1. Introduction
Computed tomography (CT) is a commonly used modality for the evaluation of bony spine abnormalities by virtue of its widespread availability, rapid acquisition times, and depiction of fine bony detail. However, its clinical utility in assessing intraspinal soft tissue abnormalities is limited. Cord compression, disc herniations, or intraspinal soft tissue tumors can be missed on CT, even by experienced radiologists, because they are often inconspicuous or invisible on CT [
1,
2]. Magnetic resonance imaging (MRI) offers superior intraspinal soft tissue and fluid depiction but is not always readily available or feasible, often due to contraindications or patient aversions to MRI, such as claustrophobia, body habitus, metallic implants, etc. [
3,
4]. Myelography, in which extrinsic iodinated contrast material is injected into the spinal canal, permits high-resolution imaging of the spinal cord and nerve roots [
5] but is invasive and is much more time-consuming than CT. In some cases, myelography is not possible due to the limited tolerance of prone positioning, bleeding concerns, or other procedural contraindications. Therefore, there is a clinical need for alternative approaches to augment spinal cord visualization accurately and robustly on routine spine CT.
Conventional CT relies on differences in X-ray attenuation among different tissue types based on atomic number, mass density, or uptake of extrinsic contrast agents. However, within the spinal canal, the ability to discern the spinal cord from surrounding cerebrospinal fluid (CSF) is markedly limited due to the high overlap in attenuation values between the spinal cord and CSF, rendering CT unreliable for visualization of the spinal cord in most practical settings. Dual-energy CT post-processing techniques can improve the specificity of differentiation among tissue types [
6], but routinely used segmentation methods based on material decomposition typically require a sufficiently large atomic number difference that is not present between the spinal cord and CSF. Dual-energy post-processing techniques have been shown to be helpful in reducing artifacts from metallic hardware [
7,
8,
9], but even in the absence of metallic hardware, beam-hardening artifacts and noise related to photon attenuation by the bony spinal elements result in wide variations in pixel values that further hinder attempts to differentiate between spinal cord and CSF on CT. Differential uptake of extrinsic iodinated contrast material can assist in segmenting different tissue types in many non-neuroradiologic imaging applications, but because normal spinal cord parenchyma is separated from the vasculature by a blood–CNS barrier, contrast administration has very little utility in improving spinal cord visualization on CT.
Machine learning approaches, such as deep convolutional neural networks (CNNs), have been used to augment medical images [
10,
11,
12], including the segmentation of medical images by tissue type. The application of deep neural networks to the automated segmentation of bony structures from soft tissue in spine CT has been reported [
13,
14,
15]. For instance, Vania et al. used two-dimensional patches of spine CT images to create a method for segmenting the bony elements of the spine from adjacent non-bony tissues, with pixel-wise sensitivity and a specificity of 97% and 99%, respectively [
13]. Another two-dimensional patch-based method was able to distinguish bone from non-bone pixels on spine CT with 91% sensitivity and 93% specificity [
14]. Often favorable accuracies are achievable for this task due to the large differences in density between bone and soft tissue pixels, but none of the above studies address the more challenging task of differentiating spinal cord voxels from CSF on CT. Automated methods have also been described for detecting the general region of the spinal cord or nerve roots by their location relative to vertebral anatomy [
16,
17], but these methods likely rely on the pixel position relative to bony structures without attempting to differentiate neural soft tissue pixels from water pixels that may occur in these locations.
In this study, we propose a patch-based machine learning method to help differentiate major tissue types in the spine, with an emphasis on differentiating the spinal cord from CSF. We applied this to lumbar spine CT exams as the transition in intraspinal contents from the spinal cord to predominantly CSF near the thoracolumbar junction serves as a suitable site for evaluating the model’s performance. A prior study reporting improved soft-tissue visualization of spine tumors using dual-energy CT post-processing [
18] lends credence to the proposition that sufficient information is contained within dual-energy CT data to differentiate among soft tissues with subtle attenuation differences on conventional CT. In this study, we test the hypothesis that machine learning-based predictive models can accurately classify small dual-energy CT patches as CSF, neural tissue (spinal cord and nerves), fat, or bone using only information from the center voxel and its immediate surrounding neighboring voxels.
4. Discussion
In this study, we developed a novel machine learning model to assist the visualization of intraspinal soft tissue structures on an unenhanced dual-energy spine CT by differentiating four major tissue classes with high accuracy. We trained and tested multiple convolutional neural networks to perform the task of classifying a small 3D patch of a CT volume using HUs at both 100 kVp and 140 kVp in the center voxel and surrounding voxels. We found that an ensemble model, consisting of six individually trained models that differ in the number of surrounding voxels they accept as input, showed 99.4% overall accuracy on labeled ROIs from a holdout test CT exam not used for training/validation. The accuracy is much higher than would be expected for this task when relying on visual assessment, especially since HUs alone cannot reliably separate water pixels from soft tissue, given their near-identical HU distributions, as shown in
Figure 4. Among the tissue types, fat and bone show the best classification. Sensitivity and specificity for fat classification were both 100.0% in the holdout test set. Our bone voxel prediction, with 100% sensitivity and 99.6% specificity in our holdout test set, is mildly more accurate than prior studies using machine learning to segment bone from adjacent soft tissues (91–97% sensitivity and 93–99% specificity [
13,
14]). For the more difficult task of identifying soft tissue and water voxels, which to our knowledge has not been performed in this setting, the ensemble model performs relatively well, with test sensitivities of 98.6% and 90.0% and specificities of 100.0% and 99.6% for the soft tissue and water classes, respectively.
