Seven electronic journal databases including Institute of Electrical and Electronics Engineers (IEEE) Xplore, PubMed, ScienceDirect, Scopus, SpringerLink, Web of Science, and Wiley Online Library were used for searching the literature on the performance of the commercial DLAS software for the PCa RT planning on 7 November 2024. The search statement: “Commercial” AND (“Deep Learning” OR “Artificial Neural Network”) AND (“Segmentation” OR “Delineation” OR “Contouring”) AND “Prostate Cancer” AND (“Radiotherapy” OR “Radiation Therapy”) was used. These keywords were based on the focus of the review. The search did not have any restrictions on publication year [13,14,22].
2.2. Article Selection
The selection of articles was handled by a single reviewer with more than 20 years’ experience in the literature review. Article selection criteria are shown in Table 1 [22,23,24,25].

Table 1.
Selection criteria for articles.
The selection criteria in Table 1 were determined based on the purpose of this review which included appraisal of methods of original studies on evaluating the performance of DLAS software for the PCa RT planning [22,23,24,25,27]. For systematic reviews of DL that covered the study methodology appraisal, the conference proceedings were considered not suitable due to their lower academic rigour in general [22,27,28]. The detail of article selection is illustrated in Figure 1 [22,23,24,25]. The main processes included article deduplication and screening of paper titles, abstracts, and full texts as per the selection criteria. Each non-duplicate paper in the search results was retained unless a decision on its exclusion could be reached. Reference lists of all included papers were checked for identification of additional articles (i.e., backward reference searching) [22,23,24,29,30].
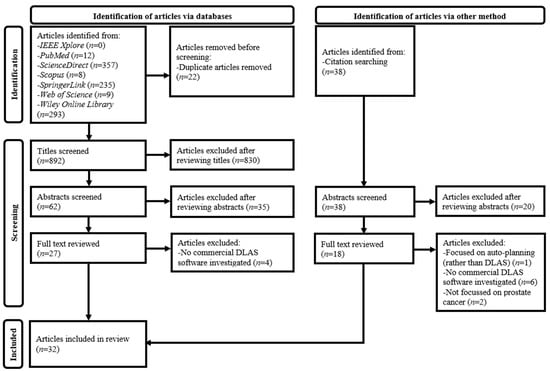
Figure 1.
Preferred reporting items for systematic reviews and meta-analyses flow diagram for this systematic review. DLAS, deep learning-based auto-segmentation; IEEE, Institute of Electrical and Electronics Engineers.
2.3. Data Extraction and Synthesis
Data extraction was conducted based on a form derived from one narrative review [12] and one critical review on the DLAS for different RT plannings [15], one systematic review of the DLAS for the PCa RT planning [14], and another systematic review focusing on commercial DLAS software for breast cancer RT planning [22]. The following data were extracted from every included paper: name and country of author; publication year; software product name and version (such as Siemens Healthineers AG AI-Rad Companion Organs RT VA20); DLAS architecture (e.g., 3-dimensional U-Net, etc.); study design (either prospective or retrospective); any involvement of multiple centres; patient/population (such as postprostatectomy PCa patients); any model finetuning; source (e.g., private: 1 Dutch centre, public: USA The Cancer Imaging Archive, etc.) and size (such as 84 patients) of dataset for development (training of model) and evaluation (testing) of software; any sample size calculation for evaluation and external testing (model evaluation with dataset different from that for its development and obtained from different source); imaging modality for DLAS (e.g., MRI, etc.); OARs (such as rectum) and CTVs (e.g., prostate, SVs, etc.) delineated; source of reference contour, i.e., ground truth (such as four expert ROs); contouring guidelines (e.g., Australia and New Zealand Faculty of Radiation Oncology Genito-Urinary Group (FROGG)) used; performances of delineation regarding geometric accuracy (expressed by metrics, e.g., DSC, etc.) for the DLAS and manual segmentation (in terms of inter-observer variation (IOV)-average performance of multiple observers), and any differences between them; subjective accuracy assessment (such as percentage of CTVs and OARs that needed no and minor adjustments); evaluation of efficiency (e.g., percentage of average segmentation time reduction, etc.); and dosimetric impact (such as DLAS and manual segmentation not having any clinically relevant dose difference for OARs) [12,14,15,22].
For papers that evaluated both DL- and atlas-based auto-segmentation software products or covered multiple cancer types, only data about the DLAS for the PCa RT planning were included. When more than one set of evaluation results were provided by a study, only mean (or median if mean unavailable) performance findings of the best model determined through the external testing and/or expert (instead of clinical) contours were extracted for the facilitation of software comparison. In addition, the following strategies for data synthesis were used when applicable: 1. calculating mean values for figures from multiple PCa patient demographic subgroups and ROs; 2. overall percentage calculation based on individual percentage figures for unusable CTVs and OARs contours as well as those requiring no and minor adjustments; and 3. calculating percentages based on reported absolute values of average segmentation time reduction, and delineated structure volume differences between manual contouring and DLAS [22,23,24,25]. The DLAS delineation time included time for data transfer, auto-segmentation, and contour review and correction [12,21]. Since the included papers used various performance evaluation approaches, resulting in a high study heterogeneity, meta-analysis was not performed due to limited value [22,27,31,32]. The quality of all included papers was assessed based on revised checklist for artificial intelligence in medical imaging (CLAIM) 2024 [33,34,35,36].
3. Results
Thirty-two articles were included in this review as per the selection criteria (Figure 1). The performances of the commercial DLAS software products evaluated by the selected studies are shown in Table 2 and Table 3 [37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68]. These studies covered a total of 12 products, namely, Carina Medical LLC INTContour [37,38,39,65,67], Elekta AB ADMIRE (Stockholm, Sweden) [40], Limbus AI Inc. Contour [41,42,43,44,45,46,66], Manteia Medical Technologies Co. AccuContour (Jian Sheng, China) [67], MIM Software Inc. Contour ProtégéAI (Cleveland, OH, USA) [47,48,67], Mirada Medical Ltd. DLCExpert [49,50,51,68], MVision.ai Contour+ [52,53,54,55,66,68], Radformation Inc. AutoContour [68], RaySearch Laboratories AB RayStation [56,57,65,68], Siemens Healthineers AG AI-Rad Companion Organs RT [58,60], syngo.via RT Image Suite [58,59] and DirectORGANS [61], Therapanacea Annotate [62,63,68], and Varian Medical Systems, Inc. Ethos (Palo Alto, CA, USA) [64].

Table 2.
Geometric accuracy of commercial deep learning-based auto-segmentation (DLAS) software products for prostate cancer radiation therapy planning.

Table 3.
Subjective accuracy, efficiency, and dosimetric evaluation results of commercial deep learning-based auto-segmentation (DLAS) software products for prostate cancer radiation therapy planning.
