Harnessing Artificial Intelligence for Automated Diagnosis
Abstract
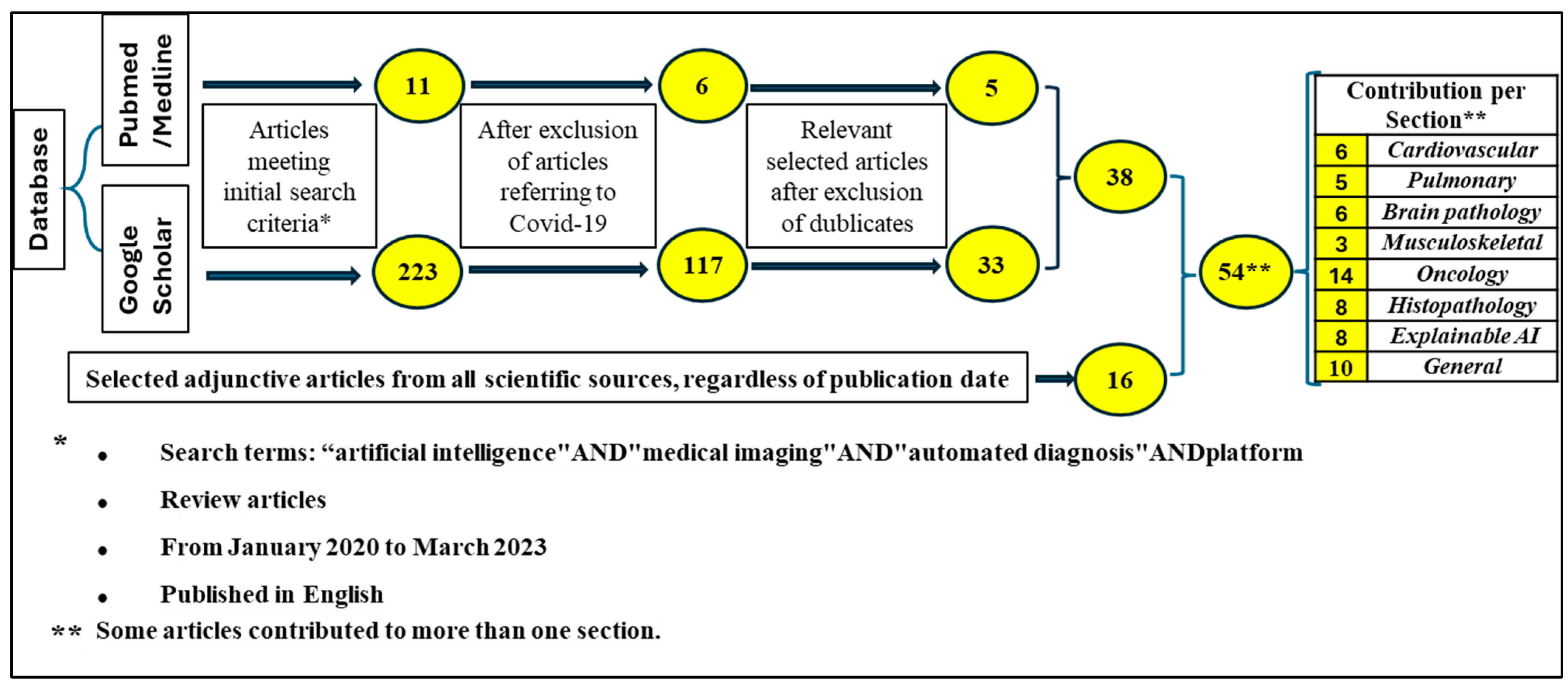
1. Introduction
2. Material and Methods
3. The Role of AI in the Diagnostic Imaging of Different Organ Systems
3.1. Cardiovascular System
3.2. Pulmonary System
3.3. Brain Pathology
3.4. Musculoskeletal
| Organ System | Disease | Medical Imaging/ DL Architecture | Outcomes | Author | Year |
|---|---|---|---|---|---|
| Cardiovascular | Coronary atherosclerosis | CCTA/ SVM algorithm | 93% sensitivity * 95% specificity ** 94% accuracy | Kang D. et al. [12] | 2015 |
| Hypertrophic cardiomyopathy | Cardiac MRI/ 3D-CNN ResNet18 | 85–89% accuracy | Linardos A. et al. [14] | 2022 | |
| Pulmonary | Pneumothorax | Chest X-ray/ CNN CheXNet | 88.87% accuracy in detection | Rajpurkar P. et al. [16] | 2017 |
| Chest X-ray/ 3 LinkNet networks: se-resnext50, se-resnext101 and SENet154 | 88.21% accuracy in classification | Groza V. et al. [17] | 2020 | ||
| Pulmonary embolism | CTPAs/ 2D segmentation U-Net model | 93% sensitivity 89% specificity | Ajmeral P. et al. [18] | 2022 | |
| Brain pathology | Multiple sclerosis | Brain MRI/ CNN architecture based on 3D convolutional layers | Distinction: 98.8% accuracy | Rocca M. et al. [20] | 2021 |
| Brain MRI/ 2D HWT and 3 CNN networks: Adam, SGS and RMSDrop | Identification: 95.4% precision 99.14% sensitivity 99.05% specificity | Alijamaat A. et al. [21] | 2021 | ||
| Tumor | Brain MRI/ 2-staged DL system | 72.7–88.9% sensitivity 84.9–96.8% specificity | Gao P. et al. [23] | 2022 | |
| Ischemic stroke | CT angiogram/ CNN, SVM, VGG-16, GoogleNet and ResNet-50, Viz-AI-Algrithm® v3.04 | 45–98% sensitivity 57–95% specificity in automated ASPECT scoring | Shafaat O. et al. [25] | 2021 | |
| Musculoskeletal | Osteoarthritis of the hip | Hip X-ray/ CNN model: VGG-16 layer network | 95% sensitivity * 90.7% specificity ** 92.8% accuracy | Xue Y. et al. [26] | 2017 |
| Femoral Intertrochanteric fractures | Hip X-ray/ Faster-RCNN | 89% sensitivity * 87%% specificity ** 88% accuracy | Liu P. et al. [27] | 2022 | |
| Distal radius fractures | Wrist X-ray ResNet18 DCNN | Fracture detection (2 tests): 97.5%/98.1% sensitivity * | Tobler P. et al. [28] | 2021 | |
| Fragment displacement (2 tests): 58.9%/73.6% accuracy | |||||
| Joint involvement (2 tests): 61.8%/65.4% accuracy | |||||
| Multiple fragments (2 tests): 84.2%/85.1% accuracy |
4. AI Algorithms for Tumor Detection in Oncology
| Tumor | Medical Imaging/ DL Architecture | Outcomes | Author | Year |
|---|---|---|---|---|
| Abnormal laryngeal cartilage | Thyroid CT/ VGG16 | 83% sensitivity 64% specificity | Santin M. et al. [30] | 2019 |
| Bone metastasis in the spine | Spine MRI/ CLSTM network | 75% sensitivity 83% specificity | Lang N. et al. [31] | 2019 |
| Bone lesions | Routine MRI/ EfficientNet and ImageNet database | Distinction benign vs. malignant 79% sensitivity 75% specificity | Eweje F. R. et al. [32] | 2021 |
| Bone metastasis | Bone scintigraphy/ ANN by EXINI diagnostics | 90% sensitivity 89% specificity | Sadik M. et al. [34] | 2008 |
