Abstract
Since their inception, deep-learning architectures have shown promising results for automatic segmentation. However, despite the technical advances introduced by fully convolutional networks, generative adversarial networks or recurrent neural networks, and their usage in hybrid architectures, automatic segmentation in the medical field is still not used at scale. One main reason is related to data scarcity and quality, which in turn generates a lack of annotated data that hinder the generalization of the models. The second main issue refers to challenges in training deep models. This process uses large amounts of GPU memory (that might exceed current hardware limitations) and requires high training times. In this article, we want to prove that despite these issues, good results can be obtained even when using a lower resource architecture, thus opening the way for more researchers to employ and use deep neural networks. In achieving the multi-organ segmentation, we are employing modern pre-processing techniques, a smart model design and fusion between several models trained on the same dataset. Our architecture is compared against state-of-the-art methods employed in a publicly available challenge and the notable results prove the effectiveness of our method.
1. Introduction
Automated medical segmentation employed on human organs from computed tomography (CT) or magnetic resonance imaging (MRI) has the potential to help radiology practitioners perform day to day activities. The manual interpretation of the images is a “tedious and exhausting work that is further impacted by the large variations in pathology between different individuals”, while the training of human experts is a complex and long-running process. All these factors indicate that the automatic segmentation of medical images, computerized delineation of human organs, or even automated diagnosis can be helpful for doctors if they provide accurate results.
1.1. Related Work
Due to their good results, deep-learning (DL) architectures are one of the most used solutions for computer vision [1] and for multi-organ segmentation. Since they were first proposed by [2], fully convolutional networks (FCN), alongside the well-known variants “2D U-Net” [3], “3D U-Net” [4] and the “V-Net” [5], have been the most used and recognized DL architectures. These DL networks are used for automatic segmentation in single or multi-organ scenarios with good results, but they are plagued by issues. The most important ones are:
- Data scarcity—annotated datasets that can be used as training data for DL architectures are hard to generate mainly because it is time intensive and costly to manually segment them by human experts;
- Data quality—data can be plagued by different issues such as noise or heterogenic intensities and contrast;
- Class imbalance—in medical image processing, organ size, appearance or location vary greatly from individual to individual. This is even more significant when there are several lesions or tumors. One important corner case issue of class imbalance is related to small organs;
- Challenges with training deep models—over-fitting (achieving a good fit of the DL model on the training/testing dataset, but not achieving the generalization to obtain correct results on new, unseen data), “reducing the time and the computational complexity of deep learning networks” [6] and lowering the high amounts of GPU memory needed in order to train models that can provide satisfactory results.
Proposed solutions to these issues are briefly summarized below.
The creation of hybrid architectures, which combine traditional deep-learning networks with generative adversarial networks (GANs), can improve the segmentation results. GANs were initially proposed by [7] and they have the ability to create new datasets that closely resemble to the initial training set. The most obvious usage of a GAN network is to try to reduce the data scarcity issue. A successful GAN hybrid approach was used by [8] in thorax segmentation by employing “generator and discriminator networks that compete against each other in an adversarial learning process” and in abdomen organ segmentation by [9], who “cascaded convolutional networks with adversarial networks to alleviate data scarcity limitations”.
Other proven hybrid architectures combine traditional DL networks with recurrent neural networks (RNNs). These networks are able to store the patterns of previous inputs and therefore can improve the segmentation results of the DL networks. Additionally, [10] designed a system consisting of U-Net networks and RNN networks in which “feature accumulation with recurrent residual convolutional layers” improves the segmentation outcome, while [11] presented a “U-Net-like network enhanced with bidirectional C-LSTM”.
One option employed to reduce training times is transfer learning. This is the ability to reuse the knowledge obtained when training a neural network and to transfer it to a new architecture [12]. Transfer learning in medical scenarios is performed either by re-using parameters from networks pre-trained on common images [12] or by fine-tuning networks that were already trained for another organ or segmentation task. Transfer learning generates better results when transferring weights from networks that have similar architectures. However, even on more differing architectures, it was proved that transfer learning is more efficient than random initialization [13].