DSC was the most common metric (used in 28 out of 32 (87.5%) papers) for the delineated structures’ geometry accuracy evaluation (Table 2) [37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,55,57,58,59,60,61,63,65,66,67,68]. A total of 26 out of these 28 (92.9%) articles used the DLAS software to delineate the OARs. Collectively, 20 OARs as well as two rectal sparing structures (balloon and spacer) were covered [37,38,39,41,42,43,44,45,46,47,48,49,50,51,52,53,55,57,58,59,60,61,63,66,67,68]. Their mean/median DSC values were: abdominopelvic cavity (0.94) [59]; anal canal (0.70–0.74) [45,63]; bladder (0.88–0.99) [37,38,39,41,42,43,44,45,46,47,48,49,50,51,52,55,57,58,59,60,61,63,66,67,68]; body (external contour) (0.99) [59]; cauda equina (0.75) [68]; left (L) (0.68–0.99) and right (R) femoral heads (0.69–0.99) [37,38,39,41,42,44,46,48,49,50,51,52,55,57,59,66,67,68]; L (0.78–0.92) and R femurs (0.78–0.92) [45,63]; L (0.91) and R pelvis (0.90) [63]; L (0.95) and R proximal femurs (0.96) [61]; large (0.49–0.76) and small bowels (0.30–0.76) [67,68]; penile bulb (0.39–0.73) [37,38,44,51,52,55,63,67]; penile root (0.54–0.71) [68]; rectum (0.51–0.95) [37,38,39,41,42,43,44,45,46,47,48,49,50,51,52,53,55,57,58,59,60,61,63,66,67,68]; sacrum (0.89) [63]; sigmoid (0.52–0.77) [68]; rectal balloon (0.89) [67]; and spacer (0.52–0.84) [51,67]. The range of the mean/median DSC for the OARs contoured by the DLAS software were 0.30 (small bowel) [67]–0.99 (bladder [42,43], body [59], and L [42,59] and R femoral heads [42]).
Based on the common DSC cutoff value, at least 0.70 for indicating clinically acceptable DLAS performance [22,44,45,51,60,68,69], 12 (abdominopelvic cavity [59], anal canal [45,63], bladder [37,38,39,41,42,43,44,45,46,47,48,49,50,51,52,55,57,58,59,60,61,63,66,67,68], body [59], cauda equina [68], L and R femurs [45,63], L and R pelvis [63], L and R proximal femurs [61], and sacrum [63]) out of 20 (60.0%) OARs were able to meet this standard (Table 2). With the same criterion (DSC ≥ 0.70) [22,44,45,51,60,68,69], at least 80.0% of the OARs contoured by the following seven DLAS products were considered clinically acceptable: Carina Medical LLC INTContour (four (bladder, L and R femoral heads, and rectum) out of five (80.0%) OARs) [37,38,39,67], Limbus AI Inc. Contour (seven (anal canal, bladder, L and R femoral heads, L and R femurs, and rectum) out of eight (87.5%) OARs) [41,42,43,44,45,46,66], Manteia Medical Technologies Co. AccuContour (all (four) OARs (bladder, L and R femoral heads, and rectum)) [67], MIM Software Inc. Contour ProtégéAI (all (four) OARs (bladder, L and R femoral heads, and rectum)) [47,48,67], Siemens Healthineers AG AI-Rad Companion Organs RT/syngo.via RT Image Suite/DirectORGANS (all (eight) OARs (abdominopelvic cavity, bladder, body, L and R femoral heads, L and R proximal femurs, and rectum)) [58,59,60,61], Therapanacea Annotate ART-Plan (13 (anal canal, bladder, bowels, L and R femoral heads, L and R femurs, L and R pelvis, penile bulb and root, rectum, and sacrum) out of 14 (92.9%) OARs) [63,68], and Radformation Inc. AutoContour (six (bladder, bowels, cauda equina, L and R femoral heads, and rectum) out of seven (85.7%) OARs) [68].
Around 60.0% of the included papers evaluated the DLAS products for delineating the CTVs by the metric, DSC (Table 2) [37,38,39,40,41,42,45,46,48,51,52,55,57,59,60,63,65,67,68]. A total of 11 products were evaluated, and they were: Carina Medical LLC INTContour [37,38,39,65,67], Elekta AB ADMIRE [40], Limbus AI Inc. Contour [41,42,45,46], Manteia Medical Technologies Co. AccuContour [67], MIM Software Inc. Contour ProtégéAI [48,67], Mirada Medical Ltd. DLCExpert [51,68], MVision.ai Contour+ [52,55,68], Radformation Inc. AutoContour [68], RaySearch Laboratories AB RayStation [57,65,68], Siemens Healthineers AG AI-Rad Companion Organs RT [60] and syngo.via RT Image Suite [59], and Therapanacea Annotate ART-Plan [63,68]. Five CTVs were covered in total [37,38,39,40,41,42,45,46,48,51,52,55,57,59,60,63,65,67,68]. Their mean/median DSC values were 0.80 (lymph nodes) [52], 0.74–0.91 (prostate) [37,39,40,41,42,45,46,48,51,52,55,57,59,60,63,65,67,68], 0.73–0.75 (prostate bed) [38,39], 0.73 (SV bed) [38], and 0.30–0.83 (SVs) [37,41,46,48,51,52,55,60,63,67,68]. All (11) products were able to delineate the prostate with the clinically acceptable outcome (DSC ≥ 0.70) [37,39,40,41,42,45,46,48,51,52,55,57,59,60,63,65,67,68]. Also, the performance of Carina Medical LLC INTContour and MVision.ai Contour+ for delineating lymph nodes [52], prostate bed [38,39], and SV bed [38] was deemed acceptable. Nonetheless, highly variable segmentation outcomes are noted for SVs [37,41,46,48,51,52,55,60,63,67,68].
In addition, Table 2 shows that only 18.8% (6/32) studies compared the performances between DLAS and manual contouring (expressed in IOV) [37,41,43,46,61,65]. Nevertheless, five (83.3%) out of these six studies revealed that the DLAS performance at least matched the one of the manual segmentation (represented in IOV), and it outperformed the observers occasionally [37,41,46,65].