| Gliomas | Endomicroscopy images/ WSL-based CNNmodel to generate DFM from CLE images | 87.5% accuracy | Izadyyazdanabadi M. et al. [35] | 2018 |
| Colorectal cancer | CT colonography/ RNN-ALGA algorithm | 97% accuracy | Sivaganesan D. [37] | 2016 |
| Primary bone tumor | Plain bone X-ray/ EfficientNet-B0 CNN architecture | 77.7% sensitivity 89.6% specificity malignant vs. not malignant | He Y. et al. [39] | 2020 |
| 82.7% sensitivity 81.8% specificity benign vs. not benign | ||||
| Cystic and lucent mandibular lesions | Panoramic radiographs/ DetectNet CNN implemented with NVIDIA DIGITS | 88% detection sensitivity | Ariji Y. et al. [40] | 2019 |
5. Computer-Aided Image Recognition in Histopathology
6. Explainable AI (XAI)
7. Standardization of Multimodal (Image and Text) Medical Reports
8. Discussion
8.1. Limitations of our Study
8.2. Limitations Encountered in Overviewed Studies
8.3. Suggestions for Further Investigation
9. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| ANN | Artificial Neural Network |
| ASPECT | Alberta Stroke Program Early CT |
| AUC | Area Under the Curve |
| CAD | Computer-Aided (or Assisted) Diagnosis |
| CAM | Class Activation Mapping |
| CLE | Confocal Laser Endomicroscopy |
| CCTA | Cardiac (or Coronary) Computed Tomography Angiography |
| CDSS | Clinical Decision Support System |
| CIU | Contextual Importance and Utility |
| CLSTM | Convolutional Long Short-Term Memory |
| CNN | Convolutional Neural Network |
| CT | Computerized (Axial) Tomography |
| CTC | Computed Tomography Colonography |
| CTPAs | Computerized Tomography Pulmonary Angiograms |
| DCNNs | Deep Convolutional Neural Networks |
| DFM | Diagnostic Feature Map |
| DIANA | DICOM Image ANalysis and Archive |
| DICOM | Digital Imaging and Communication in Medicine |
| DL | Deep Learning |
| DSC | Dice Similarity Coefficient |
| DT | Decision Tree |
| EMR | Electronic Medical Record |
| EES | Explainable Expert System |
| ET | Extra Trees |
| FL | Federation Learning |
| GANs | Generative Adversarial Networks |
| HWT | Haar Wavelet Transform |
| KFCV | K-fold Cross-Validation |
| LIME | Local Interpretable Model-Agnostic Explanations |
| LRP | Layer-Wise Relevance Propagation |
| ML | Machine Learning |
| LSTM | Long Short-Term Memory |
| MRI | Magnetic Resonance Imaging |
| NLP | Natural Language Processing |
| PACS | Picture Archive and Communication System |
| PET | Positron Emission Tomography |
| PHPs | Persistent Homology Maps |
| R-CNN | Region-Based Convolutional Neural Network |
| RNN-ALGA | Regression Neural Network-Augmented Lagrangian Genetic Algorithm |
| RF | Random Forest |
| SHAP | SHapley Additive exPlanations |
| SVM | Support Vector Machine |
| UMAP | Uniform Manifold Approximation and Projection |
| VAEs | Variational Autoencoders |
| VR | Virtual Reality |
| WSI | Whole-Slide Imaging |
| WSL | Weakly Supervised Learning |
| XAI | Explainable AI |
References