Data augmentation can alleviate some of the deep neural issues described above. Pre-processing methods are executed before the training of a neural network. Methods such as the application of a set of affine transformation, e.g., flipping, scaling, rotating, mirroring, and elastic deformation [14] to the training/testing data as well as augmenting color (grey) values [15] have proven results, and they can improve the segmentation results. Other pre-processing methods include bias/attenuation correction [16] and voxel intensity normalization [17].
Most recently, the GANs, “variational Bayes AE”, proposed by Kingma et al. [18], “adversarial data augmentation” put forward by Volpi et al. [19] and reinforcement learning as suggested by Cubuk et al. [20] have been employed to learn augmentation techniques from the existing training data.
Post-processing can be also applied to refine and smoothen the segmentations to make them more continuous or realistic. The most popular method applied in DL is the conditional random field proposed by Christ et al. [21].
One important solution for class imbalance is the use of a patch-based technique for learning. The training data are split into “multiple patches which can be either overlapping or random patches” [22]. Overlapping patches offer better training results but are computationally intensive [23] while random patches provide higher variance and improved results [24] but produce lower results for small organs as they might miss completely the areas of interest. Other important works that demonstrate the improvement capabilities of patch-wise training in 2D or 3D include [25,26].
Another solution was proposed by Dai et al. [27] in the form of a “critic network” that applies to the training phase the regular structures found in human physiology in order to correct the training data. Other proven resolution is to enlarge the network’s depth so that there are even more layers of convolutions that can learn the features [28].
Architectures that employ multi-modality approaches can alleviate class imbalance problems with solutions proposed by [29,30], while other authors used GAN networks to synthesize images from different modalities [26,31], with encouraging results.
As previously stated, an important challenge of training deep neural networks is over-fitting. Besides the obvious solutions of increasing the training and testing data (which is not easily employable as human annotated datasets are time consuming to generate) there are several other applicable techniques. These are weight regularization [32], dropout [33] or ensemble learning [34].
Another challenge is to “reduce the time and the computational complexity of deep learning networks” [6]. Important works that propose solutions are [35,36]. Other authors tried to simplify the shape of DL networks with good results obtained by [37,38].
1.2. Aim
Our aim was to prove that even an architecture that uses a lower-resource environment, can achieve good segmentation results that rank in the upper bracket of a recognized multi-organ challenge or competition. In this way, all researchers can use deep neural networks to advance the knowledge field, regardless of the hardware capabilities.
In line with this, we have imposed some constraints. The first one, was to employ a deep-learning architecture that can be trained using a maximum of 8 GB GPU. This is achievable using a medium budget video card. In this way we could prove that even by using a smaller memory capacity than present-day state-of-the-art articles (who use up to 24 GB GPUs), good results can be obtained when using a solid DL architecture.
The second constraint that we imposed to ourselves was to use a recognized, but still simple, DL network. We chose the U-Net 3D architecture that is widely used in research and has proven time and time again that is a good fit for medical segmentation.
These are hard constraints considering today’s state-of-the-art in hardware capabilities and the technical advances in DL, but even so, an architecture that uses a good design should obtain meaningful and consistent outcomes.
2. Materials and Methods
2.1. Dataset
In order to design, train and test our proposed deep-learning architecture, we selected the SegTHOR [39] challenge which addresses the problem of segmenting 4 thoracic organs at risk: esophagus, heart, aorta and trachea.
The challenge provides 40 CTs “with manual segmentation while the test set contains 20 CTs. The CT scans have 512 × 512 pixels size with in-plane resolution varying between 0.90 mm and 1.37 mm per pixel, depending on the patient. The number of slices varies from 150 to 284 with a z-resolution between 2 mm and 3.7 mm. The most frequent resolution is 0.98 × 0.98 × 2.5 mm3” [39]. Figure 1. is a visual representation of one of the CTs provided by the challenge.
