Toward Efficient Similarity Search under Edit Distance on Hybrid Architectures
Abstract
1. Introduction
2. Related Work
3. Preliminaries
3.1. Similarity Search under Edit Distance
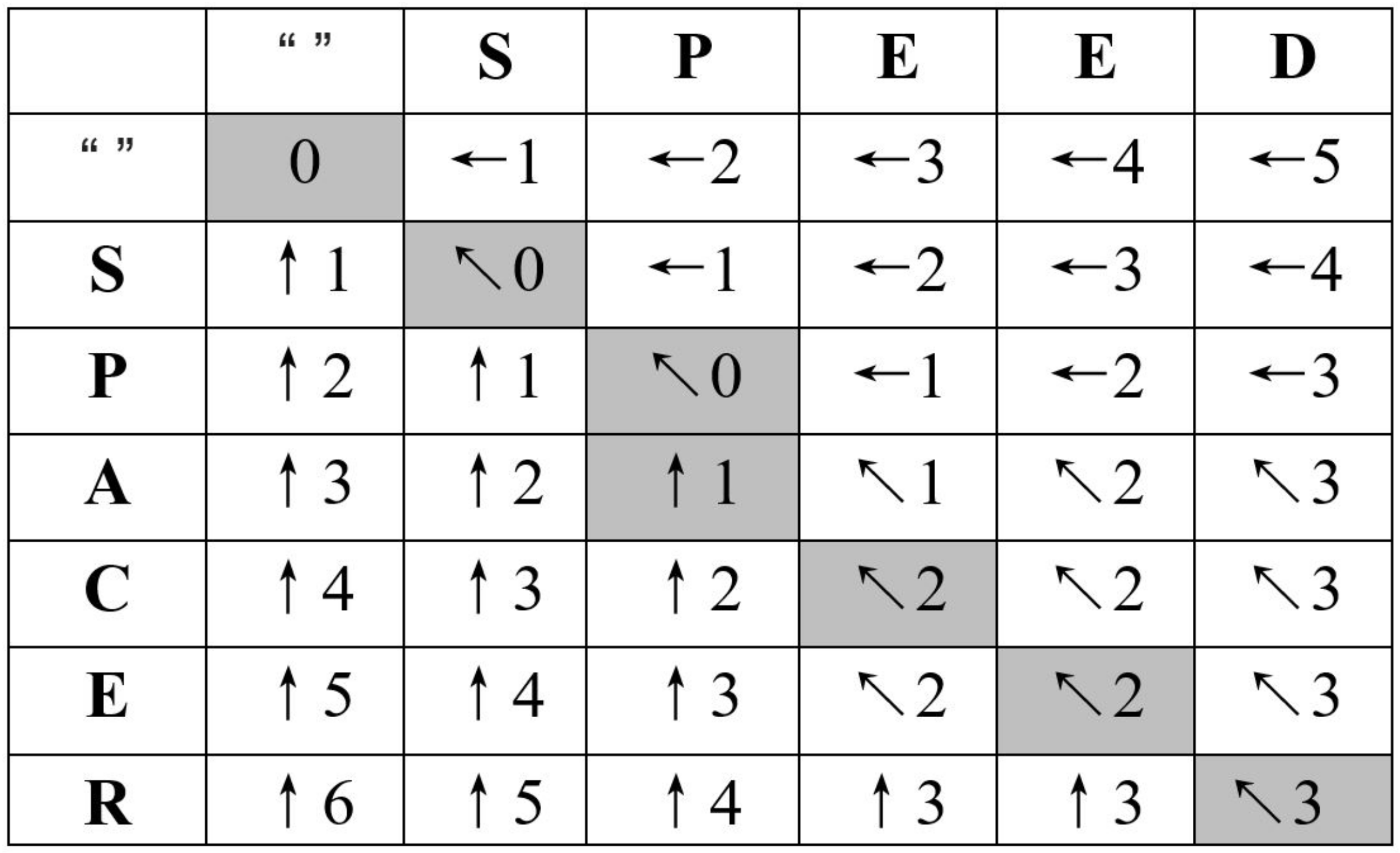
3.2. Levenshtein Distance
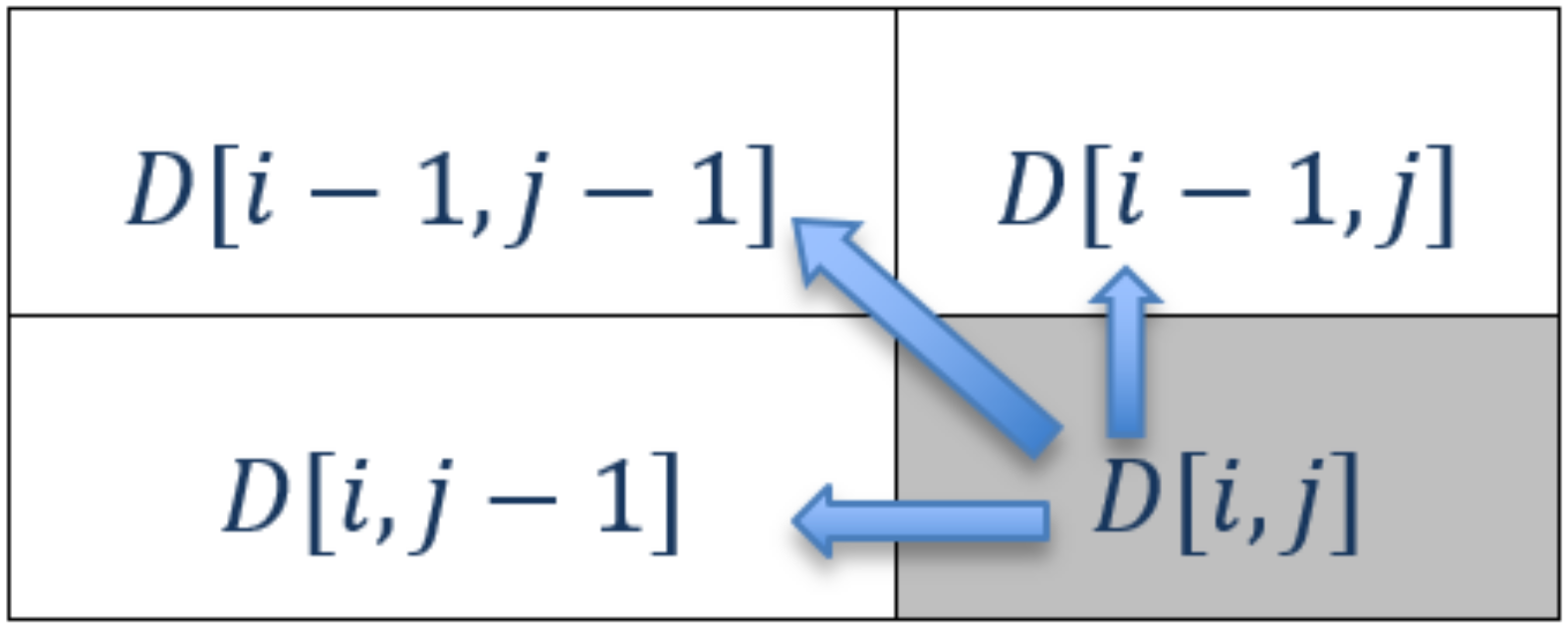
3.3. Computational Dependencies
- If it is a match case : The edit distance is the distance between two substrings that are one character shorter than the current substring, i.e., .