- Shaikh, F.; Dehmeshki, J.; Bisdas, S.; Roettger-Dupont, D.; Kubassova, O.; Aziz, M.; Awan, O. Artificial Intelligence-Based Clinical Decision Support Systems Using Advanced Medical Imaging and Radiomics. Curr. Probl. Diagn. Radiol. 2021, 50, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Lucieri, A.; Bajwa, M.N.; Dengel, A.; Ahmed, S. Achievements and challenges in explaining deep learning based computer-aided diagnosis systems. arXiv 2020, arXiv:2011.13169. [Google Scholar]
- Pandey, B.; Pandey, D.K.; Mishra, B.P.; Rhmann, W. A comprehensive survey of deep learning in the field of medical imaging and medical natural language processing: Challenges and research directions. J. King Saud Univ. Comput. Inf. Sci. 2022, 34, 5083–5099. [Google Scholar] [CrossRef]
- Gore, J.C. Artificial intelligence in medical imaging. Magn. Reson. Imaging 2020, 68, A1–A4. [Google Scholar] [CrossRef] [PubMed]
- Barragán-Montero, A.; Javaid, U.; Valdés, G.; Nguyen, D.; Desbordes, P.; Macq, B.; Willems, S.; Vandewinckele, L.; Holmström, M.; Löfman, F.; et al. Artificial intelligence and machine learning for medical imaging: A technology review. Phys. Med. 2021, 83, 242–256. [Google Scholar] [CrossRef] [PubMed]
- Diaz, O.; Kushibar, K.; Osuala, R.; Linardos, A.; Garrucho, L.; Igual, L.; Radeva, P.; Prior, F.; Gkontra, P.; Lekadir, K. Data preparation for artificial intelligence in medical imaging: A comprehensive guide to open-access platforms and tools. Phys. Med. 2021, 83, 25–37. [Google Scholar] [CrossRef]
- Boeken, T.; Feydy, J.; Lecler, A.; Soyer, P.; Feydy, A.; Barat, M.; Duron, L. Artificial intelligence in diagnostic and interventional radiology: Where are we now? Diagn. Interv. Imaging 2023, 104, 1–5. [Google Scholar] [CrossRef]
- Clunie, D.A. DICOM Format and Protocol Standardization-A Core Requirement for Digital Pathology Success. Toxicol. Pathol. 2021, 49, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Han, R.; Shao, G.; Lv, B.; Sun, K. Artificial Intelligence in Cardiovascular Atherosclerosis Imaging. J. Pers. Med. 2022, 12, 420. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Brandt, V.; Emrich, T.; Schoepf, U.J.; Dargis, D.M.; Bayer, R.R.; De Cecco, C.N.; Tesche, C. Ischemia and outcome prediction by cardiac CT based machine learning. Int. J. Cardiovasc. Imaging 2020, 36, 2429–2439. [Google Scholar] [CrossRef] [PubMed]
- Patel, B.; Makaryus, A.N. Artificial Intelligence Advances in the World of Cardiovascular Imaging. Healthcare 2022, 10, 154. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kang, D.; Dey, D.; Slomka, P.J.; Arsanjani, R.; Nakazato, R.; Ko, H.; Berman, D.S.; Li, D.; Kuo, C.C. Structured learning algorithm for detection of nonobstructive and obstructive coronary plaque lesions from computed tomography angiography. J. Med. Imaging 2015, 2, 014003. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Seetharam, K.; Bhat, P.; Orris, M.; Prabhu, H.; Shah, J.; Asti, D.; Chawla, P.; Mir, T. Artificial intelligence and machine learning in cardiovascular computed tomography. World J. Cardiol. 2021, 13, 546–555. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Linardos, A.; Kushibar, K.; Walsh, S.; Gkontra, P.; Lekadir, K. Federated learning for multi-center imaging diagnostics: A simulation study in cardiovascular disease. Sci. Rep. 2022, 12, 3551. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Iqbal, T.; Shaukat, A.; Akram, M.U.; Mustansar, Z.; Khan, A. Automatic Diagnosis of Pneumothorax from Chest Radiographs: A Systematic Literature Review. IEEE Access 2021, 9, 145817–145839. [Google Scholar] [CrossRef]