Table 3 illustrates that greater than 90.0% (29/32) papers evaluated the DLAS products with more clinically meaningful approaches (subjective accuracy, efficiency, and/or dosimetric impact assessment) [37,38,39,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,59,60,61,62,63,64,65,66,67,68]. Except for Walker et al.’s [49] and Wang et al.’s [51] studies on Mirada Medical Ltd. DLCExpert, 18 (90.0%) out of 20 articles showed that a large proportion of the structures contoured by Carina Medical LLC INTContour [37,38,39,65,67], Limbus AI Inc. Contour [42,43,45,46,66], Manteia Medical Technologies Co. AccuContour [67], MIM Software Inc. Contour ProtégéAI [48,67], Mirada Medical Ltd. DLCExpert [50], MVision.ai Contour+ [54,66], RaySearch Laboratories AB RayStation [65], Siemens Healthineers AG AI-Rad Companion Organs RT [60] and syngo.via RT Image Suite [59], Therapanacea Annotate ART-Plan [62,63], and Varian Medical Systems, Inc. Ethos [64] required no or minor adjustments. Thus, the DLAS contributed to substantial efficiency gain for the PCa RT planning [41,43,44,45,48,49,50,51,52,54,55,59,60,61,66,68]. The range of average delineation time reduction per patient reported by 16 out of the 32 (50.0%) papers was [41,43,44,45,48,49,50,51,52,54,55,59,60,61,66,68]: 5.7% (Siemens Healthineers AG AI-Rad Companion Organs RT [60])–99.8% (Therapanacea Annotate ART-Plan [68]). However, when excluding the articles not reporting the contour review and adjustment time [41,55], and those merely indicating the review and correction time [45,49,50,54,68], as per the NRG Oncology recommendation [21], the average contouring time reduction range became 5.7% (Siemens Healthineers AG AI-Rad Companion Organs RT [60])–81.1% (Limbus AI Inc. Contour [66]). For the DLAS dosimetric impact, only seven out of the 32 (21.9%) papers investigated this [37,44,45,53,56,61,64]. All but two of these articles indicated no clinically relevant (> 5%) or statistically significant dose difference to the delineated structures (except anal canal) between the manual contouring and DLAS [37,45,53,56,64].
Table 4 shows the included articles’ characteristics. CT images were used for the DLAS software evaluation in 29 out of the 32 (90.6%) studies. However, cone beam CT (CBCT) [39,46,64] and MRI [63] were also used in several (four) papers. Two of these studies finetuned the commercial software products (Carina Medical LLC INTContour and Therapanacea Annotate ART-Plan) originally designed for CT to handle the CBCT [39] and MRI image segmentation in 2024 [63]. The other two employed the models already trained with the CBCT images by the companies (Limbus AI Inc. Contour v1.5.0-D2 and Varian Medical Systems, Inc. Ethos) [46,64]. In total, the DLAS model finetuning was conducted in eight out of the 32 (25.0%) articles [37,38,39,51,53,63,65,67].

Table 4.
Characteristics of studies on commercial deep learning-based auto-segmentation (DLAS) software products for prostate cancer (PCa) radiation therapy planning.
Nearly 80.0% (25/32) of the included papers were published in the previous three years (Table 4) [37,38,39,44,45,46,48,49,50,51,53,54,55,56,57,58,59,60,61,62,63,65,66,67,68]. However, these articles had some methodological weaknesses, e.g., only two (6.3%) prospective [42,44] and four (12.5%) multi-centre studies [42,49,52,58], none with sample size calculation for testing dataset [37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68], 84.4% (27/32) of the papers with fewer than 100 patients for software testing [37,38,39,41,42,43,44,45,46,47,48,49,50,51,52,54,55,56,59,60,61,62,63,64,65,66,68] (even four studies with ≤ 10 patients [44,46,55,62]), etc. Nevertheless, 27 out of the 32 (84.4%) articles performed the external testing [38,39,40,41,42,43,44,45,47,48,49,50,52,53,54,55,57,58,59,60,61,62,64,65,66,67,68]. In addition, the model testing datasets of the included articles (except Radici et al. not specifying the source [46]) were obtained from 14 different countries [37,38,39,40,41,42,43,44,45,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68] on four continents: Asia [47,52,55,60,68], Australia [50], Europe [40,44,45,48,49,52,53,54,56,58,59,61,62,63,66], and North America [37,38,39,41,42,43,51,57,64,65,67]. The mix of the merits and deficiencies of these papers resulted in their quality scores between 40.0% and 72.0% (mean: 52.1% and median: 53.0%) [37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68]. Figure 2 shows the number of included articles meeting each revised CLAIM 2024 criterion [36].
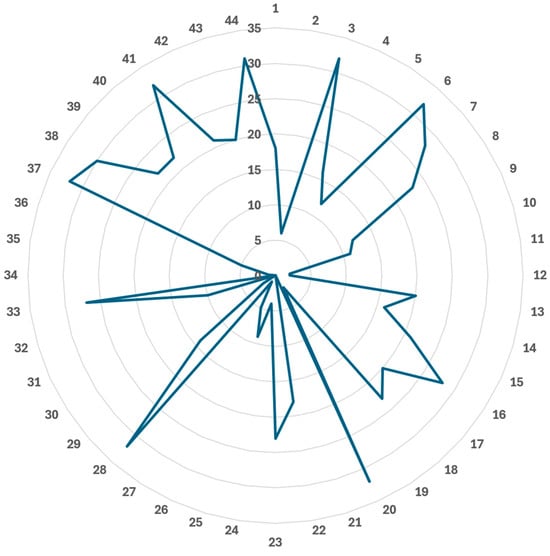
Figure 2.
Number of included papers meeting each revised CLAIM 2024 criterion. Radial axis: number of papers. Angular axis: CLAIM 2024 criterion.
4. Discussion
This paper is the first systematic review about the commercial DLAS software performance for PCa RT planning, covering 12 products and 32 articles [37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68]. Thus, it advances Almeida et al.’s [14] DLAS systematic review on the PCa RT planning which covered 28 papers on non-commercial DL models published in 2020 and Matoska et al.’s [15] critical review published in 2024 which included 12 articles about CTV DLAS for PCa with only four studies evaluating three commercial products (Carina Medical LLC INTContour, Limbus AI Inc. Contour, and Siemens Healthineers AG AI-Rad Companion Organs RT). Another strength of this review is about 80.0% of the included papers published in the previous three years. Therefore, it can provide more relevant and recent findings to inform clinical practice (Table 2, Table 3 and Table 4) [37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68].
As per Table 2 (geometry accuracy findings), the commercial DLAS products were able to delineate 12 OARs (abdominopelvic cavity [59], anal canal [45,63], bladder [37,38,39,41,42,43,44,45,46,47,48,49,50,51,52,55,57,58,59,60,61,63,66,67,68], body [59], cauda equina [68], L and R femurs [45,63], L and R pelvis [63], L and R proximal femurs [61], and sacrum [63]), and four CTVs (prostate [37,39,40,41,42,45,46,48,51,52,55,57,59,60,63,65,67,68], lymph nodes [52], prostate bed [38,39], and SV bed [38]) with the clinically acceptable outcomes (DSC ≥ 0.70) [22,44,45,51,60,68,69]. The performance of the commercial products covered in this review for the prostate (CTV) segmentation (DSC: 0.74–0.91) [37,39,40,41,42,45,46,48,51,52,55,57,59,60,63,65,67,68] is comparable to the findings of Almeida et al.’s [14] systematic review (DSC: 0.65–0.95) and Matoska et al.’s [15] critical review (DSC: 0.70–0.90). For the other CTVs as well as OARs, both of those review papers did not report the respective DSC findings except for penile bulb with a DSC value of 0.72 [14,15], which is within the corresponding DSC range: 0.39–0.73 illustrated in Table 2 [37,38,44,51,52,55,63,67]. The unsatisfactory DSC results for penile bulb can be attributed to its small size and the low soft tissue contrast of CT for its visualisation [14,38,52,55,67]. The small size of a structure and the low contrast of CT also explain the low DSC values of SVs and penile root in some settings, respectively [46,51,68]. The suboptimal DSC values of SVs as well as other structures including L and R femoral heads, large and small bowels, rectum, and sigmoid could be due to the variation in delineation guidelines for the DLAS model development and evaluation as well [41,49,52,57,67].