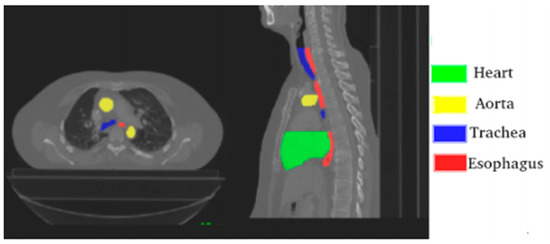
Figure 1.
Axial and sagittal planes with annotated organs; source Lambert et al. [39].
The challenge’s authors evaluate the results independently using code that is automatically run and which is also open source. The results are generated independently for each of the organ and the employed metrics are:
- The overlap dice metric (DM), “defined as 2*intersection of automatic and manual areas/(sum of automatic and manual areas)” [39];
- The Hausdorff distance (HD), “defined as max(ha,hb), where ha is the maximum distance, for all automatic contour points, to the closest manual contour point and hb is the maximum distance, for all manual contour points, to the closest automatic contour point” [39].
2.2. Proposed Deep-Learning Architecture
Our proposed architecture consists of a pipeline with four main components: a preprocessing step, a 3D U-Net [4] trained for multi-organ segmentation, four separate 3D U-Nets [4] for single organ segmentation and a fusion of results that will generate the final segmentation. The diagram of the architecture is presented in Figure 2.
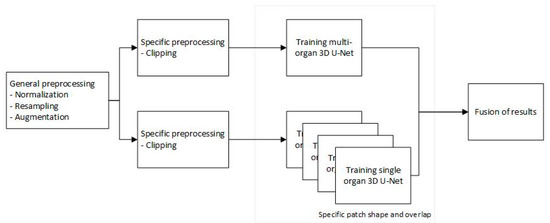
Figure 2.
Proposed pipeline.
2.2.1. Preprocessing
The initial step in the pipeline is all about pre-processing. Besides patients’ morphological differences, CT scans are produced with varying voxel sizes because the CT scanners inherently have different setups. All these factors produce different imaging artefacts which increase the complexity of segmentation. Therefore, the first step of pre-processing involves resampling the CT scans to normalize the slice thickness and also to reduce the image sizes (by a factor of 2).
The second step in pre-processing was to apply a clipping of the voxel values based on the Hounsfield scale for the organs of interest [40]. This clipping is based on Table 1 and greatly helps the deep-learning model concentrate only on the values and body locations which are relevant.

Table 1.
Hounsfield scale for the organs of interest.
In the third and final step, the images were normalized using a simple standard Z-score normalization [17].
Deep-learning results are more accurate when employing a big dataset. Conversely, a small dataset will increase the model’s tendency to overfit. Unfortunately, the SegTHOR [39] dataset has only 40 training CTs; therefore, data augmentation techniques were used. These included: scaling, rotating, elastic deformation [14], augmenting color (grey) values [15], gamma correction and adding gaussian noise. All these augmentation techniques implied that two learning cycles could be executed using the same dataset but with altered or enhanced characteristics, which reduced the data scarcity issue.
The preprocessing procedures were applied on the whole dataset.
2.2.2. Model Description
The segmentation architecture consists of 5 DL networks. For each organ a separate model was trained, while another model was trained in a multi-organ setup (all 4 organs). A standard U-Net [4] network was used in all training scenarios with a completely ordinary setup: a depth of 4, 32 convolutional filters (3 × 3 × 3) in the initial layer, 64, 128 and 256 filters in the subsequent convolutional layers, batch normalization, max pooling, RELU activation function for the hidden layers and Softmax activation function in the last output layer.
Because a medium-memory GPU was employed and due to the size of CTs, there was not enough memory to train our models on the complete data captured in a CT. Therefore, a smart patching mechanism was used. This would take random chunks of smaller sized 3D parts of a CT and feed them to the model. The size of the chunks was based on the expected organ morphology, and the patch strategy was seconded by an overlapping of patches which also matched the expected organ dimension. For the multi-organ model, a medium patch size and patch overlapping was used in order to accommodate all organ sizes.