- Rajpurkar, P.; Irvin, J.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.; Shpanskaya, K.; et al. CheXNet: Radiologist-level pneumonia detection on chest X-rays with deep learning. arXiv 2017, arXiv:1711.05225. [Google Scholar]
- Groza, V.; Kuzin, A. Pneumothorax segmentation with effective conditioned post-processing in chest X-ray. In Proceedings of the IEEE 17th International Symposium on Biomedical Imaging Workshops (ISBI Workshops), Iowa City, IA, USA, 3–7 April 2020; pp. 1–4. [Google Scholar] [CrossRef]
- Ajmera, P.; Kharat, A.; Seth, J.; Rathi, S.; Pant, R.; Gawali, M.; Kulkarni, V.; Maramraju, R.; Kedia, I.; Botchu, R.; et al. A deep learning approach for automated diagnosis of pulmonary embolism on computed tomographic pulmonary angiography. BMC Med. Imaging 2022, 22, 195. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cano-Espinosa, C.; Cazorla, M.; González, G. Computer Aided Detection of Pulmonary Embolism Using Multi-Slice Multi-Axial Segmentation. Appl. Sci. 2020, 10, 2945. [Google Scholar] [CrossRef]
- Rocca, M.A.; Anzalone, N.; Storelli, L.; Del Poggio, A.; Cacciaguerra, L.; Manfredi, A.A.; Meani, A.; Filippi, M. Deep Learning on Conventional Magnetic Resonance Imaging Improves the Diagnosis of Multiple Sclerosis Mimics. Investig. Radiol. 2021, 56, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Alijamaat, A.; NikravanShalmani, A.; Bayat, P. Multiple sclerosis identification in brain MRI images using wavelet convolutional neural networks. Int. J. Imaging Syst. Technol. 2021, 31, 778–785. [Google Scholar] [CrossRef]
- Olatunji, S.O.; Alsheikh, N.; Alnajrani, L.; Alanazy, A.; Almusairii, M.; Alshammasi, S.; Alansari, A.; Zaghdoud, R.; Alahmadi, A.; Basheer Ahmed, M.I.; et al. Comprehensible Machine-Learning-Based Models for the Pre-Emptive Diagnosis of Multiple Sclerosis Using Clinical Data: A Retrospective Study in the Eastern Province of Saudi Arabia. Int. J. Environ. Res. Public Health 2023, 20, 4261. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gao, P.; Shan, W.; Guo, Y.; Wang, Y.; Sun, R.; Cai, J.; Li, H.; Chan, W.S.; Liu, P.; Yi, L.; et al. Development and Validation of a Deep Learning Model for Brain Tumor Diagnosis and Classification Using Magnetic Resonance Imaging. JAMA Netw. Open 2022, 5, e2225608. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Naeem, A.; Anees, T.; Naqvi, R.A.; Loh, W.K. A Comprehensive Analysis of Recent Deep and Federated-Learning-Based Methodologies for Brain Tumor Diagnosis. J. Pers. Med. 2022, 12, 275. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shafaat, O.; Bernstock, J.D.; Shafaat, A.; Yedavalli, V.S.; Elsayed, G.; Gupta, S.; Sotoudeh, E.; Sair, H.I.; Yousem, D.M.; Sotoudeh, H. Leveraging artificial intelligence in ischemic stroke imaging. J. Neuroradiol. 