The DSC was the most common metric for determining the geometric accuracy of structures delineated by the DLAS software in the included studies (Table 2) [37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,55,57,58,59,60,61,63,65,66,67,68]. This is consistent with the findings of Ng’s [22] systematic review on the commercial DLAS software for breast cancer RT planning because the DSC facilitates the software performance comparison [37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,55,57,58,59,60,61,63,65,66,67,68]. Since each geometric accuracy metric has its advantages and disadvantages, more than 80% (26/32) of the included articles employed multiple geometric accuracy metrics to complement the weaknesses of individual ones [37,38,39,41,42,44,45,46,47,48,49,50,51,52,53,55,56,57,58,60,61,63,65,66,67,68]. However, discussion of their strengths and weaknesses is not within the scope of this review, and it is available elsewhere [21,71,72]. Nevertheless, all geometric metrics are considered less clinically meaningful because the main purpose of employing the DLAS is to automatically contour OARs and CTVs that meet the local clinicians’ requirements for providing effective RT treatments, leading to segmentation time reduction as a result of minimal manual adjustments for the contours [12,21,37,38,39,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,59,60,61,62,63,64,65,66,67,68].
Table 2 and Table 3 show that more than four fifths (26/32) of the included studies conducted geometric accuracy as well as subjective accuracy, efficiency, and/or dosimetric evaluations, and hence they were able to meet the NRG Oncology recommendation for more thorough assessment of the DLAS product performance. However, none of these papers used all evaluation approaches stated in Table 2 and Table 3, which is not ideal [12,21,37,38,39,41,42,43,44,45,46,48,49,50,51,52,53,55,56,59,60,61,63,64,65,66,67,68]. A good example for the comprehensive evaluation is illustrated in Almberg et al.’s [73] study on the evaluation of RaySearch Laboratories AB RayStation DLAS performance for breast cancer RT [12,21,22]. Nevertheless, these studies’ findings in Table 3 match the corresponding ones in Table 2, in general [12,21,37,38,39,41,42,43,44,45,46,48,49,50,51,52,53,55,56,59,60,61,63,64,65,66,67,68]. Thus, nine DLAS products, Carina Medical LLC INTContour [37,38,39,65,67], Limbus AI Inc. Contour [41,42,43,44,45,46,66], Manteia Medical Technologies Co. AccuContour [67], MIM Software Inc. Contour ProtégéAI [47,48,67], MVision.ai Contour+ [52,53,54,55,66,68], RaySearch Laboratories AB RayStation [56,57,65,68], Siemens Healthineers AG AI-Rad Companion Organs RT/syngo.via RT Image Suite/DirectORGANS [58,59,60,61], Therapanacea Annotate ART-Plan [62,63,68], and Varian Medical Systems, Inc. Ethos [64], should be useful for PCa RT planning in clinical settings.
Table 4 illustrates that a quarter of studies (all of which were published in the last three years) evaluated the commercial products (Carina Medical LLC INTContour [37,38,39,65,67], Mirada Medical Ltd. DLCExpert [51], MVision.ai Contour+ [53], and Therapanacea Annotate ART-Plan [63]) finetuned by additional datasets. Except for Carina Medical LLC INTContour, which provides a model finetuning function to end users (clinicians) [39], the other three products require their developers to do so through collaboration arrangements [51,53,63]. Model finetuning is an effective strategy for addressing the aforementioned issue of the commercial DLAS products, mismatch of contouring guidelines used by the developers and end users. Through re-training (finetuning) the commercial models with the end users’ datasets, the finetuned models would be able to meet their needs, resulting in better geometric and subjective accuracy, efficiency, and dosimetric outcomes [22,37,53,63,65]. For example, Duan et al. [65] used 57 intact PCa patients’ CT images to finetune the Carina Medical LLC INTContour original model. Their finetuned model outperformed the original one in terms of DSC, Hausdorff distance (HD), 95-percentile HD (HD95), 98-percentile HD (HD98), mean surface distance (MSD) and surface DSC (sDSC), and experienced ROs regarding HD95 and HD98 (p < 0.05) with no statistically significant differences for DSC, HD, MSD, and sDSC.
Another important role of DLAS model finetuning is to enable automatic MRI and CBCT image segmentation for meeting the increasing clinical demand on online adaptive RT (ART). Between treatment factions, prostate and SVs can move and deform. Hence, this requires structure re-contouring on new MRI/CBCT images acquired by linear accelerators during treatment sessions, but it is less practical if manual segmentation is performed [39,46,63,64]. It is noted that all DLAS products except Varian Medical Systems, Inc. Ethos covered in this review were originally trained with CT images only [37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68]. Although CT and CBCT images appear similar, the image contrast of CBCT is lower than the former. If the finetuning is not conducted, this will greatly affect the DLAS model performance [46]. In contrast, MRI has better soft tissue contrast which is good for DLAS [52]. However, the image characteristics of CT and MRI are very different. Again, the model finetuning is essential for such [63].
For the quality score range of all included studies, 40.0–72.0% (mean: 52.1% and median: 53.0%) (Table 4) [37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68], they are similar to the corresponding ones (range: 44.0–65.0%, mean: 53.9%, and median: 53.0%) of Ng’s [22] systematic review on the commercial DLAS software for breast cancer RT planning, and comparable to Sivanesan et al.’s [34] (median: 57.0%) and Bhandari et al.’s [35] (range: 23.8–73.8% and mean: 47.6%) included article quality scores (based on CLAIM) in their literature reviews on AI in medical imaging and MRI. Also, the issues of the papers included in this review are similar to those of the reviews by Ng [22], Sivanesan et al. [34], and Bhandari et al. [35] such as no sample size calculation in all studies and limited disclosure of details of dataset collection to assure no test set contamination [37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,74].
According to NRG Oncology [21], each clinical centre should perform its own evaluation of commercial DLAS software prior to implementation due to potential mismatch of delineation guidelines employed for software development and local clinical practice. However, such evaluation seems more important for Manteia Medical Technologies Co. AccuContour [67] and Varian Medical Systems, Inc. Ethos [64]. This is because they were only tested with small datasets (≤ 25 PCa cancer patients) in single studies, implying that the testing results might not be generalizable [15,64,67]. Also, Moazzezi et al. [64] only collected the data retrospectively, and Duan et al. [67] did not specify this.
There are two major limitations in this systematic review. Only one author with more than 20 years of experience in the literature review selected papers, extracted, and synthesised data [22,23,24,25,30]. However, as per a systematic review about methodology, this is considered appropriate when these processes are handled by an experienced single reviewer [22,23,24,25,30,75,76]. In addition, any potential bias could be further addressed through following the PRISMA guidelines [26], use of the data extraction form based on four review papers [12,14,15,22], and CLAIM [33,36]. Also, only articles written in English were included. This might affect its comprehensiveness [22,23,24,25,30]. Nevertheless, 32 papers on DLAS for PCa RT were covered in this review. This number is larger than those of Almeida et al.’s [14] and Matoska et al.’s [15] review articles. Furthermore, this paper included the studies from four continents [37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68].