- If it is a non-match case : The edit distance is one greater than the smallest edit distance of any of the three possible substring situations, i.e., .
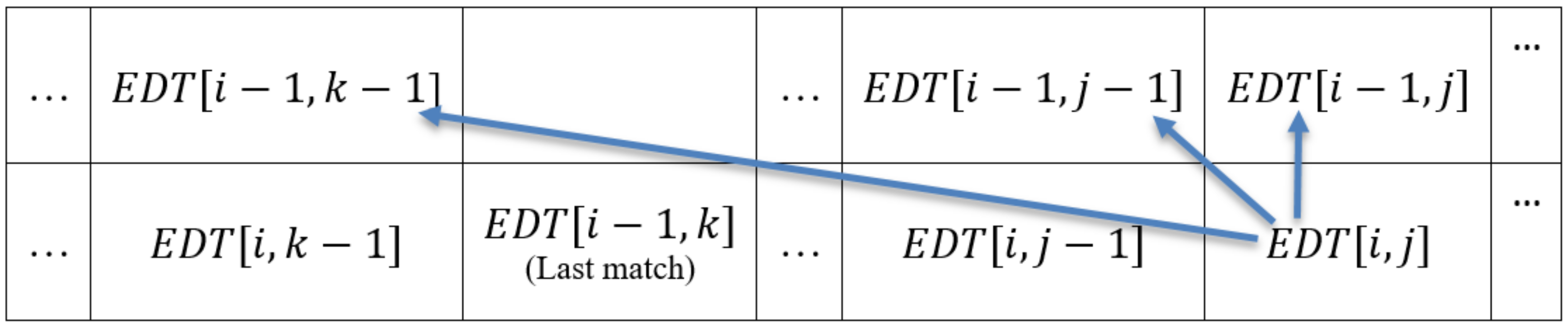
3.4. Redefining Computational Dependencies
3.5. Communication Cost
3.6. Speedup
4. Parallel Approaches for Similarity Search under Edit Distance
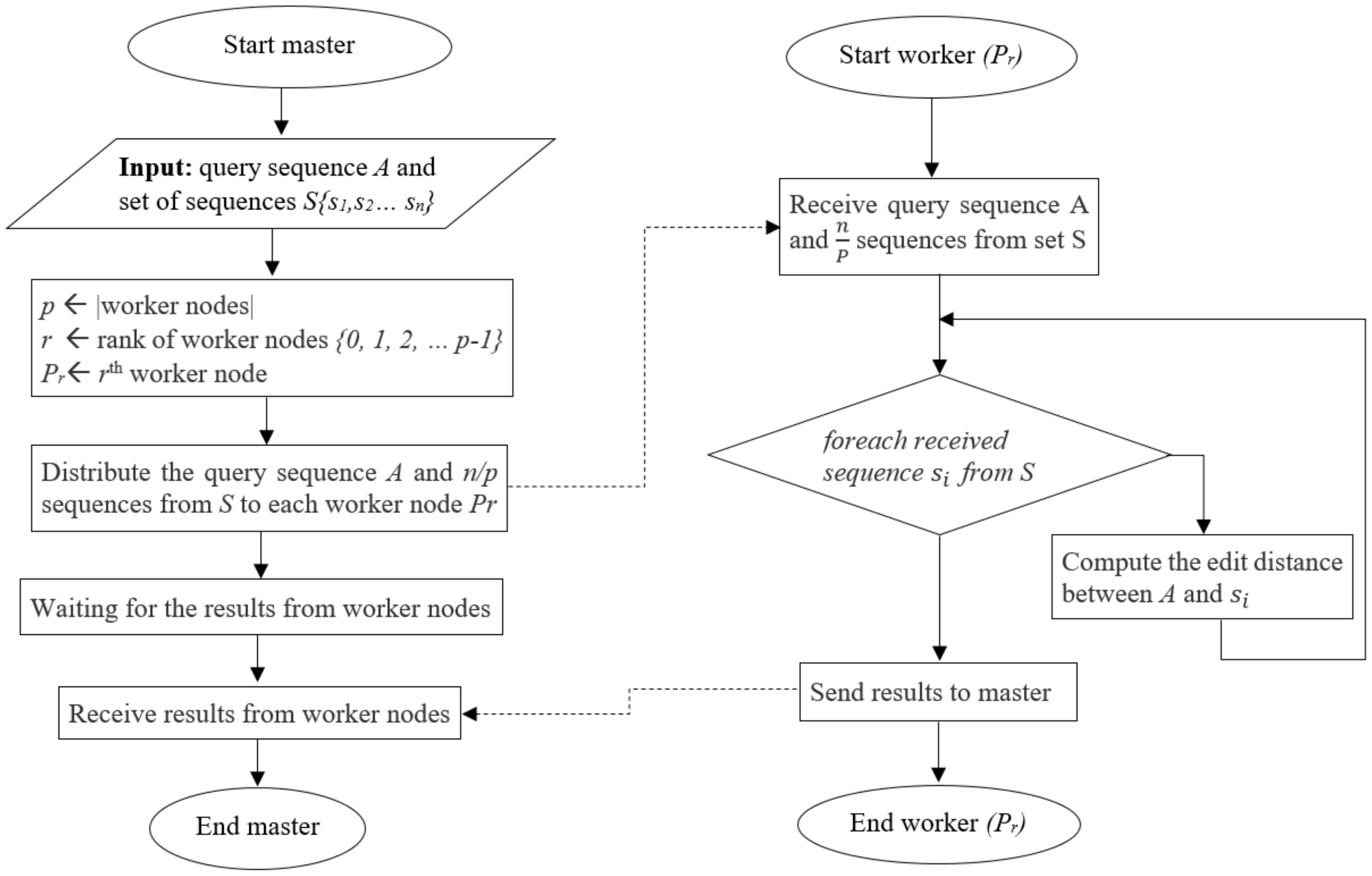
4.1. Inter-Task Parallelism
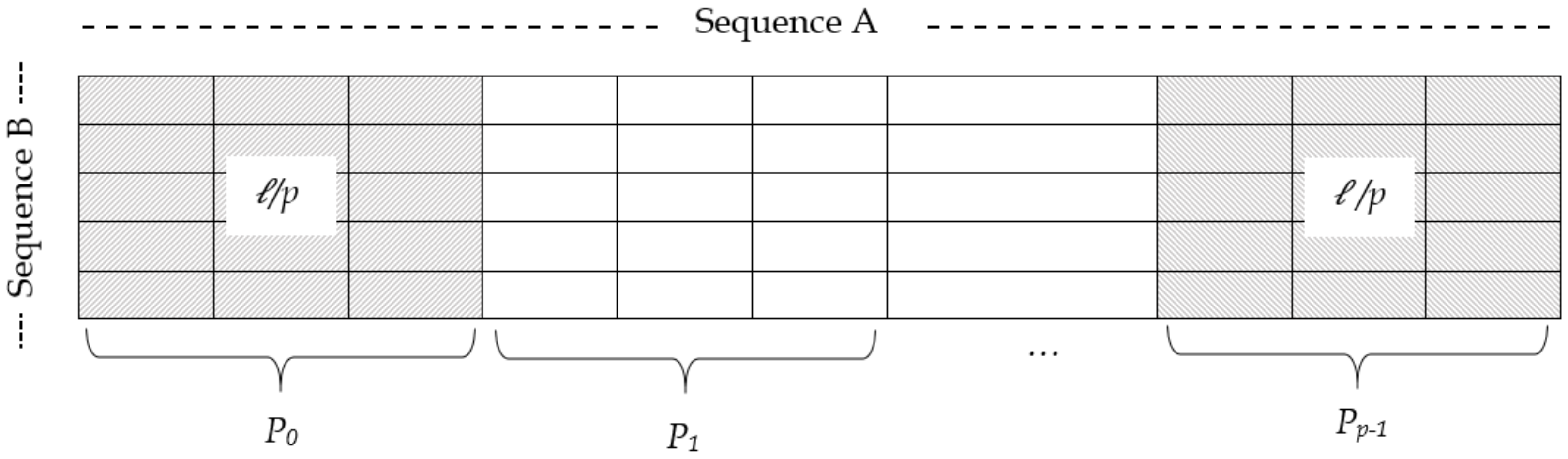
4.2. Intra-Task Parallelism Approach
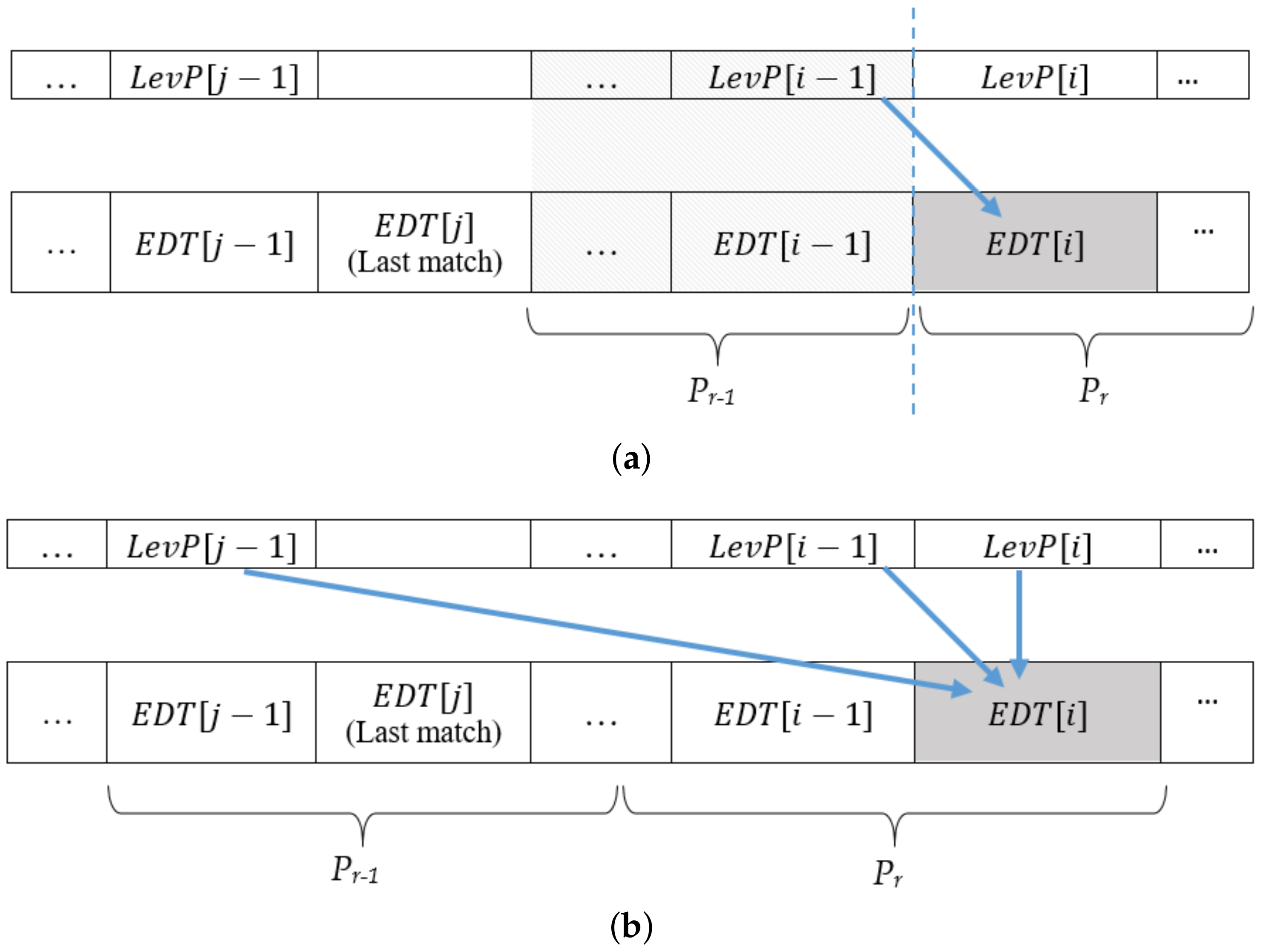
- To compute the first value (leftmost value), i.e., of the edit distance table, each processor needs the diagonal value of the previous row, say , which is not available locally and can be found from the preceding processor .
- There is a possibility that for some initial cells of a processor , the value of the last match case resides in the part of data that is assigned to one of the preceding processors.
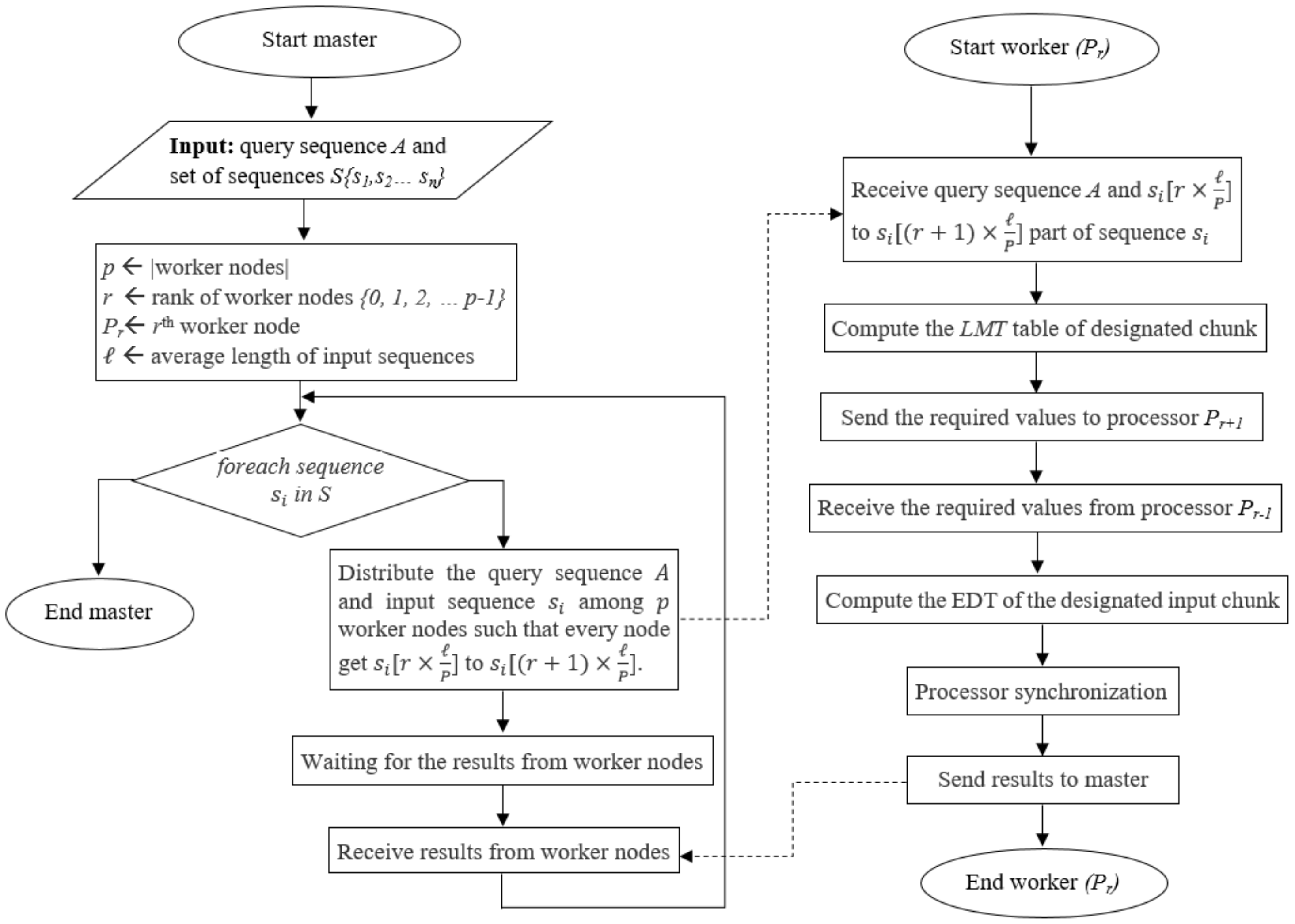
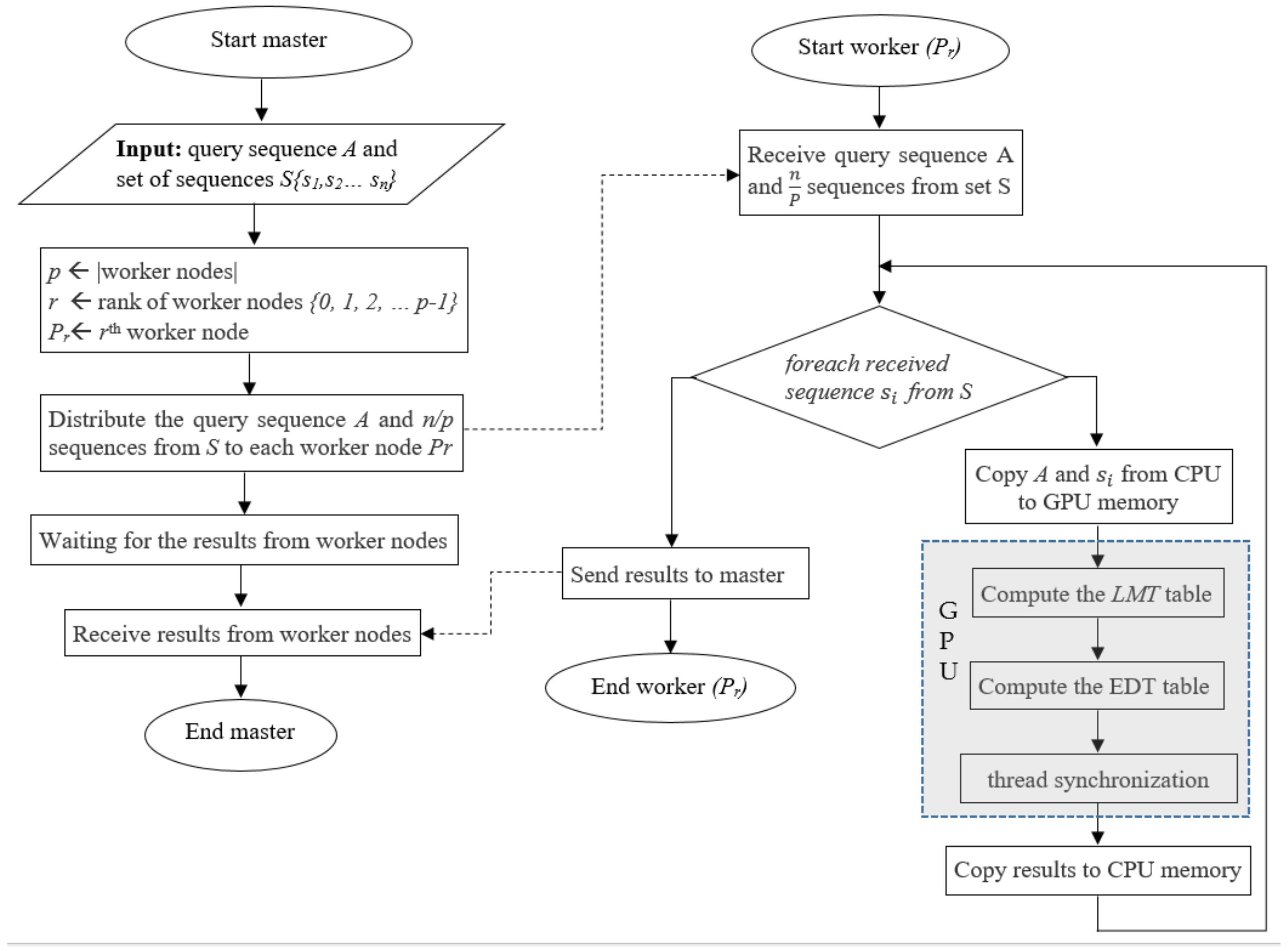
4.3. Hybrid Approach
5. Experiments and Evaluation
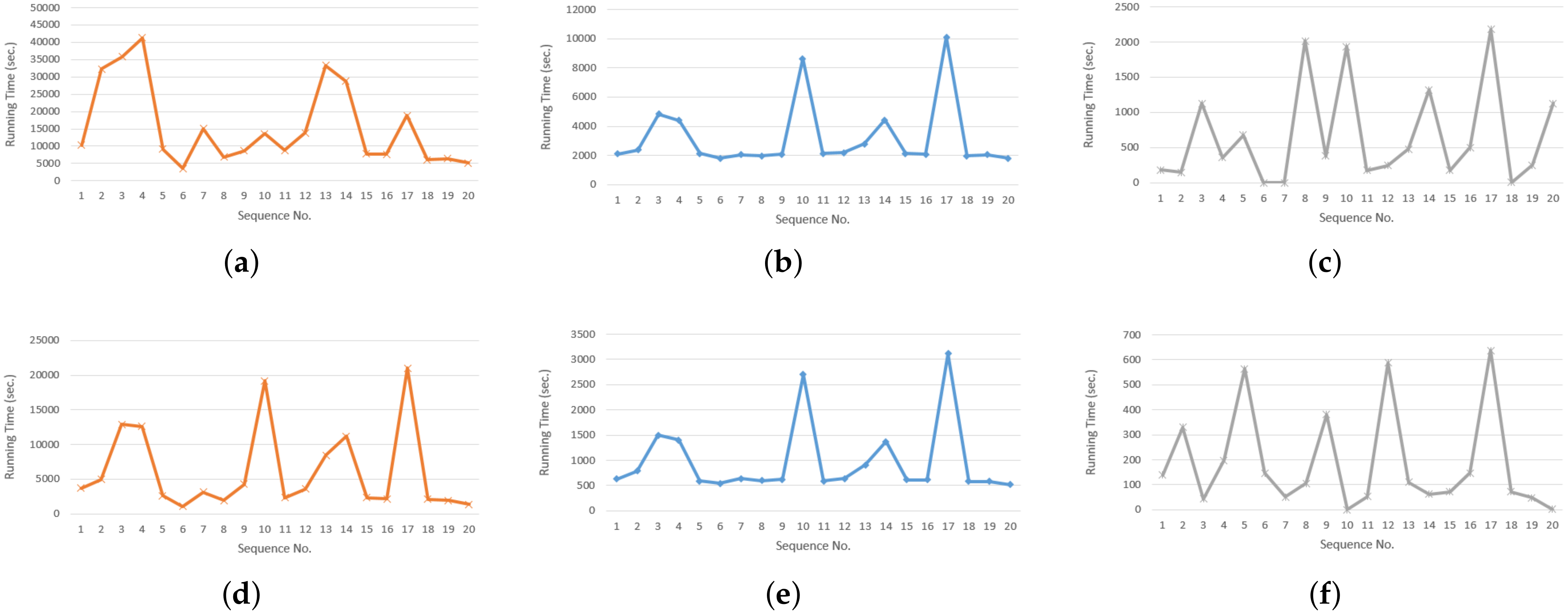
5.1. Evaluating Execution Time
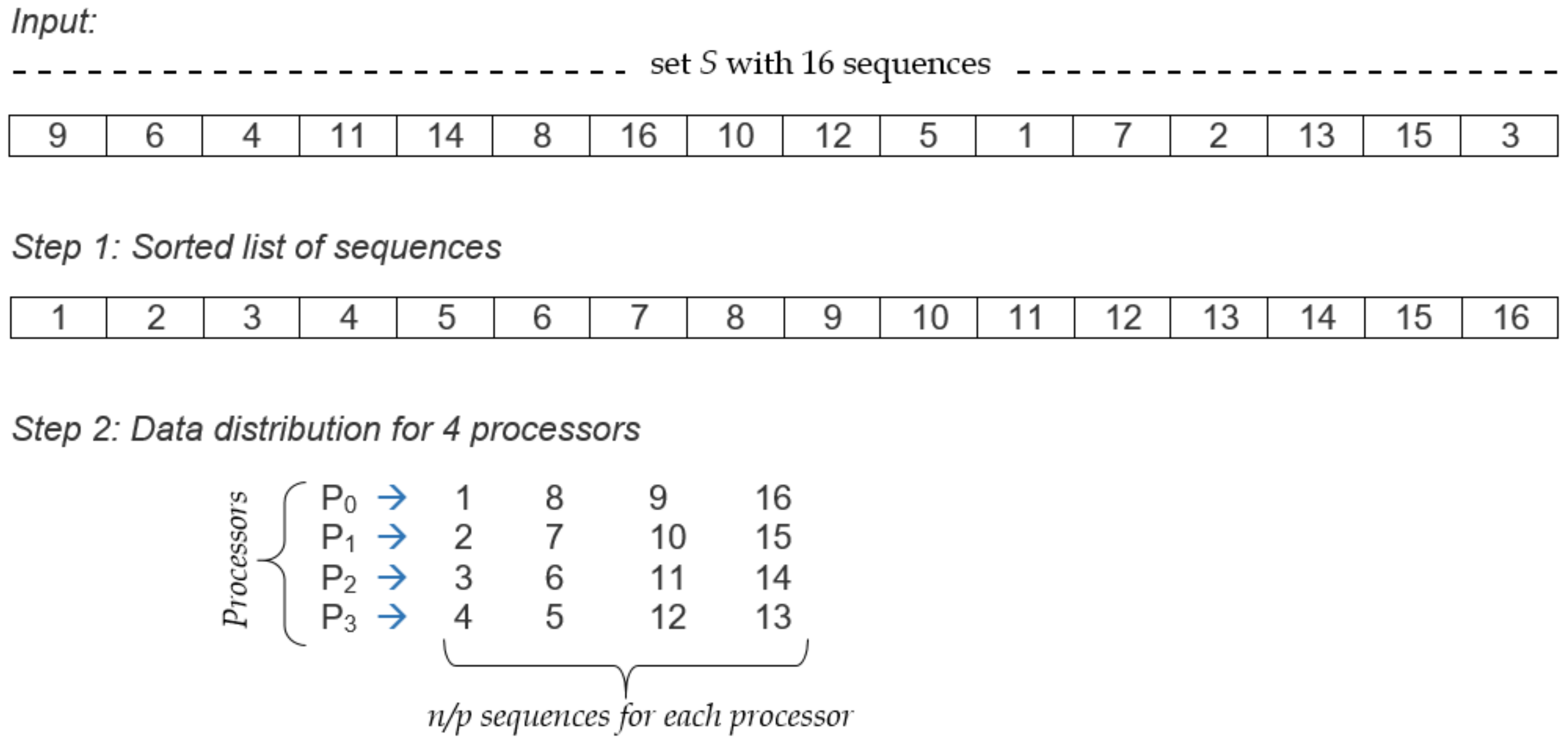
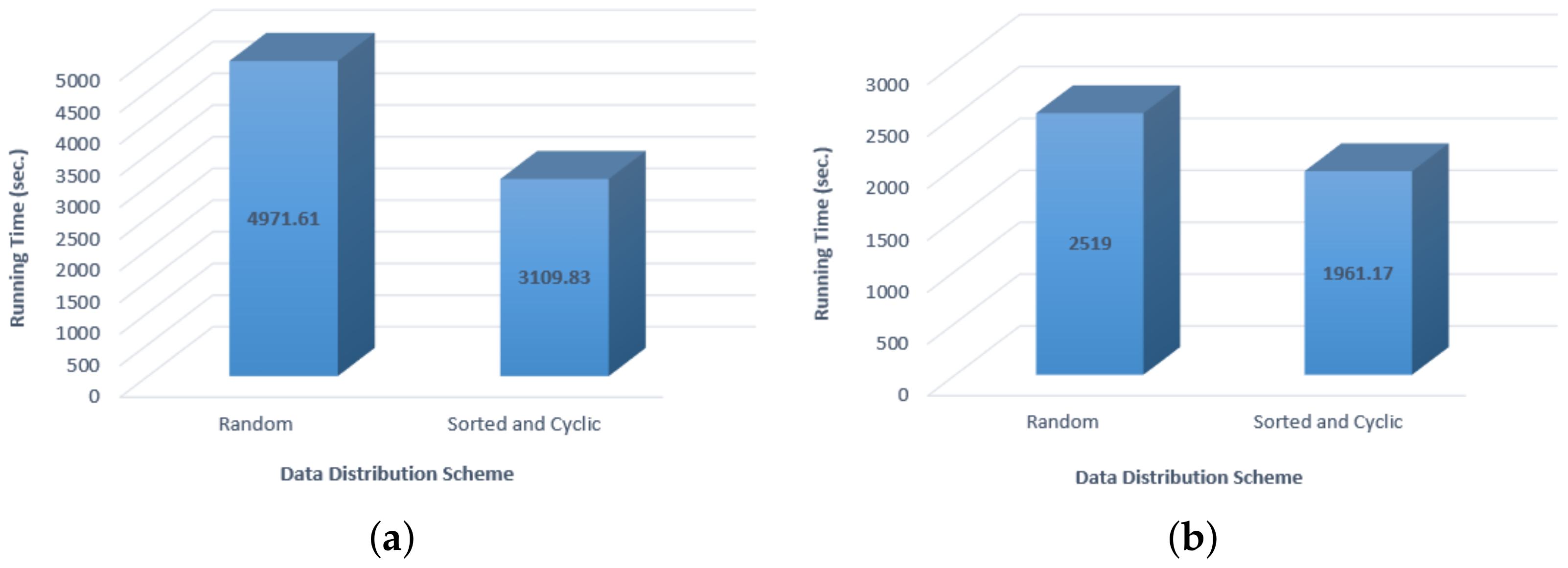
5.2. Evaluating Load Balancing
- Sort all the sequences in ascending order according to their sizes.
- From the sorted list of sequences, distribute n/p sequences to processors in a circular manner except that in every alternate step, the processor ordering will be reversed.
5.3. Evaluating Scalability
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Prasetya, D.D.; Wibawa, A.P.; Hirashima, T. The performance of text similarity algorithms. Int. J. Adv. Intell. Inform. 2018, 4, 63–69. [Google Scholar] [CrossRef]
- Levenshtein, V. Binary codes capable of correcting spurious insertions and deletion of ones. Probl. Inf. Transm. 1965, 1, 8–17. [Google Scholar]
- Wagner, R.A.; Fischer, M.J. The string-to-string correction problem. J. ACM (JACM) 1974, 21, 168–173. [Google Scholar] [CrossRef]
- Damerau, F.J. A technique for computer detection and correction of spelling errors. Commun. ACM 1964, 7, 171–176. [Google Scholar] [CrossRef]
- Jaro, M.A. Advances in record-linkage methodology as applied to matching the 1985 census of Tampa, Florida. J. Am. Stat. Assoc. 1989, 84, 414–420. [Google Scholar] [CrossRef]
- Winkler, W.E. String Comparator Metrics and Enhanced Decision Rules in the Fellegi-Sunter Model of Record Linkage; Bureau of the Censu: Washington, DC, USA, 1990.
- Smith, T.F.; Waterman, M.S. Identification of common molecular subsequences. J. Mol. Biol. 1981, 147, 195–197. [Google Scholar] [CrossRef]
- Needleman, S.B.; Wunsch, C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970, 48, 443–453. [Google Scholar] [CrossRef]
- Hirschberg, D.S. A linear space algorithm for computing maximal common subsequences. Commun. ACM 1975, 18, 341–343. [Google Scholar] [CrossRef]
- Kondrak, G. N-gram similarity and distance. In Proceedings of the International Symposium on String Processing and Information Retrieval, Buenos Aires, Argentina, 2–4 November 2005; Springer: Berlin/Heidelberg, Germany, 2005; pp. 115–126. [Google Scholar] [CrossRef]
- Khalid, M. Bulk Data Processing of Parallel String Similarity Measures. Ph.D. Dissertation, University of the Punjab, Lahore, Punjab, Pakistan, 2021. [Google Scholar]
- Minghe, Y.; Guoliang, L.D.D.; Feng, J. String similarity search and join: A survey. Front. Comput. Sci. 2016, 10, 399–417. [Google Scholar] [CrossRef]
- Nunes, L.S.; Bordim, J.L.; Nakano, K.; Ito, Y. A fast approximate string matching algorithm on GPU. In Proceedings of the 2015 Third International Symposium on Computing and Networking (CANDAR), Sapporo, Japan, 8–11 December 2015; pp. 188–192. [Google Scholar]
- Nunes, L.S.; Bordim, J.L.; Nakano, K.; Ito, Y. A memory-access-efficient implementation of the approximate string matching algorithm on GPU. In Proceedings of the 2016 Fourth International Symposium on Computing and Networking (CANDAR), Hiroshima, Japan, 22–25 November 2016; pp. 483–489. [Google Scholar]
- Chen, X.; Wang, C.; Tang, S.; Yu, C.; Zou, Q. CMSA: A heterogeneous CPU/GPU computing system for multiple similar RNA/DNA sequence alignment. BMC Bioinform. 2017, 18, 315. [Google Scholar] [CrossRef]
- Jiang, Y.; Deng, D.; Wang, J.; Li, G.; Feng, J. Efficient parallel partition-based algorithms for similarity search and join with edit distance constraints. In Proceedings of the Joint EDBT/ICDT 2013 Workshops, Genoa, Italy, 18–22 March 2013; pp. 341–348. [Google Scholar] [CrossRef]
- Zhou, J.; Guo, Q.; Jagadish, H.; Krcal, L.; Liu, S.; Luan, W.; Tung, A.K.; Yang, Y.; Zheng, Y. A generic inverted index framework for similarity search on the gpu. In Proceedings of the 2018 IEEE 34th International Conference on Data Engineering (ICDE), Paris, France, 16–19 April 2018; pp. 893–904. [Google Scholar] [CrossRef]
- Ho, T.; Oh, S.R.; Kim, H. A parallel approximate string matching under Levenshtein distance on graphics processing units using warp-shuffle operations. PLoS ONE 2017, 12, e0186251. [Google Scholar] [CrossRef]
- Groth, T.; Groppe, S.; Koppehel, M.; Pionteck, T. Parallelizing Approximate Search on Adaptive Radix Trees. In Proceedings of the SEBD, Villasimius, Sardinia, Italy, 21–24 June 2020. [Google Scholar]
- Ji, S.; Li, G.; Li, C.; Feng, J. Efficient interactive fuzzy keyword search. In Proceedings of the 18th International Conference on World Wide Web, Madrid, Spain, 20–24 April 2009; pp. 371–380. [Google Scholar]
- Chaudhuri, S.; Kaushik, R. Extending autocompletion to tolerate errors. In Proceedings of the 2009 ACM SIGMOD International Conference on Management of Data, Providence, RI, USA, 29 June–2 July 2009; pp. 707–718. [Google Scholar]
- Li, G.; Ji, S.; Li, C.; Feng, J. Efficient fuzzy full-text type-ahead search. VLDB J. 2011, 20, 617–640. [Google Scholar] [CrossRef]
- Deng, D.; Li, G.; Feng, J.; Li, W.S. Top-k string similarity search with edit-distance constraints. In Proceedings of the 2013 IEEE 29th International Conference on Data Engineering (ICDE), Brisbane, QLD, Australia, 8–12 April 2013; pp. 925–936. [Google Scholar]
- Lu, W.; Du, X.; Hadjieleftheriou, M.; Ooi, B.C. Efficiently Supporting Edit Distance Based String Similarity Search Using B+-Trees. IEEE Trans. Knowl. Data Eng. 2014, 26, 2983–2996. [Google Scholar] [CrossRef]
- Zhang, Z.; Hadjieleftheriou, M.; Ooi, B.C.; Srivastava, D. Bed-tree: An all-purpose index structure for string similarity search based on edit distance. In Proceedings of the 2010 ACM SIGMOD International Conference on Management of Data, Indianapolis, IN, USA, 6–10 June 2010; pp. 915–926. [Google Scholar]
- Farivar, R.; Kharbanda, H.; Venkataraman, S.; Campbell, R.H. An algorithm for fast edit distance computation on GPUs. In Proceedings of the 2012 Innovative Parallel Computing (InPar), San Jose, CA, USA, 13–14 May 2012; pp. 1–9. [Google Scholar] [CrossRef]
- Wang, X.; Ding, X.; Tung, A.K.; Zhang, Z. Efficient and effective knn sequence search with approximate n-grams. Proc. VLDB Endow. 2013, 7, 1–12. [Google Scholar] [CrossRef]
- Chen, W.; Chen, J.; Zou, F.; Li, Y.F.; Lu, P.; Wang, Q.; Zhao, W. Vector and line quantization for billion-scale similarity search on GPUs. Future Gener. Comput. Syst. 2019, 99, 295–307. [Google Scholar] [CrossRef]
- Johnson, J.; Douze, M.; Jégou, H. Billion-scale similarity search with gpus. IEEE Trans. Big Data 2019, 7, 535–547. [Google Scholar] [CrossRef]
- Li, C.; Wang, B.; Yang, X. VGRAM: Improving Performance of Approximate Queries on String Collections Using Variable-Length Grams. In Proceedings of the 33rd International Conference on Very Large Data Bases (VLDB), Vienna, Austria, 23–27 September 2007; Volume 7, pp. 303–314. [Google Scholar]
- Kim, M.S.; Whang, K.Y.; Lee, J.G.; Lee, M.J. n-gram/2L: A space and time efficient two-level n-gram inverted index structure. In Proceedings of the 31st International Conference on Very Large Data Bases (VLDB), Trondheim, Norway, 30 August–2 September 2005; pp. 325–336. [Google Scholar]
- Behm, A.; Ji, S.; Li, C.; Lu, J. Space-constrained gram-based indexing for efficient approximate string search. In Proceedings of the 2009 IEEE 25th International Conference on Data Engineering, Shanghai, China, 29 March–2 April 2009; pp. 604–615. [Google Scholar]
- Qin, J.; Wang, W.; Lu, Y.; Xiao, C.; Lin, X. Efficient exact edit similarity query processing with the asymmetric signature scheme. In Proceedings of the 2011 ACM SIGMOD International Conference on Management of Data, Athens, Greece, 12–16 June 2011; pp. 1033–1044. [Google Scholar]
- Wang, J.; Li, G.; Feng, J. Can we beat the prefix filtering? An adaptive framework for similarity join and search. In Proceedings of the 2012 ACM SIGMOD International Conference on Management of Data, Scottsdale, AZ, USA, 20–24 May 2012; pp. 85–96. [Google Scholar]
- Yang, X.; Wang, B.; Li, C. Cost-based variable-length-gram selection for string collections to support approximate queries efficiently. In Proceedings of the 2008 ACM SIGMOD International Conference on Management of Data, Vancouver, BC, Canada, 9–12 June 2008; pp. 353–364. [Google Scholar]
- Behm, A.; Li, C.; Carey, M.J. Answering approximate string queries on large data sets using external memory. In Proceedings of the 2011 IEEE 27th International Conference on Data Engineering, Hannover, Germany, 11–16 April 2011; pp. 888–899. [Google Scholar]
- Qin, J.; Xiao, C.; Hu, S.; Zhang, J.; Wang, W.; Ishikawa, Y.; Tsuda, K.; Sadakane, K. Efficient query autocompletion with edit distance-based error tolerance. VLDB J. 2020, 29, 919–943. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, Q. Minsearch: An efficient algorithm for similarity search under edit distance. In Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Virtual Event, 6–10 July 2020; pp. 566–576. [Google Scholar] [CrossRef]
- Yang, Z.; Yu, J.; Kitsuregawa, M. Fast algorithms for top-k approximate string matching. In Proceedings of the Twenty-Fourth AAAI Conference on Artificial Intelligence, Atlanta, GA, USA, 11–15 July 2010. [Google Scholar]
- Mishra, S.; Gandhi, T.; Arora, A.; Bhattacharya, A. Efficient edit distance based string similarity search using deletion neighborhoods. In Proceedings of the Joint EDBT/ICDT 2013 Workshops, Genoa, Italy, 18–22 March 2013; pp. 375–383. [Google Scholar] [CrossRef]
- Wang, J.; