Abstract
Accurate and efficient medicinal plant image classification is of utmost importance as these plants produce a wide variety of bioactive compounds that offer therapeutic benefits. With a long history of medicinal plant usage, different parts of plants, such as flowers, leaves, and roots, have been recognized for their medicinal properties and are used for plant identification. However, leaf images are extensively used due to their convenient accessibility and are a major source of information. In recent years, transfer learning and fine-tuning, which use pre-trained deep convolutional networks to extract pertinent features, have emerged as an extremely effective approach for image-identification problems. This study leveraged the power by three-component deep convolutional neural networks, namely VGG16, VGG19, and DenseNet201, to derive features from the input images of the medicinal plant dataset, containing leaf images of 30 classes. The models were compared and ensembled to make four hybrid models to enhance the predictive performance by utilizing the averaging and weighted averaging strategies. Quantitative experiments were carried out to evaluate the models on the Mendeley Medicinal Leaf Dataset. The resultant ensemble of VGG19+DensNet201 with fine-tuning showcased an enhanced capability in identifying medicinal plant images with an improvement of 7.43% and 5.8% compared with VGG19 and VGG16. Furthermore, VGG19+DensNet201 can outperform its standalone counterparts by achieving an accuracy of 99.12% on the test set. A thorough assessment with metrics such as accuracy, recall, precision, and the F1-score firmly established the effectiveness of the ensemble strategy.
1. Introduction
Medicinal plants are gaining popularity in the pharmaceutical industry as they are less likely to have adverse effects and are less expensive than modern pharmaceuticals. According to the World Health Organization, there are over 21,000 plant species that can potentially be utilized for medicinal purposes. It is also reported that 80% of people around the world use medicinal plants for the treatment of their primary health ailments [1].
Systematic identification and naming of plants are often carried out by professional botanists (taxonomists), who have deep knowledge of plant taxonomy [2,3]. Manually identifying plant species is a challenging and time-consuming process. Furthermore, the process is prone to errors, as every aspect of the identification is entirely based on human perception [4]. There is also a dearth of these plant identification subject matter experts, which gives rise to a situation of “taxonomic impediment” [5]. It is, therefore, important to develop an effective and reliable method for the accurate identification of these valuable plants.
Machine learning (ML) is a sub-field of artificial intelligence that solves various intricate problems in different application domains with minimal human intervention [6]. Deep learning (DL), inspired by the structure and functionality of biological neurons, is a sub-field of machine learning (ML). DL involves the training of algorithms and demands a large amount of data to achieve better identification results. Due to the advancements in hardware technology and the extensive availability of the data, DL has gained popularity in various tasks like natural language processing, game playing, and image processing, and outstanding performance is being achieved, which could be otherwise impossible for humans to discern [7].
Various research studies have revealed that researchers are showing a great interest in the automatic identification and classification of plant species using plant image specimens and by employing ML and DL techniques [8,9,10,11,12]. This automatic identification is carried out in different stages, viz. (a) image acquisition, (b) image preprocessing, (c) feature extraction, and (d) classification. Plant species identification can be performed using the different parts of plants like flowers, bark, fruits, and leaves or using the entire plant image. Researchers prefer to use leaf images for the identification process as leaves are easily identifiable parts of the plant. Leaf images are usually available for a long time during the year, while flowers and fruits are specific to a particular season only. There are more the one-hundred studies that have used plant leaf images for the automatic identification process [4].
In this research, transfer learning and ensemble learning approaches were employed to classify medicinal plant leaf images into thirty classes. The transfer learning approach uses the existing knowledge gained from one task to solve problems of a related nature [13]. This approach is extensively used in image-classification problems, particularly with convolutional neural network models pre-trained on advanced GPUs capable of categorizing objects across a wide range of 1000 classes. Transfer learning can be used in medicinal plant image classification as the approach facilitates the migration of acquired features and parameters, so it reduces the need for extensive training from scratch.
The main aim of ensemble learning is to improve the overall performance of classifiers by combining the predictions of individual neural network models. Ensemble learning has recently gained popularity in image classification using deep learning [14,15,16]. We trained VGG16, VGG19, and DenseNet201 on the Mendeley Medicinal Leaf Dataset and evaluated the efficiency of these component models. After individual evaluation of the models, an ensemble learning approach using the averaging and weighted averaging strategies was employed to make the final prediction. Some of the novel contributions of this work are as follows:
- An automated system was developed to reduce the reliance on human experts when it comes to identifying different medicinal plant species. Classically, identifying plant species often requires experts who possess knowledge of botanical characteristics, but this system aims to lessen that dependency by using technology to perform the identification task.
- In this work, we adjusted the hyperparameters, like the learning rates, batch sizes, and regularization techniques, to ensure the model performed optimally for this classification task.
- Also, transfer learning and fine-tuning are being used to extract meaningful and informative features from images of medicinal plant leaves. Instead of training a deep learning model from scratch, the pre-trained models VGG16, VGG19, and DenseNet201 were used as a starting point. These models were leveraged to enhance the ability of identifying and extracting relevant information from medicinal plant images, which were used for species identification or other related purposes.
- Also, a comparative analysis was performed in this study, including several previously state-of-the-art approaches for the identification of medicinal plants using the same dataset, leveraging the complementary features of VGG19 and DenseNet201, the ensemble approach improved the robustness and balanced the performance, making it a potent solution for medicinal plant identification.
In this section, we provide an overview of medicinal plants, highlighting their numerous benefits and the challenges associated with their identification. We also discuss existing methods used for identifying medicinal plants.
The upcoming sections will explore various related research studies shedding light on the advancements made in utilizing deep learning techniques for plant identification using their image data. Following the literature review, we describe the methodology employed and present the results obtained from our experiments. Furthermore, we discuss the significance and relevance of our proposed approach in addressing the existing challenges in medicinal plant identification.
2. Related Work
Numerous efforts have been undertaken to tackle the challenge of identifying medicinal plants from their different parts [17], employing different machine learning approaches. However, researchers and experts often rely on leaf images to accurately classify other plant species, as leaf images, are widely recognized as the most-accessible and trustworthy sources of information for plant species identification.
Various authors have relied on low-level features such as the leaf shape, color, and texture to differentiate between species. For example, the mobile application is known as Leafsnap by Kumar et al. [18] identifies plant species based on plant leaf images. The feature extraction method captures the curvature of leaf margins at multiple scales. The authors of [19] emphasized incorporating all these features, including shape, color, texture, and venation, into a comprehensive feature vector for the probabilistic neural network (PNN). By utilizing this feature vector, they achieved an impressive average accuracy of 93.75% on the publicly accessible Flavia dataset [20]. Amala Sabu et al. [21] proposed an Ayurvedic plant-identification system and extracted features with SURF and HOG-based techniques. In this study, a total of 200 images of 20 different plant leaves were collected, and K-NN was chosen as the classifier. It is evident that the studies as mentioned earlier have primarily concentrated on recognition using hand-engineered image features. However, this approach has several limitations that need to be considered. The expressiveness and ability to discern intricate patterns and subtle changes in the data may be lacking in handcrafted features. Manual feature engineering is laborious, time-consuming, and subject to bias. Handcrafted features might not be able to manage noise and variations effectively. For massive datasets, the approach can also be computationally costly and ineffective. These techniques are difficult to apply for practical applications and necessitate the need to design a method that is less affected by the environment and is well suited for the recognition of real-world plant images.