5. Conclusions
This systematic review paper covers 12 commercial DLAS products, namely Carina Medical LLC INTContour, Elekta AB ADMIRE, Limbus AI Inc. Contour, Manteia Medical Technologies Co. AccuContour, MIM Software Inc. Contour ProtégéAI, Mirada Medical Ltd. DLCExpert, MVision.ai Contour+, Radformation Inc. AutoContour, RaySearch Laboratories AB RayStation, Siemens Healthineers AG AI-Rad Companion Organs RT, syngo.via RT Image Suite and DirectORGANS, Therapanacea Annotate, and Varian Medical Systems, Inc. Ethos, for the PCa RT planning. As per the geometric accuracy evaluation findings of the included papers, collectively, these products can delineate 12 OARs (abdominopelvic cavity, anal canal, bladder, body, cauda equina, L and R femurs, L and R pelvis, L and R proximal femurs, and sacrum) and four CTVs (prostate, lymph nodes, prostate bed, and SV bed) with clinically acceptable outcomes. These geometric accuracy results match the respective subjective and dosimetric assessment findings. These result in substantial average delineation time reduction per patient, 5.7–81.1%. In addition, this review’s findings reveal that Therapanacea Annotate ART-Plan is capable of delineating 13 OARs as well as the prostate with clinically acceptable outcomes. This contributes to minimal needs of structure contour adjustment and significant segmentation time reduction. Also, it has the model finetuning potential. Although NRG Oncology has recommended that each clinical centre should perform its own evaluation of commercial DLAS software prior to implementation due to potential mismatch of delineation guidelines employed for software development and local clinical practice, such evaluation seems more important for Manteia Medical Technologies Co. AccuContour and Varian Medical Systems, Inc. Ethos because of the methodological issues of the single studies that investigated them. In the future, comprehensive evaluations (including geometric accuracy, subjective, efficiency, and dosimetric assessments) with traditional power calculations to determine the external testing dataset size and prospective data collection from multiple centres should be conducted. Also, further studies on commercial model finetuning and reviews on non-commercial DLAS software are encouraged. It is expected that this review’s results will assist clinical centres in selecting commercial DLAS products for further evaluation prior to implementation.
Funding
This work received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The author declares no conflicts of interest.
References
- Prostate Cancer Statistics. Available online: https://www.wcrf.org/preventing-cancer/cancer-statistics/prostate-cancer-statistics/ (accessed on 15 January 2025).
- Cancer in Men: Prostate Cancer is #1 for 118 Countries Globally. Available online: https://www.cancer.org/research/acs-research-news/prostate-cancer-is-number-1-for-118-countries-worldwide.html (accessed on 15 January 2025).
- Prostate Cancer: 3. Epidemiology and Aetiology. Available online: https://uroweb.org/guidelines/prostate-cancer/chapter/epidemiology-and-aetiology (accessed on 15 January 2025).
- Wasim, S.; Park, J.; Nam, S.; Kim, J. Review of current treatment intensification strategies for prostate cancer patients. Cancers 2023, 15, 5615. [Google Scholar] [CrossRef] [PubMed]
- Hekman, L.; Barrett, A.; Ross, D.; Palaganas, E.; Giridhar, P.; Elumalai, T.; Pragathee, V.; Block, A.M.; Welsh, J.S.; Harkenrider, M.M.; et al. A systematic review of clinical trials comparing radiation therapy versus radical prostatectomy in prostate cancer. Clin. Genitourin. Cancer 2024, 22, 102157. [Google Scholar] [CrossRef] [PubMed]
- Numakura, K.; Kobayashi, M.; Muto, Y.; Sato, H.; Sekine, Y.; Sobu, R.; Aoyama, Y.; Takahashi, Y.; Okada, S.; Sasagawa, H.; et al. The current trend of radiation therapy for patients with localized prostate cancer. Curr. Oncol. 2023, 30, 8092–8110. [Google Scholar] [CrossRef] [PubMed]
- Fassia, M.K.; Balasubramanian, A.; Woo, S.; Vargas, H.A.; Hricak, H.; Konukoglu, E.; Becker, A.S. Deep learning prostate MRI segmentation accuracy and robustness: A systematic review. Radiol. Artif. Intell. 2024, 6, e230138. [Google Scholar] [CrossRef]
- Nayagam, R.D.; Selvathi, D. A systematic review of deep learning methods for the classification and segmentation of prostate cancer on magnetic resonance images. Int. J. Imaging Syst. Technol. 2024, 34, e23064. [Google Scholar] [CrossRef]
- Khan, Z.; Yahya, N.; Alsaih, K.; Al-Hiyali, M.I.; Meriaudeau, F. Recent automatic segmentation algorithms of MRI prostate regions: A review. IEEE Access 2021, 9, 97878–97905. [Google Scholar] [CrossRef]
- Kalantar, R.; Lin, G.; Winfield, J.M.; Messiou, C.; Lalondrelle, S.; Blackledge, M.D.; Koh, D.M. Automatic segmentation of pelvic cancers using deep learning: State-of-the-art approaches and challenges. Diagnostics 2021, 11, 1964. [Google Scholar] [CrossRef]
- Wu, C.; Montagne, S.; Hamzaoui, D.; Ayache, N.; Delingette, H.; Renard-Penna, R. Automatic segmentation of prostate zonal anatomy on MRI: A systematic review of the literature. Insights Imaging 2022, 13, 202. [Google Scholar] [CrossRef]
- De Biase, A.; Sijtsema, N.M.; Janssen, T.; Hurkmans, C.; Brouwer, C.; van Ooijen, P. Clinical adoption of deep learning target auto-segmentation for radiation therapy: Challenges, clinical risks, and mitigation strategies. BJR Artif. Intell. 2024, 1, ubae015. [Google Scholar] [CrossRef]
- Isaksson, L.J.; Summers, P.; Mastroleo, F.; Marvaso, G.; Corrao, G.; Vincini, M.G.; Zaffaroni, M.; Ceci, F.; Petralia, G.; Orecchia, R.; et al. Automatic segmentation with deep learning in radiotherapy. Cancers 2023, 15, 4389. [Google Scholar] [CrossRef]
- Almeida, G.; Tavares, J.M.R.S. Deep learning in radiation oncology treatment planning for prostate cancer: A systematic review. J. Med. Syst. 2020, 44, 179. [Google Scholar] [CrossRef] [PubMed]