While striving to use the available memory to the maximum, the biggest possible patch sizes per model were obtained and detailed in Table 2, while the batch size was set to 2. We want to highlight that the size of the initial CTs was 512 × 512 pixels with number of slices that varied between 150 and 284.

Table 2.
Patch sizes.
The 40 CTs were split randomly into learning and testing datasets with a ratio of 80:20, and a maximum of 500 epochs were executed for each different network. Higher learning rates were used in the initial phases to obtain a good set of parameters faster, while a more discrete learning rate was used in the final phase of learning to obtain the best possible parameter values. Different loss algorithms were used based on single or multi-organ learning. Therefore, Tversky loss [41] was used for single organs networks while the Tversky enhanced with cross-entropy was employed for the multi-organ network.
2.2.3. Fusion of Results
The results from each individual network are merged thus obtaining the final segmentation result. This process starts by taking the segmentation of the multi-organ network and adding on top of them the results from each single organ network. This process followed four rules:
- Merging will start with smaller or thinner organs to offer a boost to those organs that are harder to track. The order was: trachea, esophagus, aorta, and heart;
- Voxels with the same segmentation on both multi-organ and one single organ networks are guaranteed to obey that segmentation result;
- In case of mismatch between the multi-organ network result and the single network, the segmentation result that has the most neighboring voxels with the same segmentation wins;
- In case there are several segmentation results, or a clear winner based on neighbors cannot be achieved, the multi-organ segmentation has priority. This is based on the fact that the multi-organ segmentation has all the organs while the single organ network incorporates results only for one organ type.
2.2.4. Implementation
For the implementation, the MISCNN https://github.com/frankkramer-lab/MIScnn (accessed on 1 May 2022) [42] open source library was used. This library provides 2D or 3D DL model implementation and data I/O modules. For data augmentation we used batchgenerators by MIC@DKFZ which is Python package developed by “The Division of Medical Image Computing at the German Cancer Research Center (DKFZ) and the Applied Computer Vision Lab of the Helmholtz Imaging Platform”—https://github.com/MIC-DKFZ/batchgenerators (accessed on 1 May 2022) [43].
Our complete Python implementation can be found at https://github.com/valentinogrean/Multi-organ-segmentation-low-resource-environment (accessed on 28 September 2022).
3. Results
The five DL networks that are part of the architecture were trained one by one on the same hardware with the maximum number of epochs set to 500. During the training process, if the loss did not improve for 20 epochs, the learning rate would be decreased with a factor of 10, while the minimum allowed learning rate was set to 0.00001. The training process would be considered complete if the loss would not improve for 20 epochs using the minimum learning rate. With this implementation, the maximum number of epochs was never reached, and in practice it took around 350 epochs to fully train each model. From the computational time’s perspective, a complete training of one DL network took around 24 h.
The current architecture managed to obtain as high as eighth place in the SegTHOR [39] challenge out of at least forty valid submissions.
As per the SegTHOR documentation, the results were evaluated based on the “overlap Dice metric (DM)” and the “Hausdorff distance (HD)”. Therefore, two metrics are computed for each organ, totaling eight different metrics. The final ranking is “based on the average of the 8 metrics”.
As SegTHOR is an open challenge, new submissions can be added, and they will influence the raking. In Table 3. we present our best results next to the results for the highest-ranking user at the time of writing of this article.

Table 3.
Detailed results of our best submission.
The best results were obtained using our proposed fusion strategy from Section 2.2.3. In support of this, we present, in Table 4, additional outcomes from different submissions that were obtained using other strategies. All of these results were automatically calculated by the SegTHOR challenge, making them objective elements of an ablation experimentation.

Table 4.
Comparison between different strategies.
As visual examples, we have provided Figure 3, which shows the automatic segmentations using our proposed method for some of the patients from the SegTHOR dataset. For patient 01, the ground truth is provided as this is part of the training set. However, for patient 41, only our own segmentations are provided, as the ground truth is private to the SegTHOR team and is used in ranking the submissions.
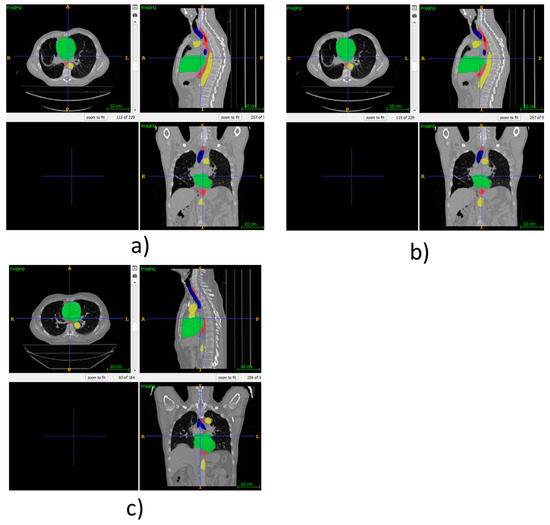
Figure 3.
(a) Ground truth SegTHOR patient 01, (b) automatic segmentation using our proposed method for SegTHOR patient 01, (c) automatic segmentation using our proposed method for SegTHOR patient 41.
4. Discussion
The architecture demonstrates that even thought a lower memory GPU was employed, results close to state of the art can be achieved. Other important contributions of the architecture are the novel patching mechanism that mimics the organ shape and employing the smart fusion of results between several deep neural networks.
The best results were obtained for esophagus and the worst for trachea. We theorized that the poor results for the trachea stemmed from its morphological structure, having the lowest values on the Hounsfield scale. This was a challenge for our models that was observed more on the multi-organ model than on the single organ network.
Secondly, the esophagus and the trachea are two neighboring organs. The target to improve the results for trachea had a negative impact on the results for the esophagus and vice versa. We still tried to boost the scores for trachea by making it the first merged organ, but with limited effect. The issue came from the fact that the multi-organ network had low accuracy on the trachea segmentation in the first place (something that we could not alleviate with our model).
We have tested the architecture on GPUs with higher total memory (16 GB and 24 GB) that allowed us to use larger patches and larger batch sizes. Although we were able to replicate the results, we could not improve them. Thus, we theorize that the patching mechanism is efficient enough to alleviate most of the issues that arise from not being able to train in one step over the complete CT data. Regardless, we still succeeded in proving that, despite the imposed constraints (medium-sized GPU with only 8 GB of RAM, standard 3D U-NET), state-of-the-art results can be obtained. These were achieved by employing intensive and smart pre-processing, clever patching, using several deep neural networks, and merging their results in a consistent way.
As an immediate improvement to our proposed method, we can mention enlarging the dataset, the inclusion of GANs to reduce the data scarcity issue, or the addition of RRNs to improve the segmentation.
Author Contributions
Conceptualization, V.O. and R.B.; methodology, R.B.; software, V.O.; validation, V.O.; formal analysis, V.O.; investigation, V.O.; resources, V.O.; data curation, V.O.; writing—original draft preparation, V.O.; writing—review and editing, R.B. and V.O.; visualization, V.O.; supervision, R.B.; project administration, V.O. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Data was obtained from SegTHOR challenge and are available at https://competitions.codalab.org/competitions/21145. The complete code can be found at https://github.com/valentinogrean/Multi-organ-segmentation-low-resource-environment.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lu, L.; Wang, X.; Carneiro, G.; Yang, L. Deep Learning and Convolutional Neural Networks for Medical Imaging and Clinical Informatics; Springer: Berlin/Heidelberg, Germany, 2019; ISBN 978-3-030-13969-8. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; IEEE: Piscataway, NJ, USA, 2015; pp. 3431–3440. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2015; Volume 9351, pp. 234–241. ISBN 978-3-319-24573-7. [Google Scholar]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2016; Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2016; Volume 9901, pp. 424–432. ISBN 978-3-319-46722-1. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Seg-mentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 565–571. [Google Scholar]
- Asgari Taghanaki, S.; Abhishek, K.; Cohen, J.P.; Cohen-Adad, J.; Hamarneh, G. Deep Semantic Segmentation of Natural and Medical Images: A Review. Artif. Intell. Rev. 2021, 54, 137–178. [Google Scholar] [CrossRef]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Networks. arXiv 2014, arXiv:1406.2661. [Google Scholar] [CrossRef]
- Dong, X.; Lei, Y.; Wang, T.; Thomas, M.; Tang, L.; Curran, W.J.; Liu, T.; Yang, X. Automatic Multiorgan Segmentation in Thorax CT Images Using U-net-GAN. Med. Phys. 2019, 46, 2157–2168. [Google Scholar] [CrossRef]
- Conze, P.-H.; Kavur, A.E.; Cornec-Le Gall, E.; Gezer, N.S.; Le Meur, Y.; Selver, M.A.; Rousseau, F. Abdominal Multi-Organ Segmentation with Cascaded Convolutional and Adversarial Deep Networks. Artif. Intell. Med. 2021, 117, 102109. [Google Scholar] [CrossRef]
- Alom, M.Z.; Yakopcic, C.; Hasan, M.; Taha, T.M.; Asari, V.K. Recurrent Residual U-Net for Medical Image Segmentation. J. Med. Imaging 2019, 6, 1. [Google Scholar] [CrossRef]
- Novikov, A.A.; Major, D.; Wimmer, M.; Lenis, D.; Buhler, K. Deep Sequential Segmentation of Organs in Volumetric Medical Scans. IEEE Trans. Med. Imaging 2019, 38, 1207–1215. [Google Scholar] [CrossRef]
- Shie, C.-K.; Chuang, C.-H.; Chou, C.-N.; Wu, M.-H.; Chang, E.Y. Transfer Representation Learning for Medical Image Analysis. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; IEEE: Piscataway, NJ, USA, 2015; pp. 711–714. [Google Scholar]
- Yosinski, J.; Clune, J.; Bengio, Y.; Lipson, H. How Transferable Are Features in Deep Neural Networks? arXiv 2014, arXiv:1411.1792. [Google Scholar]
- Simard, P.Y.; Steinkraus, D.; Platt, J.C. Best Practices for Convolutional Neural Networks Applied to Visual Document Analysis. In Proceedings of the Seventh International Conference on Document Analysis and Recognition, Edinburgh, UK, 3–6 August 2003; IEEE Computer Society: Edinburgh, UK, 2003; Volume 1, pp. 958–963. [Google Scholar]
- Golan, R.; Jacob, C.; Denzinger, J. Lung Nodule Detection in CT Images Using Deep Convolutional Neural Networks; IEEE: Piscataway, NJ, USA, 2016; pp. 243–250. [Google Scholar]
- Yang, X.; Wang, T.; Lei, Y.; Higgins, K.; Liu, T.; Shim, H.; Curran, W.J.; Mao, H.; Nye, J.A. MRI-Based Attenuation Correction for Brain PET/MRI Based on Anatomic Signature and Machine Learning. Phys. Med. Biol. 2019, 64, 025001. [Google Scholar] [CrossRef]
- Zhou, X.-Y.; Yang, G.-Z. Normalization in Training U-Net for 2-D Biomedical Semantic Segmentation. IEEE Robot. Autom. Lett. 2019, 4, 1792–1799. [Google Scholar] [CrossRef]
- Kingma, D.P.; Welling, M. Auto-Encoding Variational Bayes. arXiv 2014, arXiv:1312.6114. [Google Scholar]
- Volpi, R.; Namkoong, H.; Sener, O.; Duchi, J.C.; Murino, V.; Savarese, S. Generalizing to Unseen Domains via Adversarial Data Augmentation. In Advances in Neural Information Processing Systems; Bengio, S., Wallach, H., Larochelle, H., Grauman, K., Cesa-Bianchi, N., Garnett, R., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2018; Volume 31. [Google Scholar]
- Cubuk, E.D.; Zoph, B.; Mane, D.; Vasudevan, V.; Le, Q.V. AutoAugment: Learning Augmentation Policies from Data. arXiv 2019, arXiv:1805.09501. [Google Scholar]
- Christ, P.F.; Elshaer, M.E.A.; Ettlinger, F.; Tatavarty, S.; Bickel, M.; Bilic, P.; Rempfler, M.; Armbruster, M.; Hofmann, F.; D’Anastasi, M.; et al. Automatic Liver and Lesion Segmentation in CT Using Cascaded Fully Convolutional Neural Networks and 3D Conditional Random Fields. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2016; Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2016; Volume 9901, pp. 415–423. ISBN 978-3-319-46722-1. [Google Scholar]
- Hesamian, M.H.; Jia, W.; He, X.; Kennedy, P. Deep Learning Techniques for Medical Image Segmentation: Achievements and Challenges. J. Digit. Imaging 2019, 32, 582–596. [Google Scholar] [CrossRef] [PubMed]
- Dou, Q.; Yu, L.; Chen, H.; Jin, Y.; Yang, X.; Qin, J.; Heng, P.-A. 3D Deeply Supervised Network for Automated Segmentation of Volumetric Medical Images. Med. Image Anal. 2017, 41, 40–54. [Google Scholar] [CrossRef] [PubMed]
- Anirudh, R.; Thiagarajan, J.J.; Bremer, T.; Kim, H. Lung Nodule Detection Using 3D Convolutional Neural Networks Trained on Weakly Labeled Data. In Proceedings of the Medical Imaging 2016: Computer-Aided Diagnosis, San Diego, CA, USA, 24 March 2016; Tourassi, G.D., Armato, S.G., Eds.; SPIE: Bellingham, WA, USA, 2016; p. 978532. [Google Scholar]
- Alex, V.; Vaidhya, K.; Thirunavukkarasu, S.; Kesavadas, C.; Krishnamurthi, G. Semisupervised Learning Using Denoising Autoencoders for Brain Lesion Detection and Segmentation. J. Med. Imaging 2017, 4, 1. [Google Scholar] [CrossRef]
- Lei, Y.; Wang, T.; Liu, Y.; Higgins, K.; Tian, S.; Liu, T.; Mao, H.; Shim, H.; Curran, W.J.; Shu, H.-K.; et al. MRI-Based Synthetic CT Generation Using Deep Convolutional Neural Network. In Proceedings of the Medical Imaging 2019: Image Processing, San Diego, CA, USA, 15 March 2019; Angelini, E.D., Landman, B.A., Eds.; SPIE: Bellingham, WA, USA, 2019; p. 100. [Google Scholar]
- Dai, W.; Dong, N.; Wang, Z.; Liang, X.; Zhang, H.; Xing, E.P. SCAN: Structure Correcting Adversarial Network for Organ Segmentation in Chest X-rays. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Stoyanov, D., Taylor, Z., Carneiro, G., Syeda-Mahmood, T., Martel, A., Maier-Hein, L., Tavares, J.M.R.S., Bradley, A., Papa, J.P., Belagiannis, V., et al., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2018; Volume 11045, pp. 263–273. ISBN 978-3-030-00888-8. [Google Scholar]
- Yu, L.; Chen, H.; Dou, Q.; Qin, J.; Heng, P.-A. Automated Melanoma Recognition in Dermoscopy Images via Very Deep Residual Networks. IEEE Trans. Med. Imaging 2017, 36, 994–1004. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, R.; Deng, H.; Wang, L.; Lin, W.; Ji, S.; Shen, D. Deep Convolutional Neural Networks for Multi-Modality Isointense Infant Brain Image Segmentation. NeuroImage 2015, 108, 214–224. [Google Scholar] [CrossRef]
- Zeng, G.; Zheng, G. Multi-Stream 3D FCN with Multi-Scale Deep Supervision for Multi-Modality Isointense Infant Brain MR Image Segmentation. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 136–140. [Google Scholar]
- Liu, Y.; Lei, Y.; Wang, Y.; Shafai-Erfani, G.; Wang, T.; Tian, S.; Patel, P.; Jani, A.B.; McDonald, M.; Curran, W.J.; et al. Evaluation of a Deep Learning-Based Pelvic Synthetic CT Generation Technique for MRI-Based Prostate Proton Treatment Planning. Phys. Med. Biol. 2019, 64, 205022. [Google Scholar] [CrossRef] [PubMed]
- Ng, A. Feature Selection, L 1 vs. L 2 Regularization, and Rotational Invariance. In Proceedings of the Twenty-First International Conference on Machine Learning, Banff, AB, Canada, 4–8 July 2004. [Google Scholar] [CrossRef]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Kamnitsas, K.; Bai, W.; Ferrante, E.; McDonagh, S.; Sinclair, M.; Pawlowski, N.; Rajchl, M.; Lee, M.; Kainz, B.; Rueckert, D.; et al. Ensembles of Multiple Models and Architectures for Robust Brain Tumour Segmentation. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Crimi, A., Bakas, S., Kuijf, H., Menze, B., Reyes, M., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2018; Volume 10670, pp. 450–462. ISBN 978-3-319-75237-2. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Leroux, S.; Molchanov, P.; Simoens, P.; Dhoedt, B.; Breuel, T.; Kautz, J. IamNN: Iterative and Adaptive Mobile Neural Network for Efficient Image Classification. arXiv 2018, arXiv:1804.10123. [Google Scholar]
- Kim, Y.-D.; Park, E.; Yoo, S.; Choi, T.; Yang, L.; Shin, D. Compression of Deep Convolutional Neural Networks for Fast and Low Power Mobile Applications. arXiv 2016, arXiv:1511.06530. [Google Scholar]
- Wen, W.; Wu, C.; Wang, Y.; Chen, Y.; Li, H. Learning Structured Sparsity in Deep Neural Networks. In Proceedings of the 30th International Conference on Neural Information Processing Systems, Barcelona, Spain, 5–10 December 2016; Curran Associates Inc.: Red Hook, NY, USA, 2016; pp. 2082–2090. [Google Scholar]
- Lambert, Z.; Petitjean, C.; Dubray, B.; Kuan, S. SegTHOR: Segmentation of thoracic organs at risk in CT images. In Proceedings of the 2020 Tenth International Conference on Image Processing Theory, Tools and Applications (IPTA), Paris, France, 9–12 November 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1–6. [Google Scholar]
- International Atomic Energy Agency. Diagnostic Radiology Physics: A Handbook for Teachers and Students; Dance, D.R., Ed.; American Association of Physicists in Medicine, STI/PUB; International Atomic Energy Agency: Vienna, Austria, 2014; ISBN 978-92-0-131010-1. [Google Scholar]
- Salehi, S.S.M.; Erdogmus, D.; Gholipour, A. Tversky Loss Function for Image Segmentation Using 3D Fully Convolutional Deep Networks. In Machine Learning in Medical Imaging; Wang, Q., Shi, Y., Suk, H.-I., Suzuki, K., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2017; Volume 10541, pp. 379–387. ISBN 978-3-319-67388-2. [Google Scholar]
- Müller, D.; Kramer, F. MIScnn: A Framework for Medical Image Segmentation with Convolutional Neural Networks and Deep Learning. BMC Med. Imaging 2021, 21, 12. [Google Scholar] [CrossRef]
- Isensee, F.; Jäger, P.; Wasserthal, J.; Zimmerer, D.; Petersen, J.; Kohl, S.; Schock, J.; Klein, A.; Roß, T.; Wirkert, S.; et al. Batchgenerators—A Python Framework for Data Augmentation. Zenodo. 2020. Available online: https://zenodo.org/record/3632567#.Y0FKN3ZBxPY (accessed on 28 September 2022).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).