In recent years, there has been a notable shift towards the adoption of convolutional neural networks (CNNs). CNNs have gained prominence due to their ability to automatically learn and extract meaningful features from input images, reducing the need for extensive human intervention in the feature-extraction process. These algorithms have received much attention in the literature. Sobitha Raj et al. [22] proposed a deep learning architecture by extracting features using MobileNet and DenseNet-121. In the study, different classifiers, viz. K-nearest neighbor, multinomial logistic regression, and linear discriminant analysis were used. Barre et al. [23] devised a deep learning approach for extracting distinctive features from leaf images and, subsequently, classifying plant species. The authors revealed that the learned features obtained from a convolutional neural network (CNN) outperformed the handcrafted features. In order to automatically detect plant diseases, the researchers in [24] successfully created a robust and deep CNN model with nine layers. To enhance the learning capability of the CNN, the input data were augmented, resulting in an increased number of samples. The model was subjected to rigorous testing using various classifiers, and it achieved an impressive accuracy rate of 96%.
In computer vision tasks, several widely recognized and open-source models have gained popularity, including VGG-16, VGG-19 [25], Inception V3 [26], DenseNet [27], and ResNet-50 [28]. These state-of-the-art architectures have been extensively trained, and other researchers can utilize their parameters and weights to address problems in various domains. This allows for efficient knowledge transfer and facilitates the application of pre-trained models in different computer vision applications. Reference [11] employed transfer learning to extract features from the pre-trained networks and used artificial neural networks (ANNs), support vector machines (SVMs), and an SVM with batch optimization (SVM with BO) as the classification algorithms. The results of their study demonstrated that the model trained with the Xception network and ANN achieved an impressive accuracy of 97.5% for real-time leaf images. Transfer learning was also employed by [29] to create different combinations of pre-trained MobileNet CNNs to classify medicinal plant leaves.
3. Methods and Material
3.1. Convolutional Neural Network
In the domain of artificial intelligence, significant progress has been made in recent years, particularly in the field of computer vision. Researchers have dedicated their efforts to unlocking the potential of machines in understanding and interpreting visual information, such as images and videos. The application of deep learning in computer vision has propelled the field forward, enabling machines to analyze, interpret, and make intelligent decisions based on visual input. This advancement has been made possible through the development of convolutional neural networks (CNNs) and deep learning techniques. A schematic of a CNN can be seen in Figure 1.
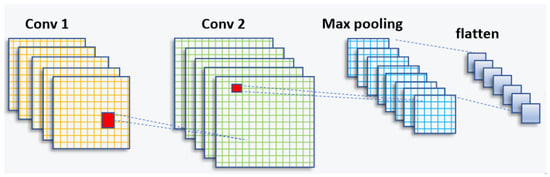
Figure 1.
CNN architecture (feature-extraction block).
The CNN structure is inspired by the human visual cortex and is widely used for automatic feature extraction from large datasets [30,31]. It consists of a series of convolutional layers, followed by sampling layers and a fully connected layer [32].
Following the convolutional layer, the ReLU activation function is applied to extract nonlinear features. The purpose of incorporating the ReLU layer is to introduce nonlinearity into the network. The mathematical representation of the ReLU function is defined as follows, as shown in Equation (1).
The pooling layer plays a crucial role in reorganizing the feature maps to effectively decrease the factors [33], the allocation of memory, and the computational budget within the framework. Each feature map undergoes pooling, with the two commonly used approaches being max pooling and average pooling, as denoted by Equation (2) and Equation (3), respectively. These pooling functions help summarize the information within the feature maps while reducing their spatial dimensions.
The pooling layer is denoted as M, and represents all parameters within this layer. The confidential criteria are computed in the fully connected layers and represented as a three-dimensional array, like 1 × 1 × c. Each component within this volume corresponds to structural scores, with c representing the categories. In a fully connected (FC) layer, all neurons are connected to neurons in the preceding layers; this is a type of interconnection. This connectivity is a characteristic feature of a typical convolutional neural network (CNN) [33,34], where all layers are sequentially interconnected, as depicted in Equation (4). This connectivity pattern allows for information flow and feature extraction across the network.
In the following, Equation (5) represents a deeper architecture of CNN with a more-significant number of fully connected layers, which may lead to vanishing or exploding gradient values. There are various techniques to deal with this issue, such as the application of shortcut connections among the layers, as can be seen for ResNet [35].
As another example of the architecture of CNNs, there is a direct connection between the whole layers in DenseNet, where the input in the current layer comes from the previous one as a standard model [36]. The formulation can be seen below.
In this study, three prominent convolutional neural network architectures, VGG16 [25], VGG19 [25], and DenseNet201 [27], were employed for the identification of medicinal plant images.
3.2. VGG-16
VGG-16 is a top-rated convolutional neural network (CNN) model that was proposed in [25]. It has achieved remarkable performance in image-detection and -classification tasks, and this network is easy to use with transfer learning. The model comprises 13 convolutional layers, five max-pooling layers, and three fully connected layers. The final layer employs the softmax activation function for classification. The architecture of VGG-16 is relatively straightforward, which takes an input tensor of size 224 × 224 with three RGB channels.
3.3. VGG-19
VGG-19 [25] is an extended version of VGG-16 designed to enhance image-recognition performance. It has achieved remarkable success in the ImageNet Challenge 2014, where the Visual Geometry Group (VGG) team secured top rankings. The VGG-19 architecture consists of 16 convolutional and three fully connected layers. Like the VGG16 network, this model also takes a 224 × 224 input image size with 3 RGB channels. The convolutional blocks serve as feature extractors, generating bottleneck features. VGG-19 exemplifies the VGG team’s commitment to advancing image-recognition capabilities.
3.4. DenseNet201
DenseNet201 [27], a deep convolutional neural network (CNN) model introduced in 2017, is known for its dense connectivity pattern. With 201 layers, it employs dense blocks, where each layer is directly connected to every other layer, promoting feature reuse and gradient flow. Transition layers are inserted to control the parameters and reduce the spatial dimensions. Batch normalization and ReLU activation enhance the training. DenseNet201 achieves high accuracy in image-recognition tasks by capturing intricate patterns and hierarchical representations. Pretrained on ImageNet, it serves as a feature extractor or can be fine-tuned for transfer learning, making it a powerful CNN model for image classification and related applications.
DenseNet201 has various advantages due to its 201 convolutional layers. These advantages include enabling feature adaptation to be used again, vanishing gradient problems, achieving optimal feature distribution, and reducing the number of parameters [33]. Let us consider an image being fed into a neural network comprising S layers with nonlinear transformation, denoted as . In this case, DenseNet201 incorporates conventional skip connections within the feed-forward network (see Figure 2). These connections allow bypassing the nonlinear alteration using an identity function, as represented as follows.