Although the mechanisms by which the model performs this discrimination task are not readily explainable, one interpretation is that low-level features, such as combinations of Hounsfield units, texture, and differences in low-energy and high-energy HUs, are sufficiently different among the four tissue types to allow the model to successfully identify the appropriate tissue class. Since the model performs the task using only small 3D volumes as input, no voxels more than three units from the center voxel in any dimension are available to the model during the classification task. Therefore, an advantage of this patch-based approach to segmentation compared to using the entire CT volume as input to a deep learning segmentation model is that the model does not rely on spatial information provided by macroscopic elements, such as the relationship to bony structures or anatomical location, to deduce a voxel’s membership in a given class. While such information could potentially improve performance, the reliance on high-level features and global spatial cues would likely introduce undesired biases (e.g., predicting water class for voxels based on their location in the lower lumbar spine) that may compromise performance in case of abnormal pathology. Another advantage is that a large number of training elements was able to be generated from a small number of CT exams, which is useful for exams like dual-energy spine CT that are not routinely requested in clinical practice. Since only homogenous single-class patches are provided to the model for training, one disadvantage of this approach is that the model performs poorly at interfaces between two or more tissue types.
Among the major implications of our study is that machine learning can distinguish between intraspinal neural tissue and CSF much better than is considered possible for radiologists reviewing the source dual-energy CT images (
Figure 4). The relatively high overlap in attenuation levels between soft tissue and water pixels (
Figure 2) precludes any conventional thresholding or window-leveling approach from separating these pixels on the original CT source images. The neural network, therefore, was charged with a much more difficult task than simply replicating what a radiologist sees; it instead performed satisfactorily in discerning subtle tissue differences imperceptible to radiologists. From a clinical perspective, our neural network model has the potential to improve the visualization of intraspinal soft tissue and may lead to better post-processing methods to assist radiologists’ interpretation of dual-energy spine CT.
To our knowledge, this patch-based machine learning strategy to segment a dual-energy CT to better visualize intraspinal CSF has not been previously described. Patch-based features have recently been used in other deep learning architectures for such tasks as brain MRI segmentation [
22]. Other deep learning techniques, such as generative adversarial networks, have been used to synthetically improve the visualization of brain MRI findings by simulating full-dose contrast enhancement in patients receiving no or reduced amounts of gadolinium [
23,
24]. In these scenarios, machine learning was applied to capture subtle relationships and patterns that predict the appearance on a more sensitive imaging technique. In our study, we created synthetic images to augment visualization of CSF on unenhanced dual-energy spine CT images, akin to a “virtual myelogram.” Our findings may lead to automated techniques that could be evaluated in subsequent studies to improve the diagnosis or detection of severe spinal stenosis or other intraspinal abnormalities. However, future clinical studies would be needed to validate the use of this or similar methods in detecting intraspinal pathology, such as cord compression, large disc herniations, or epidural tumor, before such approaches could be adopted in clinical practice.
Among the limitations of this study is that our models were trained and tested in a small number of spine CTs, all from the same scanner model and manufacturer at a single institution, thus potentially benefiting from the similarity of scan parameters. The transferability to other scan conditions was not tested in this study, and future work may need to include a greater variety of scanner types and more exam types to ensure model robustness. The generated segmentation images also illustrate some shortcomings of model performance, such as the false-positive depiction of water voxels outside the spine. This may be due in part to the decision to selectively augment intraspinal voxels to improve performance in intraspinal tissue differentiation, which is likely justified given the emphasis on intraspinal anatomy in this study. Third, the model was developed using only exams with relatively normal intraspinal anatomy; therefore, its performance in a clinical setting to facilitate diagnosis or detection of intraspinal pathology has not been tested.
Author Contributions
Conceptualization, X.V.N.; methodology, X.V.N., E.D., S.C. and L.M.P.; software, X.V.N., D.D.N. and E.D.; validation, X.V.N., D.D.N., E.D., S.C. and L.M.P.; formal analysis, X.V.N. and D.D.N.; investigation, X.V.N., D.D.N., E.D., S.C. and L.M.P.; resources, L.M.P.; data curation, X.V.N. and D.J.B.; writing—original draft preparation, X.V.N.; writing—review and editing, X.V.N., D.D.N., E.D., S.C., D.J.B. and L.M.P.; visualization, X.V.N. and D.D.N.; supervision, X.V.N. and L.M.P.; project administration, X.V.N. and L.M.P.; funding acquisition, X.V.N. and L.M.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Biomedical Sciences Institutional Review Board of the Ohio State University, along with waivers of HIPAA authorization and informed consent (protocol # 2019H0094; date of approval: 18 March 2019).
Informed Consent Statement
Patient consent was waived by the IRB due to involvement of no more than minimal risk given the retrospective design of the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to patient data not being authorized for public dissemination.
Conflicts of Interest
X.V.N. reports stock ownership and/or dividends from the following companies broadly relevant to AI services or hardware: Alphabet, Amazon, Apple, AMD, Microsoft, and NVIDIA. E.D. and L.M.P have patents relevant to AI in medical imaging but they are unrelated to the topic of this manuscript. The authors declare that they have no other conflict of interest.