- Matoska, T.; Patel, M.; Liu, H.; Beriwal, S. Review of deep learning based autosegmentation for clinical target volume: Current status and future directions. Adv. Radiat. Oncol. 2024, 9, 101470. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.K.C.; Leung, V.W.S.; Hung, R.H.M. Clinical evaluation of deep learning and atlas-based auto-contouring for head and neck radiation therapy. Appl. Sci. 2022, 12, 11681. [Google Scholar] [CrossRef]
- Erdur, A.C.; Rusche, D.; Scholz, D.; Kiechle, J.; Fischer, S.; Llorián-Salvador, Ó.; Buchner, J.A.; Nguyen, M.Q.; Etzel, L.; Weidner, J.; et al. Deep learning for autosegmentation for radiotherapy treatment planning: State-of-the-art and novel perspectives. Strahlenther. Onkol. 2025, 201, 236–254. [Google Scholar] [CrossRef]
- Sun, Z.; Ng, C.K.C. Artificial intelligence (enhanced super-resolution generative adversarial network) for calcium deblooming in coronary computed tomography angiography: A feasibility study. Diagnostics 2022, 12, 991. [Google Scholar] [CrossRef]
- Sun, Z.; Ng, C.K.C. Finetuned super-resolution generative adversarial network (artificial intelligence) model for calcium deblooming in coronary computed tomography angiography. J. Pers. Med. 2022, 12, 1354. [Google Scholar] [CrossRef]
- Leung, V.W.S.; Ng, C.K.C.; Lam, S.K.; Wong, P.T.; Ng, K.Y.; Tam, C.H.; Lee, T.C.; Chow, K.C.; Chow, Y.K.; Tam, V.C.W.; et al. Computed tomography-based radiomics for long-term prognostication of high-risk localized prostate cancer patients received whole pelvic radiotherapy. J. Pers. Med. 2023, 13, 1643. [Google Scholar] [CrossRef]
- Rong, Y.; Chen, Q.; Fu, Y.; Yang, X.; Al-Hallaq, H.A.; Wu, Q.J.; Yuan, L.; Xiao, Y.; Cai, B.; Latifi, K.; et al. NRG Oncology assessment of artificial intelligence deep learning-based auto-segmentation for radiation therapy: Current developments, clinical considerations, and future directions. Int. J. Radiat. Oncol. Biol. Phys. 2024, 119, 261–280. [Google Scholar] [CrossRef]
- Ng, C.K.C. Performance of commercial deep learning-based auto-segmentation software for breast cancer radiation therapy planning: A systematic review. Multimodal Technol. Interact. 2024, 8, 114. [Google Scholar] [CrossRef]
- Ng, C.K.C. Artificial intelligence for radiation dose optimization in pediatric radiology: A systematic review. Children 2022, 9, 1044. [Google Scholar] [CrossRef]
- Ng, C.K.C. Diagnostic performance of artificial intelligence-based computer-aided detection and diagnosis in pediatric radiology: A systematic review. Children 2023, 10, 525. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.K.C. Generative adversarial network (generative artificial intelligence) in pediatric radiology: A systematic review. Children 2023, 10, 1372. [Google Scholar] [CrossRef] [PubMed]
- PRISMA Statement. Available online: https://www.prisma-statement.org/ (accessed on 17 January 2025).
- Aggarwal, R.; Sounderajah, V.; Martin, G.; Ting, D.S.W.; Karthikesalingam, A.; King, D.; Ashrafian, H.; Darzi, A. Diagnostic accuracy of deep learning in medical imaging: A systematic review and meta-analysis. NPJ Digit. Med. 2021, 4, 65. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Liu, Y.; Zhang, X.; Ye, Y.; Yin, G.; Johnson, B.A. Deep learning in remote sensing applications: A meta-analysis and review. ISPRS J. Photogramm. Remote Sens. 2019, 152, 166–177. [Google Scholar] [CrossRef]
- Shah, K.A.; Ng, C.K.C. Workplace violence in medical radiation science: A systematic review. Radiography 2024, 30, 440–447. [Google Scholar] [CrossRef]
- Ng, C.K.C. A review of the impact of the COVID-19 pandemic on pre-registration medical radiation science education. Radiography 2022, 28, 222–231. [Google Scholar] [CrossRef]
- Vasey, B.; Ursprung, S.; Beddoe, B.; Taylor, E.H.; Marlow, N.; Bilbro, N.; Watkinson, P.; McCulloch, P. Association of clinician diagnostic performance with machine learning-based decision support systems: A systematic review. JAMA Netw. Open. 2021, 4, e211276. [Google Scholar] [CrossRef]
- Imrey, P.B. Limitations of meta-analyses of studies with high heterogeneity. JAMA Netw. Open. 2020, 3, e1919325. [Google Scholar] [CrossRef]
- Mongan, J.; Moy, L.; Kahn, C.E., Jr. Checklist for artificial intelligence in medical imaging (CLAIM): A guide for authors and reviewers. Radiol. Artif. Intell. 2020, 2, e200029. [Google Scholar] [CrossRef]
- Sivanesan, U.; Wu, K.; McInnes, M.D.F.; Dhindsa, K.; Salehi, F.; van der Pol, C.B. Checklist for artificial intelligence in medical imaging reporting adherence in peer-reviewed and preprint manuscripts with the highest Altmetric Attention Scores: A meta-research study. Can. Assoc. Radiol. J. 2023, 74, 334–342. [Google Scholar] [CrossRef]
- Bhandari, A.; Scott, L.; Weilbach, M.; Marwah, R.; Lasocki, A. Assessment of artificial intelligence (AI) reporting methodology in glioma MRI studies using the Checklist for AI in Medical Imaging (CLAIM). Neuroradiology 2023, 65, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Tejani, A.S.; Klontzas, M.E.; Gatti, A.A.; Mongan, J.T.; Moy, L.; Park, S.H.; Kahn, C.E. Jr; CLAIM 2024 Update Panel. Checklist for Artificial Intelligence in Medical Imaging (CLAIM): 2024 update. Radiol. Artif. Intell. 2024, 6, e240300. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Bernard, M.; Downes, L.; Willows, B.; Feng, X.; Mourad, W.F.; St Clair, W.; Chen, Q. Evaluating the clinical acceptability of deep learning contours of prostate and organs-at-risk in an automated prostate treatment planning process. Med. Phys. 2022, 49, 2570–2581. [Google Scholar] [CrossRef] [PubMed]
- Hobbis, D.; Yu, N.Y.; Mund, K.W.; Duan, J.; Rwigema, J.M.; Wong, W.W.; Schild, S.E.; Keole, S.R.; Feng, X.; Chen, Q.; et al. First report on physician assessment and clinical acceptability of custom-retrained artificial intelligence models for clinical target volume and organs-at-risk auto-delineation for postprostatectomy patients. Pract. Radiat. Oncol. 2023, 13, 351–362. [Google Scholar] [CrossRef]
- Tegtmeier, R.C.; Kutyreff, C.J.; Smetanick, J.L.; Hobbis, D.; Laughlin, B.S.; Toesca, D.A.S.; Clouser, E.L.; Rong, Y. Custom-trained deep learning-based auto-segmentation for male pelvic iterative CBCT on c-arm linear accelerators. Pract. Radiat. Oncol. 2024, 14, e383–e394. [Google Scholar] [CrossRef]
- Jenkins, A.; Mullen, T.S.; Johnson-Hart, C.; Green, A.; McWilliam, A.; Aznar, M.; van Herk, M.; Vasquez Osorio, E. Novel methodology to assess the effect of contouring variation on treatment outcome. Med. Phys. 2021, 48, 3234–3242. [Google Scholar] [CrossRef]
- Wong, J.; Fong, A.; McVicar, N.; Smith, S.; Giambattista, J.; Wells, D.; Kolbeck, C.; Giambattista, J.; Gondara, L.; Alexander, A. Comparing deep learning-based auto-segmentation of organs at risk and clinical target volumes to expert inter-observer variability in radiotherapy planning. Radiother. Oncol. 2020, 144, 152–158. [Google Scholar] [CrossRef]
- Wong, J.; Huang, V.; Wells, D.; Giambattista, J.; Giambattista, J.; Kolbeck, C.; Otto, K.; Saibishkumar, E.P.; Alexander, A. Implementation of deep learning-based auto-segmentation for radiotherapy planning structures: A workflow study at two cancer centers. Radiat. Oncol. 2021, 16, 101. [Google Scholar] [CrossRef]
- Zabel, W.J.; Conway, J.L.; Gladwish, A.; Skliarenko, J.; Didiodato, G.; Goorts-Matthews, L.; Michalak, A.; Reistetter, S.; King, J.; Nakonechny, K.; et al. Clinical evaluation of deep learning and atlas-based auto-contouring of bladder and rectum for prostate radiation therapy. Pract. Radiat. Oncol. 2021, 11, e80–e89. [Google Scholar] [CrossRef]
- Radici, L.; Ferrario, S.; Borca, V.C.; Cante, D.; Paolini, M.; Piva, C.; Baratto, L.; Franco, P.; La Porta, M.R. Implementation of a commercial deep learning-based auto segmentation software in radiotherapy: Evaluation of effectiveness and impact on workflow. Life 2022, 12, 2088. [Google Scholar] [CrossRef]
- Hoque, S.M.H.; Pirrone, G.; Matrone, F.; Donofrio, A.; Fanetti, G.; Caroli, A.; Rista, R.S.; Bortolus, R.; Avanzo, M.; Drigo, A.; et al. Clinical use of a commercial artificial intelligence-based software for autocontouring in radiation therapy: Geometric performance and dosimetric impact. Cancers 2023, 15, 5735. [Google Scholar] [CrossRef] [PubMed]
- Radici, L.; Piva, C.; Casanova Borca, V.; Cante, D.; Ferrario, S.; Paolini, M.; Cabras, L.; Petrucci, E.; Franco, P.; La Porta, M.R.; et al. Clinical evaluation of a deep learning CBCT auto-segmentation software for prostate adaptive radiation therapy. Clin. Transl. Radiat. Oncol. 2024, 47, 100796. [Google Scholar] [CrossRef] [PubMed]
- Urago, Y.; Okamoto, H.; Kaneda, T.; Murakami, N.; Kashihara, T.; Takemori, M.; Nakayama, H.; Iijima, K.; Chiba, T.; Kuwahara, J.; et al. Evaluation of auto-segmentation accuracy of cloud-based artificial intelligence and atlas-based models. Radiat. Oncol. 2021, 16, 175. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, G.; Mangili, P.; Deantoni, C.; Fodor, A.; Broggi, S.; Castriconi, R.; Ubeira Gabellini, M.G.; Del Vecchio, A.; Di Muzio, N.G.; Fiorino, C. Real-world validation of artificial intelligence-based computed tomography auto-contouring for prostate cancer radiotherapy planning. Phys. Imaging Radiat. Oncol. 2023, 28, 100501. [Google Scholar] [CrossRef]
- Walker, Z.; Bartley, G.; Hague, C.; Kelly, D.; Navarro, C.; Rogers, J.; South, C.; Temple, S.; Whitehurst, P.; Chuter, R. Evaluating the effectiveness of deep learning contouring across multiple radiotherapy centres. Phys. Imaging Radiat. Oncol. 2022, 24, 121–128. [Google Scholar] [CrossRef]
- Gibbons, E.; Hoffmann, M.; Westhuyzen, J.; Hodgson, A.; Chick, B.; Last, A. Clinical evaluation of deep learning and atlas-based auto-segmentation for critical organs at risk in radiation therapy. J. Med. Radiat. Sci. 2023, 70, 15–25. [Google Scholar] [CrossRef]
- Wang, Y.; Boyd, G.; Zieminski, S.; Kamran, S.C.; Zietman, A.L.; Miyamoto, D.T.; Kirk, M.C.; Efstathiou, J.A. A pair of deep learning auto-contouring models for prostate cancer patients injected with a radio-transparent versus radiopaque hydrogel spacer. Med. Phys. 2023, 50, 3324–3337. [Google Scholar] [CrossRef]
- Kiljunen, T.; Akram, S.; Niemelä, J.; Löyttyniemi, E.; Seppälä, J.; Heikkilä, J.; Vuolukka, K.; Kääriäinen, O.S.; Heikkilä, V.P.; Lehtiö, K.; et al. A deep learning-based automated CT segmentation of prostate cancer anatomy for radiation therapy planning-A retrospective multicenter study. Diagnostics 2020, 10, 959. [Google Scholar] [CrossRef]
- Elisabeth Olsson, C.; Suresh, R.; Niemelä, J.; Akram, S.U.; Valdman, A. Autosegmentation based on different-sized training datasets of consistently-curated volumes and impact on rectal contours in prostate cancer radiation therapy. Phys. Imaging Radiat. Oncol. 2022, 22, 67–72. [Google Scholar] [CrossRef]
- Strolin, S.; Santoro, M.; Paolani, G.; Ammendolia, I.; Arcelli, A.; Benini, A.; Bisello, S.; Cardano, R.; Cavallini, L.; Deraco, E.; et al. How smart is artificial intelligence in organs delineation? Testing a CE and FDA-approved deep-learning tool using multiple expert contours delineated on planning CT images. Front. Oncol. 2023, 13, 1089807. [Google Scholar] [CrossRef]
- Miura, H.; Ishihara, S.; Kenjo, M.; Nakao, M.; Ozawa, S.; Kagemoto, M. Evaluation of the accuracy of automated segmentation based on deep learning for prostate cancer patients. Med. Dosim. 2025, 50, 91–95. [Google Scholar] [CrossRef] [PubMed]
- De Kerf, G.; Claessens, M.; Raouassi, F.; Mercier, C.; Stas, D.; Ost, P.; Dirix, P.; Verellen, D. A geometry and dose-volume based performance monitoring of artificial intelligence models in radiotherapy treatment planning for prostate cancer. Phys. Imaging Radiat. Oncol. 2023, 28, 100494. [Google Scholar] [CrossRef]