2022, 49, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Zhang, R.; Deng, Y.; Chen, K.; Jiang, T. A preliminary examination of the diagnostic value of deep learning in hip osteoarthritis. PLoS ONE 2017, 12, e0178992. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, P.; Lu, L.; Chen, Y.; Huo, T.; Xue, M.; Wang, H.; Fang, Y.; Xie, Y.; Xie, M.; Ye, Z. Artificial intelligence to detect the femoral intertrochanteric fracture: The arrival of the intelligent-medicine era. Front. Bioeng. Biotechnol. 2022, 10, 927926. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tobler, P.; Cyriac, J.; Kovacs, B.K.; Hofmann, V.; Sexauer, R.; Paciolla, F.; Stieltjes, B.; Amsler, F.; Hirschmann, A. AI-based detection and classification of distal radius fractures using low-effort data labeling: Evaluation of applicability and effect of training set size. Eur. Radiol. 2021, 31, 6816–6824. [Google Scholar] [CrossRef] [PubMed]
- Tufail, A.B.; Ma, Y.K.; Kaabar, M.K.A.; Martínez, F.; Junejo, A.R.; Ullah, I.; Khan, R. Deep Learning in Cancer Diagnosis and Prognosis Prediction: A Minireview on Challenges, Recent Trends, and Future Directions. Comput. Math. Methods Med. 2021, 2021, 9025470. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Santin, M.; Brama, C.; Théro, H.; Ketheeswaran, E.; El-Karoui, I.; Bidault, F.; Gillet, R.; Gondim Teixeira, P.; Blum, A. Detecting abnormal thyroid cartilages on CT using deep learning. Diagn. Interv. Imaging 2019, 100, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Lang, N.; Zhang, Y.; Zhang, E.; Zhang, J.; Chow, D.; Chang, P.; Yu, H.J.; Yuan, H.; Su, M.Y. Differentiation of spinal metastases originated from lung and other cancers using radiomics and deep learning based on DCE-MRI. Magn. Reson. Imaging 2019, 64, 4–12. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Eweje, F.R.; Bao, B.; Wu, J.; Dalal, D.; Liao, W.H.; He, Y.; Luo, Y.; Lu, S.; Zhang, P.; Peng, X.; et al. Deep Learning for Classification of Bone Lesions on Routine MRI. EBioMedicine 2021, 68, 103402. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, M.D.; Ahmed, S.R.; Choy, E.; Lozano-Calderon, S.A.; Kalpathy-Cramer, J.; Chang, C.Y. Artificial intelligence applied to musculoskeletal oncology: A systematic review. Skeletal. Radiol. 2022, 51, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Sadik, M.; Hamadeh, I.; Nordblom, P.; Suurkula, M.; Höglund, P.; Ohlsson, M.; Edenbrandt, L. Computer-assisted interpretation of planar whole-body bone scans. J. Nucl. Med. 2008, 49, 1958–1965. [Google Scholar] [CrossRef] [PubMed]
- Izadyyazdanabadi, M.; Belykh, E.; Cavallo, C.; Zhao, X.; Gandhi, S.; Moreira, L.B.; Eschbacher, J.; Nakaji, P.; Preul, M.C.; Yang, Y. Weakly-supervised learning-based feature localization for confocal laser endomicroscopy glioma images. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2018: 21st International Conference, Granada, Spain, 16–20 September 2018; Proceedings, Part II 11. Springer International Publishing: Berlin/Heidelberg, Germany, 2018; pp. 300–308. [Google Scholar]