References
- Tins, B.J. Imaging investigations in Spine Trauma: The value of commonly used imaging modalities and emerging imaging modalities. J. Clin. Orthop. Trauma 2017, 8, 107–115. [Google Scholar] [CrossRef]
- Nkusi, A.E.; Muneza, S.; Hakizimana, D.; Nshuti, S.; Munyemana, P. Missed or Delayed Cervical Spine or Spinal Cord Injuries Treated at a Tertiary Referral Hospital in Rwanda. World Neurosurg. 2016, 87, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Andre, J.B.; Bresnahan, B.W.; Mossa-Basha, M.; Hoff, M.N.; Smith, C.P.; Anzai, Y.; Cohen, W.A. Toward Quantifying the Prevalence, Severity, and Cost Associated with Patient Motion During Clinical MR Examinations. J. Am. Coll. Radiol. 2015, 12, 689–695. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, X.V.; Tahir, S.; Bresnahan, B.W.; Andre, J.B.; Lang, E.V.; Mossa-Basha, M.; Mayr, N.A.; Bourekas, E.C. Prevalence and Financial Impact of Claustrophobia, Anxiety, Patient Motion, and Other Patient Events in Magnetic Resonance Imaging. Top. Magn. Reson. Imaging 2020, 29, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Pomerantz, S.R. Myelography: Modern technique and indications. Handb. Clin. Neurol. 2016, 135, 193–208. [Google Scholar] [CrossRef] [PubMed]
- Patino, M.; Prochowski, A.; Agrawal, M.D.; Simeone, F.J.; Gupta, R.; Hahn, P.F.; Sahani, D.V. Material Separation Using Dual-Energy CT: Current and Emerging Applications. Radiographics 2016, 36, 1087–1105. [Google Scholar] [CrossRef]
- Mallinson, P.I.; Coupal, T.M.; McLaughlin, P.D.; Nicolaou, S.; Munk, P.L.; Ouellette, H.A. Dual-Energy CT for the Musculoskeletal System. Radiology 2016, 281, 690–707. [Google Scholar] [CrossRef]
- Yun, S.Y.; Heo, Y.J.; Jeong, H.W.; Baek, J.W.; Choo, H.J.; Shin, G.W.; Kim, S.T.; Jeong, Y.G.; Lee, J.Y.; Jung, H.S. Dual-energy CT angiography-derived virtual non-contrast images for follow-up of patients with surgically clipped aneurysms: A retrospective study. Neuroradiology 2019, 61, 747–755. [Google Scholar] [CrossRef]
- Long, Z.; DeLone, D.R.; Kotsenas, A.L.; Lehman, V.T.; Nagelschneider, A.A.; Michalak, G.J.; Fletcher, J.G.; McCollough, C.H.; Yu, L. Clinical Assessment of Metal Artifact Reduction Methods in Dual-Energy CT Examinations of Instrumented Spines. AJR Am. J. Roentgenol. 2019, 212, 395–401. [Google Scholar] [CrossRef]
- Nguyen, X.V.; Oztek, M.A.; Nelakurti, D.D.; Brunnquell, C.L.; Mossa-Basha, M.; Haynor, D.R.; Prevedello, L.M. Applying Artificial Intelligence to Mitigate Effects of Patient Motion or Other Complicating Factors on Image Quality. Top. Magn. Reson. Imaging 2020, 29, 175–180. [Google Scholar] [CrossRef]
- Richardson, M.L.; Garwood, E.R.; Lee, Y.; Li, M.D.; Lo, H.S.; Nagaraju, A.; Nguyen, X.V.; Probyn, L.; Rajiah, P.; Sin, J.; et al. Noninterpretive Uses of Artificial Intelligence in Radiology. Acad. Radiol. 2020, 28, 1225–1235. [Google Scholar] [CrossRef] [PubMed]
- Lakhani, P.; Prater, A.B.; Hutson, R.K.; Andriole, K.P.; Dreyer, K.J.; Morey, J.; Prevedello, L.M.; Clark, T.J.; Geis, J.R.; Itri, J.N.; et al. Machine Learning in Radiology: Applications Beyond Image Interpretation. J. Am. Coll. Radiol. 2018, 15, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Vania, M.; Mureja, D.; Lee, D. Automatic spine segmentation from CT images using Convolutional Neural Network via redundant generation of class labels. J. Comput. Des. Eng. 2019, 6, 224–232. [Google Scholar] [CrossRef]
- Qadri, S.F.; Ai, D.; Hu, G.; Ahmad, M.; Huang, Y.; Wang, Y.; Yang, J. Automatic Deep Feature Learning via Patch-Based Deep Belief Network for Vertebrae Segmentation in CT Images. Appl. Sci. 2018, 9, 69. [Google Scholar] [CrossRef]
- Khandelwal, P.; Collins, D.L.; Siddiqi, K. Spine and Individual Vertebrae Segmentation in Computed Tomography Images Using Geometric Flows and Shape Priors. Front. Comput. Sci. 2021, 3, 66. [Google Scholar] [CrossRef]
- Fan, G.; Liu, H.; Wu, Z.; Li, Y.; Feng, C.; Wang, D.; Luo, J.; Wells, W.M.; He, S. Deep Learning-Based Automatic Segmentation of Lumbosacral Nerves on CT for Spinal Intervention: A Translational Study. AJNR Am. J. Neuroradiol. 2019, 40, 1074–1081. [Google Scholar] [CrossRef]
- Diniz, J.O.B.; Diniz, P.H.B.; Valente, T.L.A.; Silva, A.C.; Paiva, A.C. Spinal cord detection in planning CT for radiotherapy through adaptive template matching, IMSLIC and convolutional neural networks. Comput. Methods Programs Biomed. 2019, 170, 53–67. [Google Scholar] [CrossRef]
- Kraus, M.; Weiss, J.; Selo, N.; Flohr, T.; Notohamiprodjo, M.; Bamberg, F.; Nikolaou, K.; Othman, A.E. Spinal dual-energy computed tomography: Improved visualisation of spinal tumorous growth with a noise-optimised advanced monoenergetic post-processing algorithm. Neuroradiology 2016, 58, 1093–1102. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Xie, S.; Yang, T.; Wang, X.; Lin, Y. Hyper-class augmented and regularized deep learning for fine-grained image classification. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 2645–2654. [Google Scholar] [CrossRef]
- Zeiler, M.D. ADADELTA: An Adaptive Learning Rate Method. arXiv 2012, arXiv:1212.5701. [Google Scholar]
- Lee, B.; Yamanakkanavar, N.; Choi, J.Y. Automatic segmentation of brain MRI using a novel patch-wise U-net deep architecture. PLoS ONE 2020, 15, e0236493. [Google Scholar] [CrossRef] [PubMed]
- Gong, E.; Pauly, J.M.; Wintermark, M.; Zaharchuk, G. Deep learning enables reduced gadolinium dose for contrast-enhanced brain MRI. J. Magn. Reson. Imaging 2018, 48, 330–340. [Google Scholar] [CrossRef] [PubMed]
- Kleesiek, J.; Morshuis, J.N.; Isensee, F.; Deike-Hofmann, K.; Paech, D.; Kickingereder, P.; Kothe, U.; Rother, C.; Forsting, M.; Wick, W.; et al. Can Virtual Contrast Enhancement in Brain MRI Replace Gadolinium?: A Feasibility Study. Investig. Radiol. 2019, 54, 653–660. [Google Scholar] [CrossRef] [PubMed]
Figure 1.
Representative manual ROI selection of ‘water’ and ‘soft tissue’ class voxels. In this example of ‘water’ ROI selection (A), the location of a pocket of ventral subarachnoid CSF identified on MRI at the L2 level was selected on CT (rectangle) and applied to multiple consecutive slices to create a 3D ROI in the CT volume for purposes of subsequent subsampling for the ‘water’ class. For this example of intraspinal ‘soft tissue’ ROI selection (B), the conus medullaris visualized in the imaged lower thoracic spine on MRI was targeted on CT (rectangle) to similarly create a 3D ROI. Note that there is no readily apparent difference between conus and CSF based on visual inspection of the CT images alone.
Figure 2.
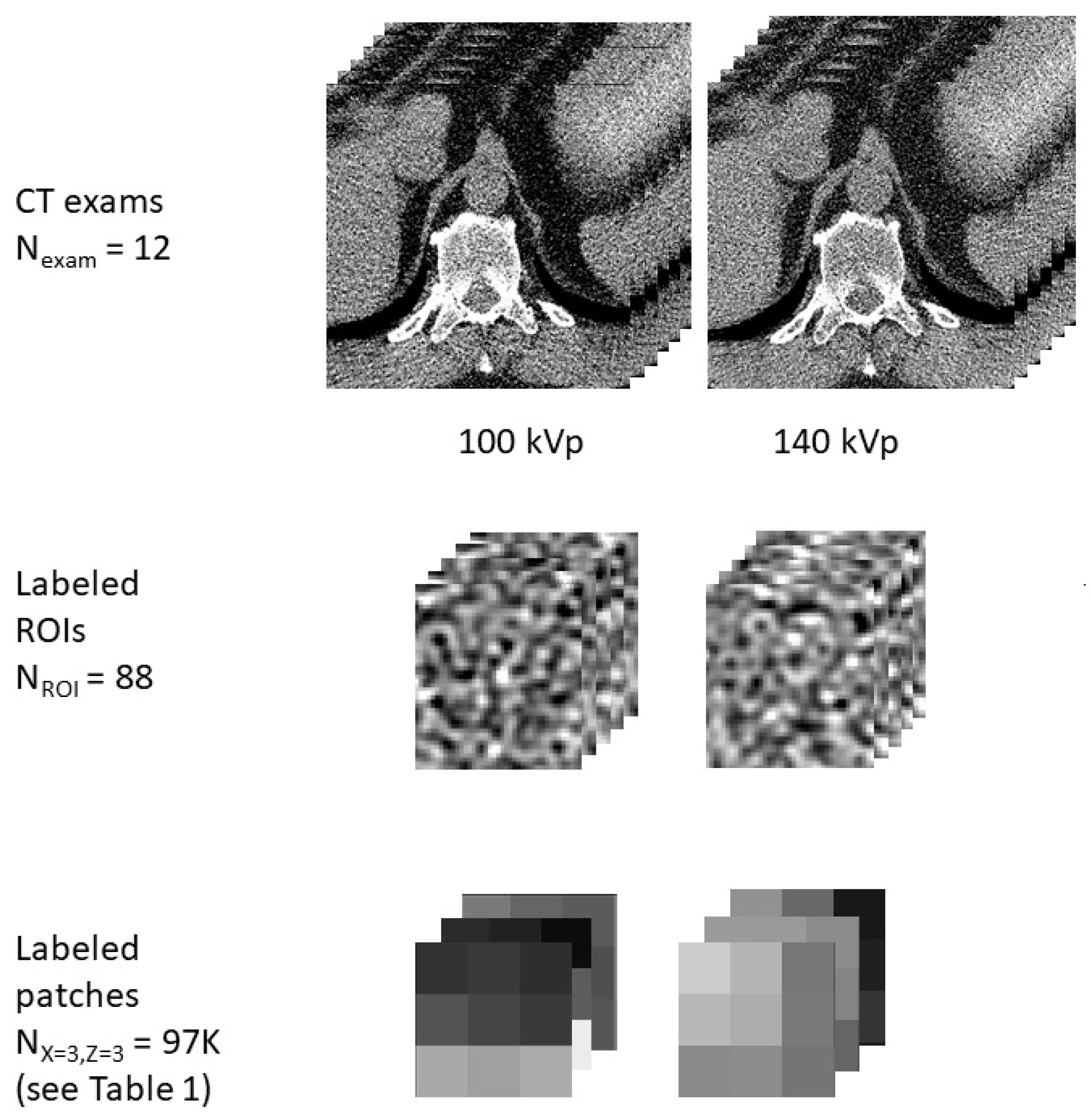
Illustrative representation of examples of data used in this study in the form of CT exams, ROIs, and patches. The study includes data aggregated from 12 CT exams, 88 ROIs of various tissue classes, and thousands of patches of varying sizes, with a representative patch shown for a patch size of X = 3, Z = 3.
Figure 3.