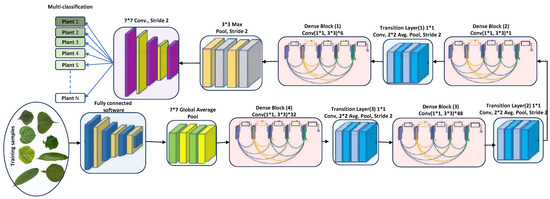
Figure 2.
DenseNet201 architecture (feature-extraction block). The star symbol (*) denotes multiplication.
Further, DenseNet offers a distinct benefit in that the gradient can flow directly through the identity function from the primary layers to the last layer. On the other hand, dense networks utilize direct end-to-end connections to maximize the details within each layer. In this case, the s- layer receives all the data from the preceding layer, as follows.
DenseNet’s architectural design incorporates a particular process for downsampled data, which occurs within dense blocks. These blocks are divided into transition layers, each including a 1 × 1 convolutional layer (CONV), an average pooling layer, and batch normalization (BN). The elements from the transition layer progressively disseminate to the dense layers, making the network less intricate. In an effort to enhance network utility, the average pooling layer was wholly converted into a 2 × 2 max pooling layer. Each convolutional layer is preceded by batch normalization. The network’s growth rate, denoted by the hyperparameter k, allows DenseNet to yield state-of-the-art results. However, the hyperparameters should be adjusted depending on the complexity and nonlinearity of the data characteristics [37]. The conventional pooling layers were removed, and the proposed detection layers were fully amalgamated and connected to the classification layers for detection purposes. DenseNet264 encompasses even more-complex network designs than the 201-layer network. However, due to its narrower network footprint, the 201-layer structure was deemed suitable for the plant leaf detection tasks. DenseNet201, despite a smaller growth rate, still performs excellently because its design employs feature maps as a network-wide mechanism. The architecture of DenseNet201 is depicted in Figure 2.
DenseNet201, as a deep learning architecture, possesses distinct advantages and disadvantages. The benefits of DenseNet201 are as follows. Firstly, it effectively addresses the vanishing gradient problem by establishing direct connections between layers. This facilitates smoother gradient flow during training, leading to improved optimization based on gradients. Secondly, DenseNet201 excels in distributing features evenly across layers due to its dense connections. This enhances the model’s overall representational capabilities. Additionally, DenseNet201 enables the reuse of feature maps at various depths, promoting efficient information flow and potentially strengthening the network’s learning capacity. Lastly, it reduces the number of parameters compared to conventional deep learning architectures by eliminating the need for redundant feature learning in individual layers. This results in a more-efficient utilization of the parameters in the model. Deep learning methods on single leaf sheets for plant identification are not a replacement for approaches dealing with segmentation and identification in scattered environments. Instead, they serve the specific purpose of simplifying the data collection, enabling in-depth species-specific analysis, and providing a foundational step for more-complex plant identification tasks. These methods are valuable and contribute to developing more robust, comprehensive plant identification systems.
3.5. Ensemble Learning Approach
Ensemble learning offers a compelling approach to elevate the performance of classifiers by encompassing diverse techniques, prominently including bagging, boosting, and stacking [38]. Ensemble learning considers either a homogeneity or heterogeneity approach. In the case of homogeneity, a single base classifier is trained on different datasets, whereas for heterogeneity, additional classifiers are trained on a shared dataset. The resultant ensemble, by harnessing the collective wisdom of its components, generates predictions through a synthesis of approaches such as averaging, weighted averaging, and voting. These mechanisms draw from the individual outputs furnished by the base classifiers. In our pursuit of automating medicinal leaf detection, we embraced the heterogeneous ensemble paradigm, where the weighted average and average play a pivotal role in amalgamating the contributions of diverse classifiers to culminate in the ultimate and refined outcome.
3.6. Proposed Ensemble Learning for Medicinal Plant Leaf Identification
The proposed ensemble learning approach illustrated in Figure 3 was tailored for the precise and reliable identification of medicinal plant leaves. The approach capitalizes on the collective intelligence of multiple convolutional neural network (CNN) models, offering a holistic and robust strategy for accurate leaf classification. The framework initiation encompasses the individual training of three prominent CNN models: VGG16, VGG19, and DenseNet201. Central to our strategy is the utilization of transfer learning, a technique where the final layers of the aforementioned CNN models are replaced with custom pooling and dense layers. This adaptation facilitates the alignment of models with the unique attributes of our medicinal leaf dataset, enabling them to discriminate between various leaf species effectively. Each model undergoes extensive training for 100 epochs to ensure thorough convergence and optimal performance.
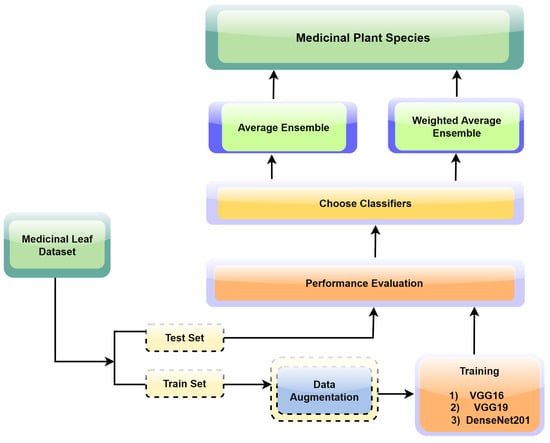
Figure 3.
The process flow of the proposed research study.
Our ensemble approach employs two techniques for combining predictions from multiple neural network models, including VGG16, VGG19, and DenseNet201. Averaging involves computing the elementwise average of predictions from each model for a given input image, reducing the impact of model variations and errors to enhance prediction reliability. Furthermore, our approach employs weighted averaging by assigning model-specific weights, represented as (wi), based on individual performance metrics calculated on the validation data. The weights are determined by dividing each model’s accuracy on the validation set by the sum of the accuracies of all models within the ensemble. These normalized weights, where ∑wi = 1, are then applied when making predictions on the test set. The weights assigned to the component models are 0.3294, 0.3229, and 0.3475 for VGG16, VGG19, and DenseNet201, respectively. This strategy optimizes ensemble performance by prioritizing models that demonstrate excellence in classifying medicinal plant images on previously unseen data.
The subsequent sections discuss the component models, ensemble learning approach, experimental results, and discussions, shedding light on the efficacy and contribution to the field. The applied steps of the proposed ensemble model can be listed as follows:
- (1)
- Data loading and splitting: Collect “Mendeley Data–Medicinal Leaf Dataset” (1835 images, 30 species), and split into training (70%) and testing (30%).
- (2)
- Model selection: Choose VGG16, VGG19, and DenseNet201 as the base models.
- (3)
- Image standardization: Resize images to 224 × 224 px, which is compatible with the input size expected by the CNN models.