- Kanwar, A.; Merz, B.; Claunch, C.; Rana, S.; Hung, A.; Thompson, R.F. Stress-testing pelvic autosegmentation algorithms using anatomical edge cases. Phys. Imaging Radiat. Oncol. 2023, 25, 100413. [Google Scholar] [CrossRef] [PubMed]
- Marschner, S.; Datar, M.; Gaasch, A.; Xu, Z.; Grbic, S.; Chabin, G.; Geiger, B.; Rosenman, J.; Corradini, S.; Niyazi, M.; et al. A deep image-to-image network organ segmentation algorithm for radiation treatment planning: Principles and evaluation. Radiat. Oncol. 2022, 17, 129. [Google Scholar] [CrossRef] [PubMed]
- Pera, Ó.; Martínez, Á.; Möhler, C.; Hamans, B.; Vega, F.; Barral, F.; Becerra, N.; Jimenez, R.; Fernandez-Velilla, E.; Quera, J.; et al. Clinical validation of Siemens’ Syngo.via automatic contouring system. Adv. Radiat. Oncol. 2023, 8, 101177. [Google Scholar] [CrossRef]
- Yamauchi, R.; Itazawa, T.; Kobayashi, T.; Kashiyama, S.; Akimoto, H.; Mizuno, N.; Kawamori, J. Clinical evaluation of deep learning and atlas-based auto-segmentation for organs at risk delineation. Med. Dosim. 2024, 49, 167–176. [Google Scholar] [CrossRef]
- Berenato, S.; Williams, M.; Woodley, O.; Möhler, C.; Evans, E.; Millin, A.E.; Wheeler, P.A. Novel dosimetric validation of a commercial CT scanner based deep learning automated contour solution for prostate radiotherapy. Phys. Med. 2024, 122, 103339. [Google Scholar] [CrossRef]
- Künzel, L.A.; Nachbar, M.; Hagmüller, M.; Gani, C.; Boeke, S.; Wegener, D.; Paulsen, F.; Zips, D.; Thorwarth, D. Clinical evaluation of autonomous, unsupervised planning integrated in MR-guided radiotherapy for prostate cancer. Radiother. Oncol. 2022, 168, 229–233. [Google Scholar] [CrossRef]
- Nachbar, M.; Lo Russo, M.; Gani, C.; Boeke, S.; Wegener, D.; Paulsen, F.; Zips, D.; Roque, T.; Paragios, N.; Thorwarth, D. Automatic AI-based contouring of prostate MRI for online adaptive radiotherapy. Z. Med. Phys. 2024, 34, 197–207. [Google Scholar] [CrossRef]
- Moazzezi, M.; Rose, B.; Kisling, K.; Moore, K.L.; Ray, X. Prospects for daily online adaptive radiotherapy via ethos for prostate cancer patients without nodal involvement using unedited CBCT auto-segmentation. J. Appl. Clin. Med. Phys. 2021, 22, 82–93. [Google Scholar] [CrossRef]
- Duan, J.; Tegtmeier, R.C.; Vargas, C.E.; Yu, N.Y.; Laughlin, B.S.; Rwigema, J.M.; Anderson, J.D.; Zhu, L.; Chen, Q.; Rong, Y. Achieving accurate prostate auto-segmentation on CT in the absence of MR imaging. Radiother. Oncol. 2025, 202, 110588. [Google Scholar] [CrossRef] [PubMed]
- Bordigoni, B.; Trivellato, S.; Pellegrini, R.; Meregalli, S.; Bonetto, E.; Belmonte, M.; Castellano, M.; Panizza, D.; Arcangeli, S.; De Ponti, E. Automated segmentation in pelvic radiotherapy: A comprehensive evaluation of ATLAS-, machine learning-, and deep learning-based models. Phys. Med. 2024, 125, 104486. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Vargas, C.E.; Yu, N.Y.; Laughlin, B.S.; Toesca, D.S.; Keole, S.; Rwigema, J.C.M.; Wong, W.W.; Schild, S.E.; Feng, X.; et al. Incremental retraining, clinical implementation, and acceptance rate of deep learning auto-segmentation for male pelvis in a multiuser environment. Med. Phys. 2023, 50, 4079–4091. [Google Scholar] [CrossRef] [PubMed]
- Doolan, P.J.; Charalambous, S.; Roussakis, Y.; Leczynski, A.; Peratikou, M.; Benjamin, M.; Ferentinos, K.; Strouthos, I.; Zamboglou, C.; Karagiannis, E. A clinical evaluation of the performance of five commercial artificial intelligence contouring systems for radiotherapy. Front. Oncol. 2023, 13, 1213068. [Google Scholar] [CrossRef]
- Van Dijk, L.V.; Van den Bosch, L.; Aljabar, P.; Peressutti, D.; Both, S.; Steenbakkers, R.J.H.M.; Langendijk, J.A.; Gooding, M.J.; Brouwer, C.L. Improving automatic delineation for head and neck organs at risk by deep learning contouring. Radiother. Oncol. 2020, 142, 115–123. [Google Scholar] [CrossRef]
- Gay, H.A.; Jin, J.Y.; Chang, A.J.; Ten Haken, R.K. Utility of normal tissue-to-tumor a/b Ratio when evaluating isodoses of isoeffective radiation therapy treatment plans. Int. J. Radiat. Oncol. Biol. Phys. 2012, 85, e81–e87. [Google Scholar] [CrossRef]
- Lin, H.; Xiao, H.; Dong, L.; Teo, K.B.; Zou, W.; Cai, J.; Li, T. Deep learning for automatic target volume segmentation in radiation therapy: A review. Quant. Imaging Med. Surg. 2021, 11, 4847–4858. [Google Scholar] [CrossRef]
- Mackay, K.; Bernstein, D.; Glocker, B.; Kamnitsas, K.; Taylor, A. A review of the metrics used to assess auto-contouring systems in radiotherapy. Clin. Oncol. 2023, 35, 354–369. [Google Scholar] [CrossRef]
- Almberg, S.S.; Lervåg, C.; Frengen, J.; Eidem, M.; Abramova, T.M.; Nordstrand, C.S.; Alsaker, M.D.; Tøndel, H.; Raj, S.X.; Wanderås, A.D. Training, validation, and clinical implementation of a deep-learning segmentation model for radiotherapy of loco-regional breast cancer. Radiother. Oncol. 2022, 173, 62–68. [Google Scholar] [CrossRef]
- Chai, Y. Letter to the editor regarding the article “comparison of transfer learning models in pelvic tilt and rotation measurement in pediatric anteroposterior pelvic radiographs”. J. Imaging Inform. Med. 2024, 37, 1259–1260. [Google Scholar] [CrossRef]
- Waffenschmidt, S.; Knelangen, M.; Sieben, W.; Bühn, S.; Pieper, D. Single screening versus conventional double screening for study selection in systematic reviews: A methodological systematic review. BMC Med. Res. Methodol. 2019, 19, 132. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Ng, C.K.C.; Dos Reis, C.S. Synchrotron radiation computed tomography versus conventional computed tomography for assessment of four types of stent grafts used for endovascular treatment of thoracic and abdominal aortic aneurysms. Quant. Imaging Med. Surg. 2018, 8, 609–620. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).