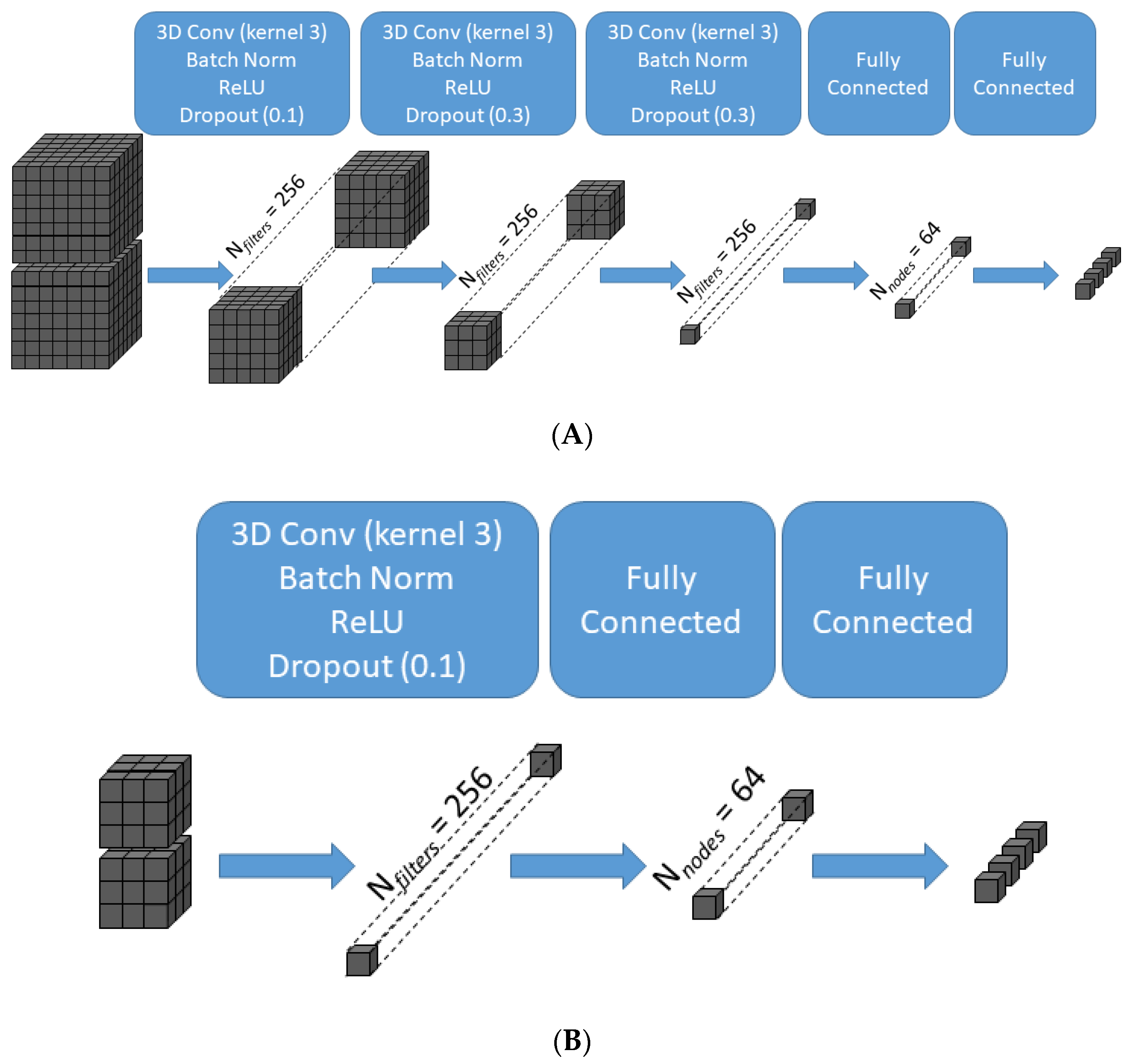
Schematic illustration of the transformation of data across the layers of the convolutional neural network for two representative patch sizes. For the largest patch size (X = 7, Z = 7) (A), the 7 × 7 × 7 × 2 input is transformed after three successive 3D convolutions of kernel size 3 × 3 × 3 without padding into a 256-element array of scalar values prior to the fully connected layers, which in turn generate the final 4-class classifier output. For the smallest patch size (X = 3, Z = 3) (B), one 3D convolution of kernel size 3 × 3 × 3 without padding is sufficient to generate a 256-element array of scalar values. 3D Conv = three-dimensional convolutional layer; Batch Norm = batch normalization layer; ReLU = rectified linear unit activation function.
Figure 4.
Histogram of average center Hounsfield units (HU) of all 3 × 3 × 3 × 2 patches generated from the labeled ROIs. Although center pixels in patches from bone ROIs, not unexpectedly, show higher HUs on average than water and soft tissue patches, and fat patches show lower HUs, the tissue classes show substantial overlap in HUs. In particular, the histograms for soft tissue and water are nearly superimposable. Therefore, no HU thresholding could separate all four classes with high accuracy.
Figure 5.
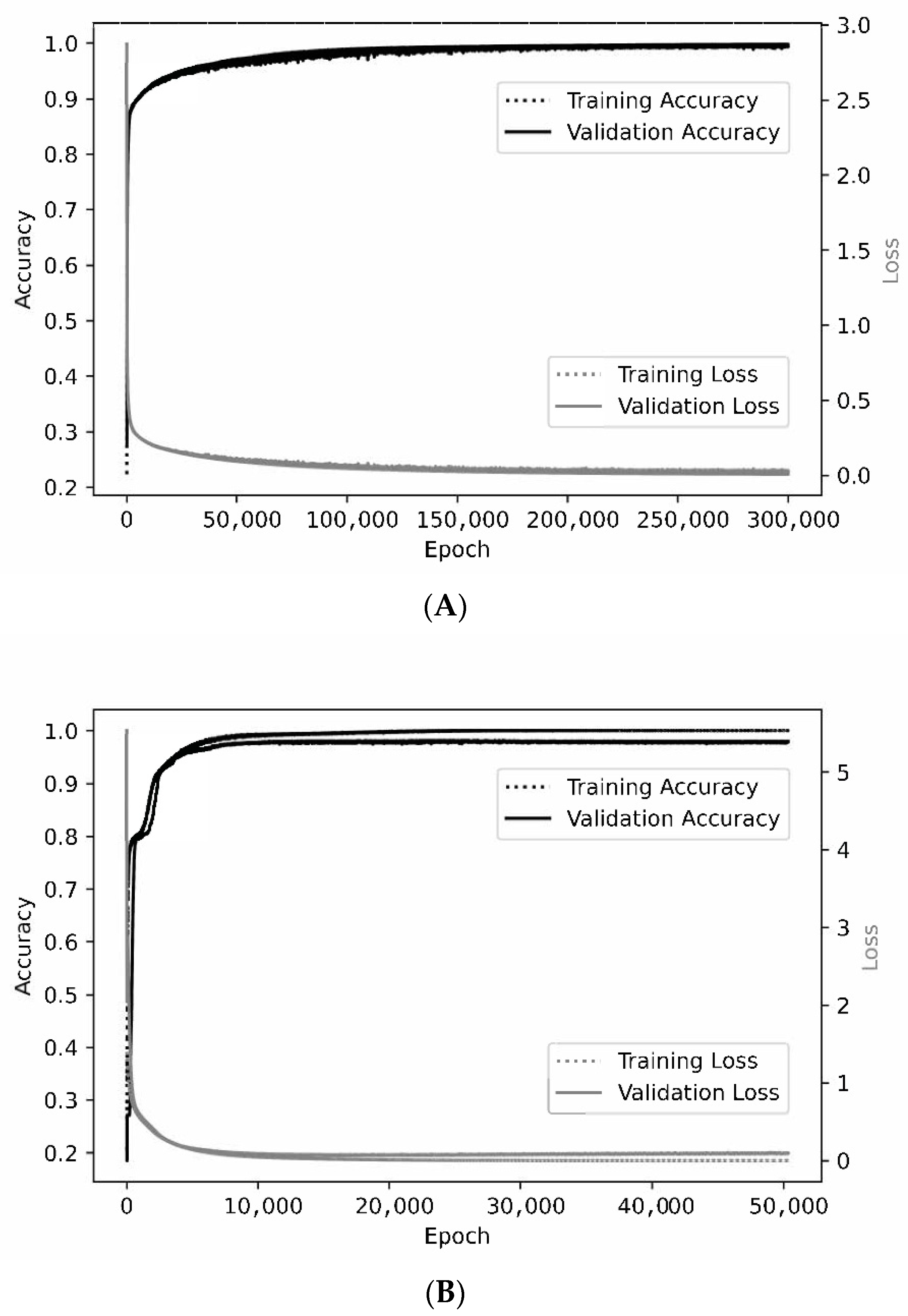
Representative learning curves. For the smallest patch size of 3 × 3 × 3 × 2 (A), the training and validation curves are nearly superimposed when assessing either accuracy or loss. At a patch size of 7 × 7 × 7 × 2 (B), optimal training accuracy and loss are slightly better than for validation.
Figure 6.
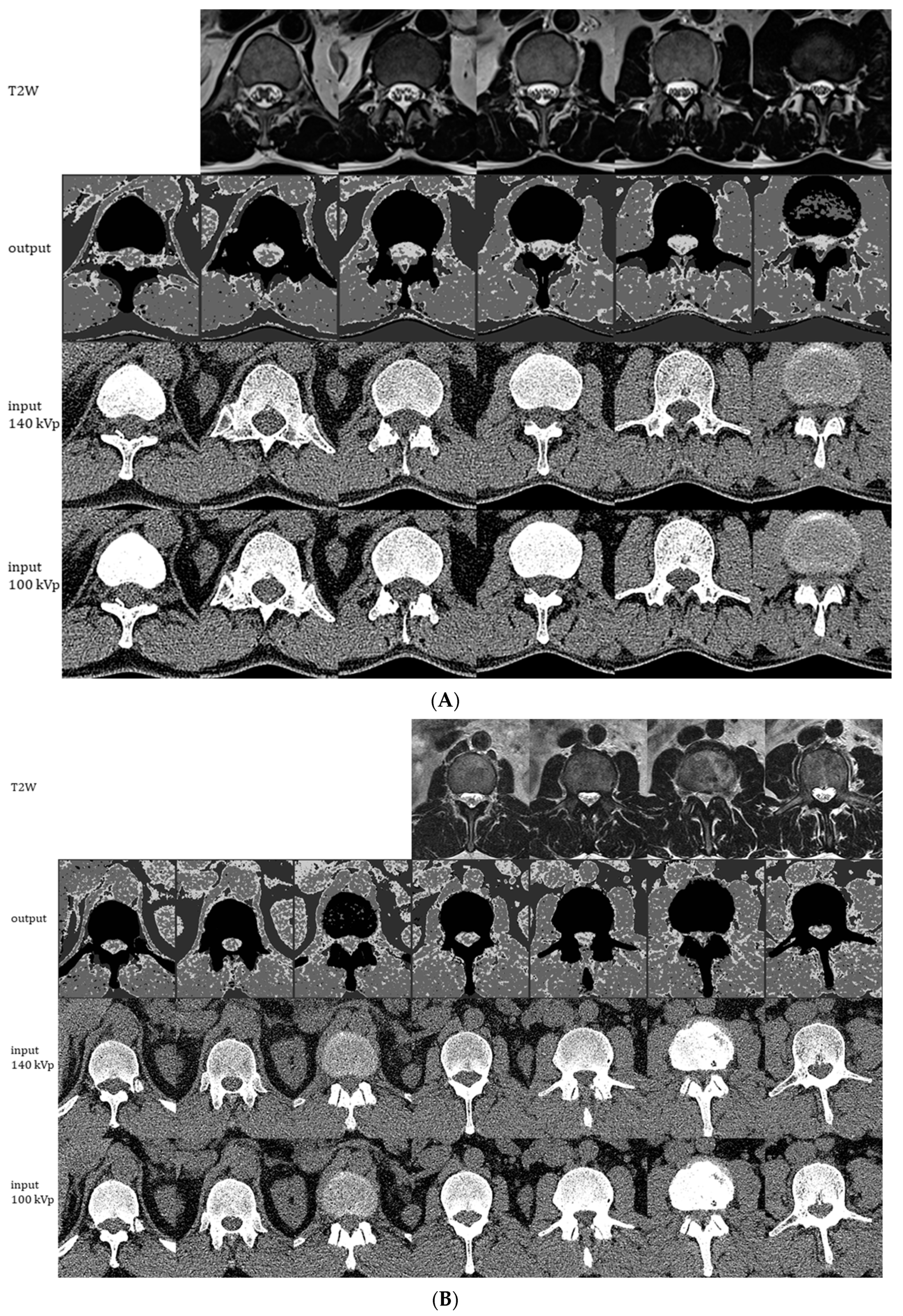
Representative images generated by the ensemble model as applied to 1 of the 11 CT exams used for individual model training/validation (A) and to the holdout CT exam that was not used for training (B). The montages show generated 4-scale output images (2nd row), original 0.75 mm CT images at 140 kVp (3rd row), and original 0.75 mm CT images at 100 kVp (4th row). Each column of CT images shown represents slices at 2.25 cm intervals. Corresponding 5 mm axial T2W MRI images are shown in the 1st row for comparison.
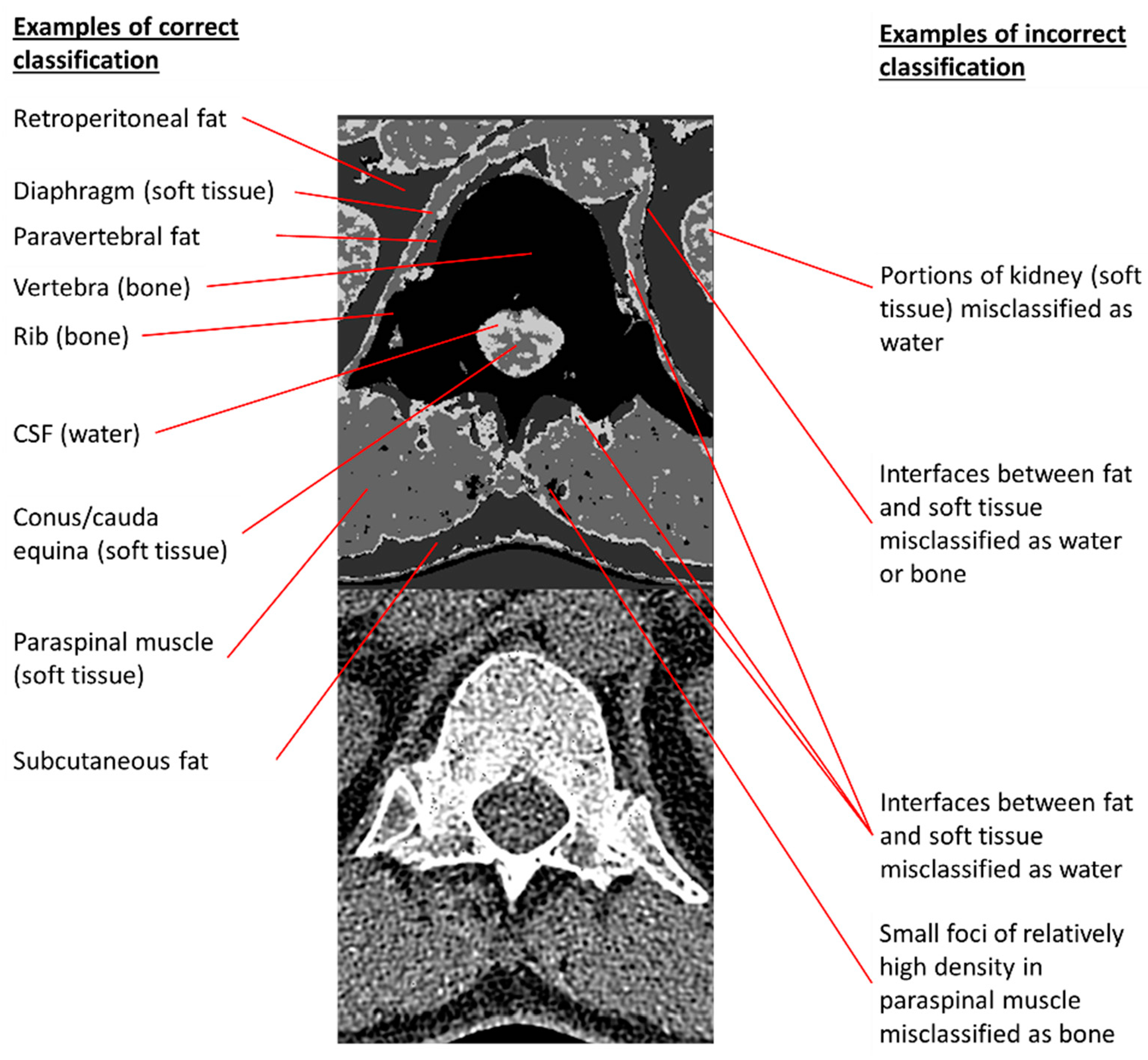
Figure 7.
Examples of correct and incorrect tissue classifications in a representative output image taken from the second column of
Figure 6A. The 140 kVp CT source image is placed below the annotated output image to permit comparisons of annotated pixels to their corresponding CT appearances.
Table 1.