- (4)
- Data augmentation: Enhance the model learning and diversity by applying random rotations, flips, translations, and adjustments to brightness or contrast. This exposure to image variations during training improves the model’s generalization.
- (5)
- Batch generation: Divide the dataset into smaller subsets of images, which are then fed into the CNN model during training. This approach enhances the computational efficiency by processing a portion of the dataset at a time, rather than the entire dataset at once.
- (6)
- Training and transfer learning: The models were trained individually employing transfer learning with softmax activation [39] for classification using the Adam optimizer and categorical cross-entropy.
- (7)
- Validation models: For each trained model, the prediction was performed by calculating the class probabilities on the test set.
- (8)
- Hybridization models: The ensemble models were created by combining the results generated by individual classifiers, using the averaging and weighted averaging strategies. Using the three components, VGG16, VGG19, and DenseNet201, the feasible ensemble models were created, validated, and compared to select the best-performing model. The proposed learning framework can be seen in Figure 4.
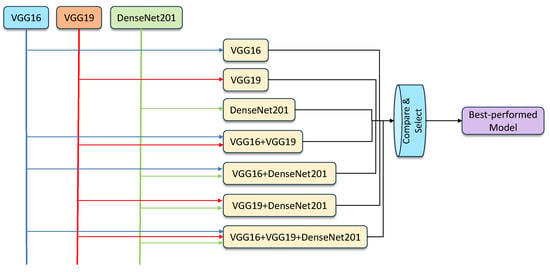 Figure 4. An overview of the proposed ensemble model framework.
Figure 4. An overview of the proposed ensemble model framework.
3.7. Hyperparameter Tuning Using Grid Search
Grid search, a commonly used methodology in machine learning for optimizing hyperparameters, involves exhaustively exploring a pre-determined grid of hyperparameter values in order to identify the combination that maximizes the performance of the model [37,40]. Let us delve into an illustration of using a parameter sweep to fine-tune the hyperparameters of the convolutional neural network (CNN) architecture, a highly esteemed CNN model known for its intricate design and exceptional accomplishments in image-classification endeavors.
In this particular case, hyperparameters such as the learning rate, batch size, and weight decay can be refined. To begin, we construct a grid encompassing a wide range of potential values for each hyperparameter. For example, we might consider learning rates that span from 0.001 to 0.1, batch sizes that range from 32 to 128, and weight decay values that encompass 0.0001 to 0.01. By utilizing the technique of parameter sweep, we systematically generated all possible combinations from these configurations of the hyperparameters. As an illustration, a combination could consist of a learning rate of 0.001, a batch size of 32, and a weight decay of 0.0001. Subsequently, we trained the CNN model using the designated values of the hyperparameters. The process of training usually involves dividing the dataset into training and validation sets, transmitting the data through the network, calculating the loss, and adjusting the model’s weights through backpropagation.
After training the model, we carefully analyzed its performance on a separate test set. This repetitive process was carried out for all combinations of the hyperparameters, and we selected the combination that achieved the best performance on the validation set. The determination of the optimal performance can be based on metrics such as accuracy, precision, recall, or the F1-score, depending on the specific classification task. Once we had identified the most-promising combination of hyperparameters through a parameter sweep, we trained the CNN model again using this configuration on the entire training dataset. Finally, we assessed the performance of the optimized model on a distinct test dataset to evaluate its ability to generalize to new, unseen data. By thoroughly exploring various combinations of hyperparameters, the parameter sweep technique enabled us to discover the optimal configuration for the CNN model, thereby enhancing its performance in image-classification endeavors.
3.8. Dataset Description
To start the process of plant species identification, the first step involved collecting the dataset. A qualitative study was conducted to identify suitable medicinal leaf dataset sources and determine the dataset format. The selection of a dataset is influenced by several factors, including the nature of the problem, the availability of data, the diversity of the data, and the relevance to the application. In order to generalize our approach, we used the benchmark dataset of medicinal plant leaf classification, i.e., Mendeley Medicinal Leaf [41]. The dataset, representing images from 30 different medicinal plants, was selected for this study. The selected dataset contained a total of 1835 leaf images. A system’s performance is affected by factors like the dataset size, class distribution, and data quality. To address this concern, data preprocessing was employed to clean the dataset. Figure 5 depicts some of the leaf samples of the Mendeley Medicinal Leaf Dataset.
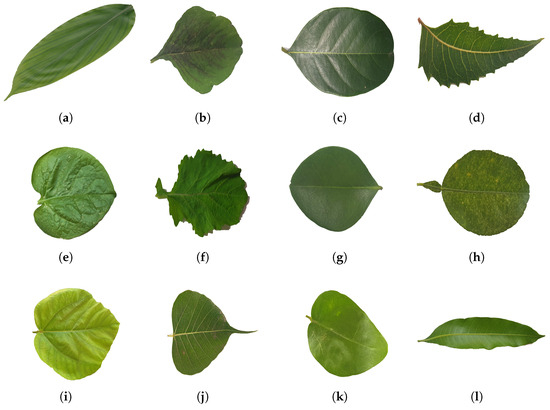
Figure 5.
Showcases a sample of the medicinal leaf dataset images, illustrating the diverse range of plant images incorporated for classification purposes. (a) Alpinia Galanga (Rasna), (b) Amaranthus Viridis (Arive-Dantu), (c) Artocarpus Heterophyllus (jackfruit), (d) Azadirachta Indica (neem), (e) Basella Alba (Basale), (f) Brassica Juncea (Indian mustard), (g) Carissa Carandas (Karanda), (h) Citrus Limon (lemon), (i) Ficus Auriculata (Roxburgh fig), (j) Ficus Religiosa (peepal tree), (k) Jasminum (jasmine), (l) Mangifera Indica (mango).
3.9. Data Preprocessing
Data preprocessing plays an important role in preparing raw data for machine learning models, ensuring that the data are in a suitable format for effective training and analysis. The dataset collected was meticulously divided into distinct training and testing subsets, adhering to the widely adopted 70–30 ratio. This partitioning ensured an optimal allocation of data for both model development and rigorous evaluation. During the component model training phase, an additional step of further subdivision was introduced. Within the training subset, 20% of the data were strategically reserved for validation. This segregation gave us a dedicated validation set, essential for gauging the convergence and generalization capabilities of our evolving models. During the training phase of the component models (VGG16, VGG19, and DenseNet201) and ensemble models, images were resized to a uniform size of 224 × 224 px. This preprocessing step ensured that all images were of the same dimensions, allowing them to be fed into the neural networks consistently. The images were then normalized to ensure the pixel values fell within a specific range, which aids in stable and efficient training. To enhance the robustness and prevent overfitting, data augmentation techniques were applied to the training dataset. These techniques involve generating new instances by applying transformations to the existing images. Various transformations, such as random rotation by up to 20, zooming by 5%, horizontal and vertical shifting by 5%, shearing at a 5% angle, and horizontal flipping, were performed. The “nearest” fill mode handled new pixels resulting from the transformations. These transformations helped increase the diversity of the training data, enabling the model to learn a wider range of features and generalize better to unseen data.