- Liang, F.; Wang, S.; Zhang, K.; Liu, T.J.; Li, J.N. Development of artificial intelligence technology in diagnosis, treatment, and prognosis of colorectal cancer. World J. Gastrointest. Oncol. 2022, 14, 124–152. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sivaganesan, D. Wireless Distributive Personal Communication for Early Detection of Collateral Cancer Using Optimized Machine Learning Methodology. Wireless Pers. Commun. 2017, 94, 2291–2302. [Google Scholar] [CrossRef]
- Soomro, M.H.; De Cola, G.; Conforto, S.; Schmid, M.; Giunta, G.; Guidi, E.; Neri, E.; Caruso, D.; Ciolina, M.; Laghi, A. Automatic segmentation of colorectal cancer in 3D MRI by combining deep learning and 3D level-set algorithm-a preliminary study. In Proceedings of the 2018 IEEE 4th Middle East Conference on Biomedical Engineering (MECBME), Gammarth, Tunisia, 28–30 March 2018; pp. 198–203. [Google Scholar] [CrossRef]
- He, Y.; Pan, I.; Bao, B.; Halsey, K.; Chang, M.; Liu, H.; Peng, S.; Sebro, R.A.; Guan, J.; Yi, T.; et al. Deep learning-based classification of primary bone tumors on radiographs: A preliminary study. EBioMedicine 2020, 62, 103121. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ariji, Y.; Yanashita, Y.; Kutsuna, S.; Muramatsu, C.; Fukuda, M.; Kise, Y.; Nozawa, M.; Kuwada, C.; Fujita, H.; Katsumata, A.; et al. Automatic detection and classification of radiolucent lesions in the mandible on panoramic radiographs using a deep learning object detection technique. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2019, 128, 424–430. [Google Scholar] [CrossRef] [PubMed]
- Sirinukunwattana, K.; Raza, S.E.A.; Tsang, Y.W.; Snead, D.R.; Cree, I.A.; Rajpoot, N.M. Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images. IEEE Trans. Med. Imaging 2016, 35, 1196–1206. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Luo, X.; Wang, G.; Gilmore, H.; Madabhushi, A. A deep convolutional neural network for segmenting and classifying epithelial and stromal regions in histopathological images. Neurocomputing 2016, 191, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Arunachalam, H.B.; Mishra, R.; Daescu, O.; Cederberg, K.; Rakheja, D.; Sengupta, A.; Leonard, D.; Hallac, R.; Leavey, P. Viable and necrotic tumor assessment from whole slide images of osteosarcoma using machine-learning and deep-learning models. PLoS ONE 2019, 14, e0210706. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Xue, P.; Ji, H.; Cui, W.; Dong, E. Deep model with Siamese network for viable and necrotic tumor regions assessment in osteosarcoma. Med. Phys. 2020, 47, 4895–4905. [Google Scholar] [CrossRef] [PubMed]
- Chaber, R.; Arthur, C.J.; Łach, K.; Raciborska, A.; Michalak, E.; Bilska, K.; Drabko, K.; Depciuch, J.; Kaznowska, E.; Cebulski, J. Predicting Ewing Sarcoma Treatment Outcome Using Infrared Spectroscopy and Machine Learning. Molecules 2019, 24, 1075. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yamamoto, Y.; Tsuzuki, T.; Akatsuka, J.; Ueki, M.; Morikawa, H.; Numata, Y.; Takahara, T.; Tsuyuki, T.; Tsutsumi, K.; Nakazawa, R.; et al. Automated acquisition of explainable knowledge from unannotated histopathology images. Nat. Commun. 2019, 10, 5642. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Allahqoli, L.; Laganà, A.S.; Mazidimoradi, A.; Salehiniya, H.; Günther, V.; Chiantera, V.; Karimi Goghari, S.; Ghiasvand, M.M.; Rahmani, A.; Momenimovahed, Z.; et al. Diagnosis of Cervical Cancer and Pre-Cancerous Lesions by Artificial Intelligence: A Systematic Review. Diagnostics 2022, 12, 2771. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Dai, W.; Zhou, Q.; Li, C.; Li, S.C.; Wang, C.; Tang, J.; Wu, X.; Wu, R. AI-assisted system improves the work efficiency of cytologists via excluding cytology-negative slides and accelerating the slide interpretation. Front. Oncol. 2023, 13, 1290112. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Band, S.S.; Yarahmadi, A.; Hsu, C.C.; Biyari, M.; Sookhak, M.; Ameri, R.; Dehzangi, I.; Chronopoulos, A.T.; Liang, H.W. Application of explainable artificial intelligence in medical health: A systematic review of interpretability methods. Inform. Med. Unlocked 2023, 40, 101286. [Google Scholar] [CrossRef]
- Erfannia, L.; Alipour, J. How does cloud computing improve cancer information management? A systematic review. Inform. Med. Unlocked 2022, 33, 101095. [Google Scholar] [CrossRef]
- Cresswell, K.; Rigby, M.; Magrabi, F.; Scott, P.; Brender, J.; Craven, C.K.; Wong, Z.S.; Kukhareva, P.; Ammenwerth, E.; Georgiou, A.; et al. The need to strengthen the evaluation of the impact of Artificial Intelligence-based decision support systems on healthcare provision. Health Policy 2023, 136, 104889. [Google Scholar] [CrossRef] [PubMed]
- Groot, O.Q.; Bongers, M.E.R.; Karhade, A.V.; Kapoor, N.D.; Fenn, B.P.; Kim, J.; Verlaan, J.J.; Schwab, J.H. Natural language processing for automated quantification of bone metastases reported in free-text bone scintigraphy reports. Acta Oncol. 2020, 59, 1455–1460. [Google Scholar] [CrossRef] [PubMed]
- Yi, T.; Pan, I.; Collins, S.; Chen, F.; Cueto, R.; Hsieh, B.; Hsieh, C.; Smith, L.J.; Yang, L.; Liao, W.; et al. DICOM Image ANalysis and Archive (DIANA): An Open-Source System for Clinical AI Applications. J. Digit. Imaging 2021, 34, 1405–1413. [Google Scholar] [CrossRef]
- Maleki, F.; Muthukrishnan, N.; Ovens, K.; Reinhold, C.; Forghani, R. Machine Learning Algorithm Validation: From Essentials to Advanced Applications and Implications for Regulatory Certification and Deployment. Neuroimaging Clin. N. Am. 2020, 30, 433–445. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zachariadis, C.B.; Leligou, H.C. Harnessing Artificial Intelligence for Automated Diagnosis. Information 2024, 15, 311. https://doi.org/10.3390/info15060311
Zachariadis CB, Leligou HC. Harnessing Artificial Intelligence for Automated Diagnosis. Information. 2024; 15(6):311. https://doi.org/10.3390/info15060311
Chicago/Turabian StyleZachariadis, Christos B., and Helen C. Leligou. 2024. "Harnessing Artificial Intelligence for Automated Diagnosis" Information 15, no. 6: 311. https://doi.org/10.3390/info15060311
APA StyleZachariadis, C. B., & Leligou, H. C. (2024). Harnessing Artificial Intelligence for Automated Diagnosis. Information, 15(6), 311. https://doi.org/10.3390/info15060311