Composition of the labeled dataset used for training and individual model testing. Entries in each cell represent numbers of non-overlapping X by X by Z by 2 tensors in the labeled dataset.
| | | Class Labels | |
|---|
| X | Z | Bone | Soft Tissue | Fat | Water | Total |
|---|
| 3 | 3 | 29,380 | 33,343 | 21,018 | 13,373 | 97,114 |
| 3 | 5 | 16,996 | 19,779 | 12,599 | 7840 | 57,214 |
| 3 | 7 | 5861 | 6667 | 4187 | 2783 | 19,498 |
| 5 | 5 | 11,146 | 13,850 | 8671 | 5270 | 38,937 |
| 5 | 7 | 3837 | 4669 | 2876 | 1873 | 13,255 |
| 7 | 7 | 1879 | 2258 | 1435 | 912 | 6484 |
Table 2.
Confusion matrices and detailed test performance characteristics a at different combinations of X and Z.
| | | Ground Truth | Prediction | | | | | | |
|---|
| X | Z | | Bone | Soft Tissue | Fat | Water | Total | Sens | Spec | Prec | F1 | Acc |
|---|
| 3 | 3 | bone | 5856 | 6 | 1 | 13 | 5876 | 99.7% | 100.0% | 99.9% | 99.8% | 99.9% |
| | | soft tissue | 0 | 6665 | 0 | 4 | 6669 | 99.9% | 99.8% | 99.7% | 99.8% | 99.9% |
| | | fat | 0 | 0 | 4204 | 0 | 4204 | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| | | water | 4 | 17 | 0 | 2653 | 2674 | 99.2% | 99.9% | 99.4% | 99.3% | 99.8% |
| | | TOTAL | 5860 | 6688 | 4205 | 2670 | 19,423 | | | | | 99.8% |
| 3 | 5 | bone | 3331 | 18 | 8 | 42 | 3399 | 98.0% | 99.9% | 99.8% | 98.9% | 99.4% |
| | | soft tissue | 0 | 3947 | 0 | 9 | 3956 | 99.8% | 99.3% | 98.7% | 99.2% | 99.4% |
| | | fat | 1 | 0 | 2519 | 0 | 2520 | 100.0% | 99.9% | 99.7% | 99.8% | 99.9% |
| | | water | 5 | 36 | 0 | 1527 | 1568 | 97.4% | 99.5% | 96.8% | 97.1% | 99.2% |
| | | TOTAL | 3337 | 4001 | 2527 | 1578 | 11,443 | | | | | 99.0% |
| 3 | 7 | bone | 2163 | 16 | 9 | 42 | 2230 | 97.0% | 99.9% | 99.9% | 98.4% | 99.1% |
| | | soft tissue | 1 | 2748 | 0 | 21 | 2770 | 99.2% | 98.6% | 97.6% | 98.4% | 98.8% |
| | | fat | 0 | 0 | 1734 | 0 | 1734 | 100.0% | 99.9% | 99.5% | 99.7% | 99.9% |
| | | water | 2 | 52 | 0 | 1000 | 1054 | 94.9% | 99.1% | 94.1% | 94.5% | 98.5% |
| | | TOTAL | 2166 | 2816 | 1743 | 1063 | 7788 | | | | | 98.2% |
| 5 | 5 | bone | 1151 | 8 | 0 | 13 | 1172 | 98.2% | 100.0% | 99.9% | 99.1% | 99.4% |
| | | soft tissue | 0 | 1329 | 0 | 5 | 1334 | 99.6% | 99.3% | 98.6% | 99.1% | 99.4% |
| | | fat | 0 | 0 | 837 | 0 | 837 | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| | | water | 1 | 11 | 0 | 545 | 557 | 97.8% | 99.5% | 96.8% | 97.3% | 99.2% |
| | | TOTAL | 1152 | 1348 | 837 | 563 | 3900 | | | | | 99.0% |
| 5 | 7 | bone | 733 | 11 | 1 | 22 | 767 | 95.6% | 100.0% | 100.0% | 97.7% | 98.7% |
| | | soft tissue | 0 | 929 | 0 | 5 | 934 | 99.5% | 98.7% | 97.6% | 98.5% | 98.9% |
| | | fat | 0 | 0 | 575 | 0 | 575 | 100.0% | 100.0% | 99.8% | 99.9% | 100.0% |
| | | water | 0 | 12 | 0 | 363 | 375 | 96.8% | 98.8% | 93.1% | 94.9% | 98.5% |
| | | TOTAL | 733 | 952 | 576 | 390 | 2651 | | | | | 98.1% |
| 7 | 7 | bone | 364 | 3 | 0 | 9 | 376 | 96.8% | 99.7% | 99.2% | 98.0% | 98.8% |
| | | soft tissue | 0 | 450 | 0 | 2 | 452 | 99.6% | 98.8% | 97.8% | 98.7% | 99.1% |
| | | fat | 0 | 0 | 287 | 0 | 287 | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| | | water | 3 | 7 | 0 | 172 | 182 | 94.5% | 99.0% | 94.0% | 94.2% | 98.4% |
| | | TOTAL | 367 | 460 | 287 | 183 | 1297 | | | | | 98.1% |
Table 3.
Confusion matrix and detailed test performance characteristics a of the final ensemble model.
| Ground Truth | Prediction b | | | | | | |
|---|
| | Bone | Soft Tissue | Fat | Water | Total | Sens | Spec | Prec | F1 | Acc |
|---|
| bone | 280 | 0 | 0 | 0 | 280 | 100.0% | 99.6% | 99.6% | 99.8% | 99.8% |
| soft tissue | 0 | 144 | 0 | 2 | 146 | 98.6% | 100.0% | 100.0% | 99.3% | 99.6% |
| fat | 0 | 0 | 96 | 0 | 96 | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| water | 1 | 0 | 0 | 9 | 10 | 90.0% | 99.6% | 81.8% | 85.7% | 99.4% |
| TOTAL | 281 | 144 | 96 | 11 | 532 | | | | | 99.4% |
| Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).