4. Evaluation Metrics
Evaluation metrics play a vital role in optimizing classifiers for the accurate detection and classification of medicinal plant images. In this study, we evaluated the trained models on the basis of commonly used performance metrics in this domain, which include accuracy, sensitivity (recall), precision, and the F1-score. Accuracy measures the proximity between predicted and target values, while sensitivity focuses on the ratio of correctly identified positive instances. True negatives (TNs) and true positives (TPs) represent correctly classified negative and positive instances, respectively, contributing to successful classification and detection. False negatives (FNs) and false positives (FPs) denote misclassifications. These metrics guide the fine-tuning of classifiers to achieve optimal performance in medicinal plant image classification. The formulae for the calculation of the metrics used in this study are shown in Equations (9)–(12).
4.1. Accuracy
Accuracy is a metric that calculates the proportion of correctly predicted values out of the total number of instances evaluated.
4.2. Recall
Recall, also known as sensitivity, quantifies the proportion of positive values that are accurately classified.
4.3. Precision
Precision assesses the accuracy of positive predictions within the predicted values of the positive class.
4.4. F1-Score
The F1-score quantifies the balanced performance of a classifier by taking into account both the recall and precision rates through their harmonic average.
The classification reports and confusion matrices were analyzed to evaluate the performance of the classifiers. Similar procedures were carried out for the other feature sets, allowing a comparison of the classifiers’ performance.
5. Experimental Results
5.1. Classification Outcomes of Component Deep Neural Networks
Employing transfer learning played a pivotal role, where the pre-trained VGG16, VGG19, and DenseNet201 deep neural network architectures were fine-tuned to classify medicinal plants. In this work, we initially trained VGG16, VGG16, and DenseNet201 individually for 100 epochs using transfer learning. The batch size was 32. Activation function = softmax, and optimizer = Adam. Then, the models were evaluated, and ensemble models were created and evaluated. The hyperparameters were adjusted using grid search, employed to optimize the results and shown in Table 1.

Table 1.
The tuned hyperparameters of the proposed classification model.
It is evident from Table 2 and Figure 6 that DenseNet201 displayed an impressive training accuracy of 100% and a validation accuracy of 94.64%. Notably, DenseNet201 exhibited remarkable generalization, achieving an exceptional test accuracy of 98.93%. In Figure 7, the training and validation accuracy and loss for DensNet201 are graphically illustrated, and Figure 8 displays the distribution of the predicted classes against the actual classes. These results underscore the capacity of transfer learning to adapt models to the specific characteristics of the medicinal plant dataset, yielding robust and accurate classifiers for medicinal leaf image classification. The VGG16 and VGG19 architectures have relatively small receptive fields, which limited their ability to capture long-range dependencies in the data and led to comparatively lower performance than DenseNet201. Moreover, VGG16 and VGG19 are deep architectures with 16 and 19 layers, respectively. While depth can be beneficial for capturing complex patterns in data, it also makes training and inference computationally expensive. As deeper and more-complex architectures have been developed, such as ResNet and Inception, they have demonstrated improved performance with fewer parameters and computational requirements.

Table 2.
Training, test, and validation accuracy of VGG16, VGG19, and DenseNet201.
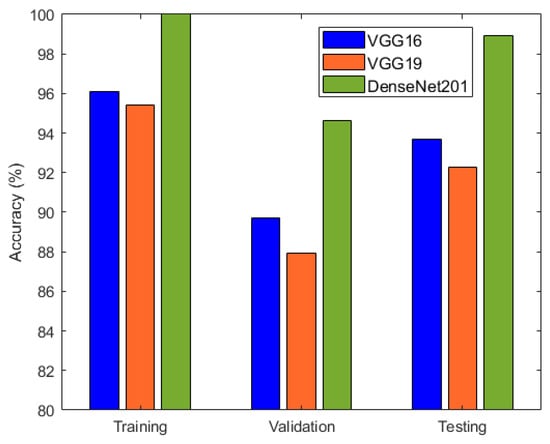
Figure 6.
Accuracy comparison of VGG16, VGG19, and DenseNet201.
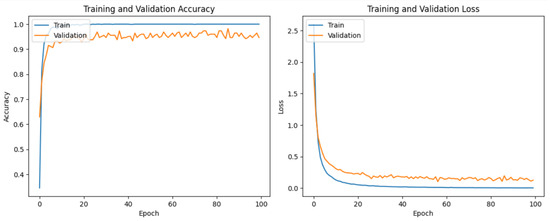
Figure 7.
The training and validation loss and accuracy of DenseNet201.
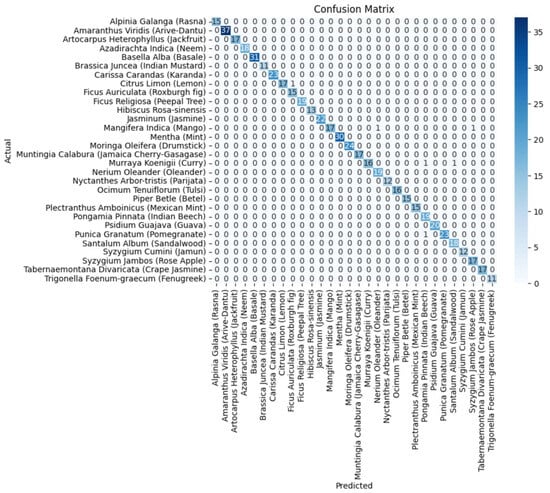
Figure 8.
The confusion matrix of the DenseNet201 component model.
The precision, recall, and F1-score results of the individual deep neural network models are summarized in Table 3 and visualized in Figure 9. These metrics provide insights into the models’ performance in terms of correctly identifying positive cases, capturing actual positive instances, and achieving a balance between precision and recall. Among the models, DenseNet201 exhibited the highest precision, recall, and F1-score, indicating its excellence in classification accuracy.

Table 3.
Precision, recall, and F1-score obtained by the component deep neural networks.
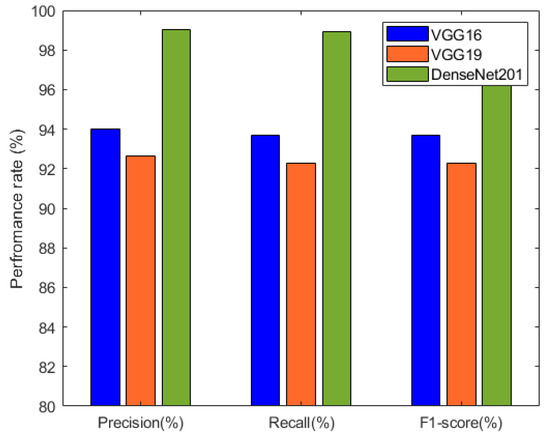
Figure 9.
Performance comparison between VGG16, VGG19, and DenseNet201 based on precision, recall, and F1-score.
5.2. Ensemble Approaches for Improved Classification Performance
After individually evaluating the component deep neural network models, the exploration turned towards ensemble techniques for further refinement. Specifically, two ensemble approaches, namely averaging and weighted averaging, were employed to combine the strengths of the individual models. This culminated in the creation of diverse ensemble models that revealed enhanced classification capabilities. Table 4 and Figure 10 present the accuracy outcomes of the four developed ensemble models, VGG19 + DenseNet201, VGG16 + VGG19, VGG16 + DenseNet201, and VGG16 + VGG19 + DenseNet201, based on the average performance strategy.

Table 4.
Accuracy outcomes of different ensembled deep neural networks using averaging ensemble approach.
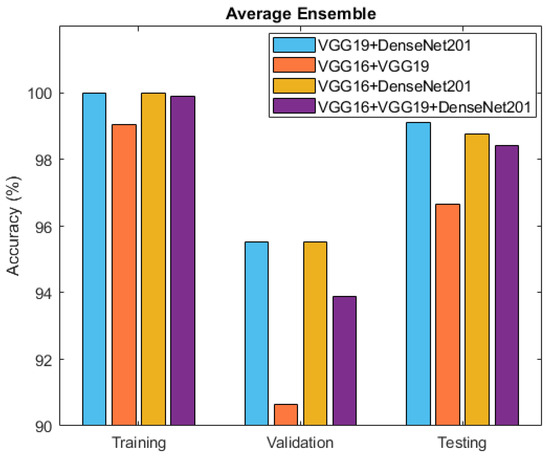
Figure 10.
Accuracy comparison between ensemble models: VGG19 + DenseNet201, VGG16 + VGG19, VGG16 + DenseNet201, and VGG16 + VGG19 + DenseNet201.
We can see the statistical analysis of the weighted average ensemble model for the four developed approaches based on the training, validation, and testing classification accuracy in Table 5.

Table 5.
Accuracy outcomes of different ensembled deep neural networks using weighted average ensemble.
In Figure 11, the confusion matrix for the ensemble of VGG19+DenseNet201 is presented, which highlights the effectiveness of the ensembled approach using the average ensemble strategy. The diagonal values of a confusion matrix represent the number of data points where the predicted label matches the true label, indicating correct predictions. They signify instances where the classifier identifies the positive class correctly. In contrast, the off-diagonal elements of the confusion matrix denote the misclassifications made by the classifier. These errors can be false positives or false negatives, depending on whether the predicted label is incorrect for the positive class or the negative class, respectively. In other words, a higher value on the diagonal of the confusion matrix indicates better performance of the classifier, as it suggests a larger number of correct predictions. The ultimate goal is to maximize the values on the diagonal while minimizing the off-diagonal elements.
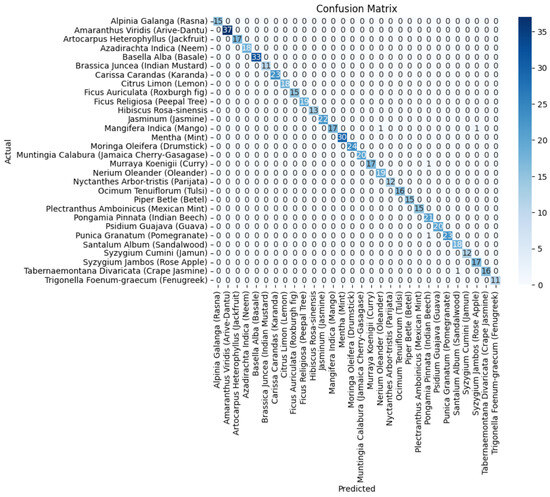
Figure 11.
The confusion matrix of the average ensemble of VGG19+DenseNet201.
6. Discussion
6.1. Experimental Analysis
In our experimental analysis, we examined the effectiveness of various pre-trained models, VGG16, VGG19, and DenseNet201. For each of these models, we excluded the top layers to utilize their pre-trained weights and the acquired feature representations. We created new models by incorporating a flattening layer to transform the extracted features into a high-dimensional vector, followed by the addition of a dense layer. These models were then compiled using the Adam optimizer and categorical cross-entropy loss function. Table 2 encapsulates the accuracy outcomes achieved by the three distinct deep neural network architectures: VGG16, VGG19, and DenseNet201. Notably, among these architectures, DenseNet201 stood out as a model of exceptional performance. It effortlessly attained a perfect training accuracy of 100%, underscoring its capability to internalize intricate nuances within the training dataset. This mastery in learning translated cohesively to the validation phase, where DenseNet201 achieved an impressive accuracy of 94.64%. Moreover, during the rigorous test phase, the model showcased a striking test accuracy of 98.93%, reinforcing its prowess in accurately classifying diverse and previously unseen medicinal plant samples.
The metrics provided in Table 3 highlight the precision, recall, and F1-score of three prominent deep neural network architectures: VGG16, VGG19, and DenseNet201. Among these models, DenseNet201 emerged as a standout performer. Impressively, it achieved a remarkable precision of 99.01%, reflecting its capability to accurately identify positive cases while minimizing false positives. The strength of DenseNet201 became evident in the recall as well, with a value of 98.94%. This signifies its effectiveness in capturing a substantial portion of actual positive cases. The F1-score of 98.93% for DenseNet201 affirmed its harmonious balance between precision and recall, showcasing its overall excellence in classification accuracy.
The accuracy outcomes attained through the distinct ensemble deep learning approaches are summarized in Table 4 and Table 5. Among the strategies applied to the individual deep neural network models, it is evident that the most-effective solution arose from combining VGG19 and DenseNet201 utilizing the averaging technique. With a perfect training accuracy of 100%, impressive validation accuracy of 95.52%, and exceptional test accuracy of 99.12%, this ensemble demonstrated the ability to master intricate patterns, generalize well to new data, and achieve precise classification. A comparative analysis of this study with the previous state-of-the-art approaches for the identification of medicinal plants using the same dataset was conducted, and it can be observed from Table 6 that leveraging the complementary features of VGG19 and DenseNet201, the ensemble approach improved the robustness and balanced the performance, making it a potent solution for medicinal plant identification. Its success underscores the significance of ensemble techniques in enhancing complex classification tasks and holds promising applications in diverse fields.

Table 6.
Comparative analysis of the proposed study with the previously state-of-the-art approaches for the identification of medicinal plant images.
6.2. Challenges
In our research, we acknowledge two key limitations. First, our dataset comprises a relatively small selection of 1835 leaf images from 30 distinct medicinal plants, potentially impacting the system’s applicability to a wider range of medicinal plant species. To address this constraint and bolster the system’s generalizability, we propose future work to focus on expanding the dataset with a more-diverse array of plant species and an increased number of images per species. Secondly, our approach exclusively relies on leaf images for plant species identification, which might not be sufficient for certain plant species requiring identification based on other parts like flowers, fruits, or roots. To address this limitation, a promising future direction involves the development of a multi-modal system that incorporates images of various plant parts to ensure comprehensive and accurate identification. These limitations underscore the need for ongoing research and development efforts within the field of medicinal plant identification.
7. Conclusions
In the contemporary world, traditional medicines have gained importance due to the high costs and potential negative impacts of allopathic medicines. Medicinal plant classification is usually carried out by plant taxonomists; however, the human-centric nature of this approach can introduce inaccuracies or errors in judgment. Artificial intelligence methodologies have found extensive application in automating plant recognition processes. This research paper investigated the application of ensemble learning to elevate the accuracy of medicinal plant identification. In this work, transfer learning was employed to extract valuable features from medicinal plant leaf images. The pre-trained models VGG16, VGG19, and DenseNet201 were used as the base models, which boosted the capability of the proposed approach to identify and extract pertinent information from the images. The dataset consisted of medicinal leaf images sourced from a published collection on Mendeley. To construct ensemble models based on convolutional neural networks (CNNs), we compared three leading CNN architectures: VGG16, VGG19, and DenseNet201. By employing transfer learning, a trio of three-component classifiers was harnessed without their upper layers. This adaptation allowed them to discern crucial attributes within the medicinal leaf images, and these attributes were then integrated into the dense layers. Subsequently, these classifiers underwent training on a dataset comprising 30 diverse classes of medicinal leaves using a softmax classifier.
Following the individual assessment of the component deep neural network models, the medicinal plant identification approach was enhanced through ensemble techniques employing the averaging and weighted averaging strategies. Upon assessing the ensemble models, it became evident that the most-potent approach emerged from the fusion of VGG19 and DenseNet201, employing the averaging method. This ensemble displayed remarkable attributes, including an impeccable training accuracy of 100%, a notable validation accuracy of 95.52%, and an outstanding test accuracy of 99.12%. These results collectively underscore the capacity of the ensemble learning approach to proficiently capture intricate patterns, generalize effectively to novel data, and achieve accurate classification.
Our future plans involve collaboration with domain experts to ensure the high-quality collection and annotation of our dataset. We also aim to develop intuitive user interfaces and adapt the model for real-time applications.
Author Contributions
Conceptualization, M.A.H., T.A. and A.M.U.D.K.; methodology, A.M.U.D.K. and T.A.; validation, M.A.H.; formal analysis, M.A.H.; investigation, M.A.H., T.A. and M.N.; data curation, M.A.H. and T.A.; writing—original draft, A.M.U.D.K. and M.A.H.; writing—review and editing, A.M.U.D.K., M.N. and M.A.H.; visualization, A.M.U.D.K., M.A.H., T.A. and M.N.; supervision, T.A. and A.M.U.D.K.; project administration, T.A.; funding acquisition, M.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in this article.
Acknowledgments
The authors would like to acknowledge Baba Ghulam Shah Badshah University for their valuable support. Also, the authors would like to thank the Center for Artificial Intelligence Research and Optimisation, Torrens University Australia, for supporting the APC of this publication.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Introduction and Importance of Medicinal Plants and Herbs. Available online: https://www.nhp.gov.in/introduction-and-importance-of-medicinal-plants-and-herbs_mtl (accessed on 9 November 2023).
- Azlah, M.A.F.; Chua, L.S.; Rahmad, F.R.; Abdullah, F.I.; Wan Alwi, S.R. Review on techniques for plant leaf classification and recognition. Computers 2019, 8, 77. [Google Scholar] [CrossRef]
- Tripathi, K.; Khan, F.A.; Khanday, A.M.U.D.; Nisa, K.U. The classification of medical and botanical data through majority voting using artificial neural network. Int. J. Inf. Technol. 2023, 15, 3271–3283. [Google Scholar] [CrossRef]
- Wäldchen, J.; Mäder, P. Plant species identification using computer vision techniques: A systematic literature review. Arch. Comput. Methods Eng. 2018, 25, 507–543. [Google Scholar]
- De Carvalho, M.R.; Bockmann, F.A.; Amorim, D.S.; Brandão, C.R.F.; de Vivo, M.; de Figueiredo, J.L.; Britski, H.A.; de Pinna, M.C.; Menezes, N.A.; Marques, F.P.; et al. Taxonomic impediment or impediment to taxonomy? A commentary on systematics and the cybertaxonomic-automation paradigm. Evol. Biol. 2007, 34, 140–143. [Google Scholar] [CrossRef]
- Rabani, S.T.; Khanday, A.M.U.D.; Khan, Q.R.; Hajam, U.A.; Imran, A.S.; Kastrati, Z. Detecting suicidality on social media: Machine learning at rescue. Egypt. Inform. J. 2023, 24, 291–302. [Google Scholar]
- Sarker, I.H. Deep learning: A comprehensive overview on techniques, taxonomy, applications and research directions. SN Comput. Sci. 2021, 2, 420. [Google Scholar] [CrossRef] [PubMed]
- Husin, Z.; Shakaff, A.; Aziz, A.; Farook, R.; Jaafar, M.; Hashim, U.; Harun, A. Embedded portable device for herb leaves recognition using image processing techniques and neural network algorithm. Comput. Electron. Agric. 2012, 89, 18–29. [Google Scholar] [CrossRef]
- Gokhale, A.; Babar, S.; Gawade, S.; Jadhav, S. Identification of medicinal plant using image processing and machine learning. In Proceedings of the Applied Computer Vision and Image Processing; Proceedings of ICCET 2020; Springer: Berlin/Heidelberg, Germany, 2020; Volume 1, pp. 272–282. [Google Scholar]
- Puri, D.; Kumar, A.; Virmani, J.; Kriti. Classification of leaves of medicinal plants using laws texture features. Int. J. Inf. Technol. 2019, 14, 1–12. [Google Scholar] [CrossRef]
- Roopashree, S.; Anitha, J. DeepHerb: A vision based system for medicinal plants using xception features. IEEE Access 2021, 9, 135927–135941. [Google Scholar] [CrossRef]
- Sachar, S.; Kumar, A. Deep ensemble learning for automatic medicinal leaf identification. Int. J. Inf. Technol. 2022, 14, 3089–3097. [Google Scholar] [CrossRef]
- Pan, S.J.; Yang, Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 2009, 22, 1345–1359. [Google Scholar] [CrossRef]
- Nakata, N.; Siina, T. Ensemble Learning of Multiple Models Using Deep Learning for Multiclass Classification of Ultrasound Images of Hepatic Masses. Bioengineering 2023, 10, 69. [Google Scholar] [CrossRef] [PubMed]
- Kuzinkovas, D.; Clement, S. The detection of COVID-19 in chest x-rays using ensemble cnn techniques. Information 2023, 14, 370. [Google Scholar] [CrossRef]
- D’Angelo, M.; Nanni, L. Deep Learning-Based Human Chromosome Classification: Data Augmentation and Ensemble. Information 2023, 14, 389. [Google Scholar] [CrossRef]
- Nazarenko, D.; Kharyuk, P.; Oseledets, I.; Rodin, I.; Shpigun, O. Machine learning for LC–MS medicinal plants identification. Chemom. Intell. Lab. Syst. 2016, 156, 174–180. [Google Scholar] [CrossRef]
- Kumar, N.; Belhumeur, P.N.; Biswas, A.; Jacobs, D.W.; Kress, W.J.; Lopez, I.C.; Soares, J.V. Leafsnap: A computer vision system for automatic plant species identification. In Proceedings of the Computer Vision–ECCV 2012: 12th European Conference on Computer Vision, Part II 12, Florence, Italy, 7–13 October 2012; Springer: Berlin/Heidelberg, Germany, 2012; pp. 502–516. [Google Scholar]
- Kadir, A.; Nugroho, L.E.; Susanto, A.; Santosa, P.I. Leaf classification using shape, color, and texture features. arXiv 2013, arXiv:1401.4447. [Google Scholar]
- Wu, S.G.; Bao, F.S.; Xu, E.Y.; Wang, Y.X.; Chang, Y.F.; Xiang, Q.L. A leaf recognition algorithm for plant classification using probabilistic neural network. In Proceedings of the 2007 International Symposium on Signal Processing and Information Technology, Giza, Egypt, 15–18 December 2007; pp. 11–16. [Google Scholar]
- Sabu, A.; Sreekumar, K.; Nair, R.R. Recognition of Ayurvedic medicinal plants from leaves: A computer vision approach. In Proceedings of the 2017 4th International Conference on Image Information Processing (ICIIP), Shimla, India, 21–23 December 2017; pp. 1–5. [Google Scholar]
- Sundara Sobitha Raj, A.P.; Vajravelu, S.K. DDLA: Dual deep learning architecture for classification of plant species. IET Image Process. 2019, 13, 2176–2182. [Google Scholar] [CrossRef]
- Barré, P.; Stöver, B.C.; Müller, K.F.; Steinhage, V. LeafNet: A computer vision system for automatic plant species identification. Ecol. Inform. 2017, 40, 50–56. [Google Scholar] [CrossRef]
- Geetharamani, G.; Pandian, A. Identification of plant leaf diseases using a nine-layer deep convolutional neural network. Comput. Electr. Eng. 2019, 76, 323–338. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Duong-Trung, N.; Quach, L.D.; Nguyen, M.H.; Nguyen, C.N. A combination of transfer learning and deep learning for medicinal plant classification. In Proceedings of the 2019 4th International Conference on Intelligent Information Technology, New York, NY, USA, 16–17 November 2019; pp. 83–90. [Google Scholar]
- Prashar, N.; Sangal, A. Plant disease detection using deep learning (convolutional neural networks). In Proceedings of the 2nd International Conference on Image Processing and Capsule Networks: ICIPCN 2021 2, Bidar, India, 16–17 December 2016; Springer: Berlin/Heidelberg, Germany, 2022; pp. 635–649. [Google Scholar]
- Khanday, A.M.U.D.; Bhushan, B.; Jhaveri, R.H.; Khan, Q.R.; Raut, R.; Rabani, S.T. Nnpcov19: Artificial neural network-based propaganda identification on social media in COVID-19 era. Mob. Inf. Syst. 2022, 2022, 1–10. [Google Scholar] [CrossRef]
- Patil, S.S.; Patil, S.H.; Azfar, F.N.; Pawar, A.M.; Kumar, S.; Patel, I. Medicinal plant identification using convolutional neural network. In Proceedings of the AIP Conference Proceedings; AIP Publishing: Long Island, NY, USA, 2023; Volume 2890. [Google Scholar]
- Akhtar, M.J.; Mahum, R.; Butt, F.S.; Amin, R.; El-Sherbeeny, A.M.; Lee, S.M.; Shaikh, S. A Robust Framework for Object Detection in a Traffic Surveillance System. Electronics 2022, 11, 3425. [Google Scholar] [CrossRef]
- Dadgar, S.; Neshat, M. Comparative hybrid deep convolutional learning framework with transfer learning for diagnosis of lung cancer. In Proceedings of the International Conference on Soft Computing and Pattern Recognition; Springer: Berlin/Heidelberg, Germany, 2022; pp. 296–305. [Google Scholar]
- Leonidas, L.A.; Jie, Y. Ship classification based on improved convolutional neural network architecture for intelligent transport systems. Information 2021, 12, 302. [Google Scholar] [CrossRef]
- Ahsan, M.; Naz, S.; Ahmad, R.; Ehsan, H.; Sikandar, A. A deep learning approach for diabetic foot ulcer classification and recognition. Information 2023, 14, 36. [Google Scholar] [CrossRef]
- Neshat, M.; Lee, S.; Momin, M.M.; Truong, B.; van der Werf, J.H.; Lee, S.H. An effective hyper-parameter can increase the prediction accuracy in a single-step genetic evaluation. Front. Genet. 2023, 14, 1104906. [Google Scholar] [CrossRef]
- Al-Kaltakchi, M.T.; Mohammad, A.S.; Woo, W.L. Ensemble System of Deep Neural Networks for Single-Channel Audio Separation. Information 2023, 14, 352. [Google Scholar] [CrossRef]
- Neshat, M.; Ahmedb, M.; Askarid, H.; Thilakaratnee, M.; Mirjalilia, S. Hybrid Inception Architecture with Residual Connection: Fine-tuned Inception-ResNet Deep Learning Model for Lung Inflammation Diagnosis from Chest Radiographs. arXiv 2023, arXiv:2310.02591. [Google Scholar]
- Alibrahim, H.; Ludwig, S.A. Hyperparameter optimization: Comparing genetic algorithm against grid search and bayesian optimization. In Proceedings of the 2021 IEEE Congress on Evolutionary Computation (CEC), Krakow, Poland, 28 June–1 July 2021; pp. 1551–1559. [Google Scholar]
- Medicinal Leaf Dataset—Mendeley Data. Available online: https://data.mendeley.com/datasets/nnytj2v3n5/1 (accessed on 9 December 2023).
- Patil, S.S.; Patil, S.H.; Pawar, A.M.; Patil, N.S.; Rao, G.R. Automatic Classification of Medicinal Plants Using State-Of-The-Art Pre-Trained Neural Networks. J. Adv. Zool. 2022, 43, 80–88. [Google Scholar] [CrossRef]
- Ayumi, V.; Ermatita, E.; Abdiansah, A.; Noprisson, H.; Jumaryadi, Y.; Purba, M.; Utami, M.; Putra, E.D. Transfer Learning for Medicinal Plant Leaves Recognition: A Comparison with and without a Fine-Tuning Strategy. Int. J. Adv. Comput. Sci. Appl. 2022, 13, 138–144. [Google Scholar] [CrossRef]
- Almazaydeh, L.; Alsalameen, R.; Elleithy, K. Herbal leaf recognition using mask-region convolutional neural network (mask R-CNN). J. Theor. Appl. Inf. Technol. 2022, 100, 3664–3671. [Google Scholar]
- Ghosh, S.; Singh, A.; Kumar, S. Identification of medicinal plant using hybrid transfer learning technique. Indones. J. Electr. Eng. Comput. Sci. 2023, 31, 1605–1615. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).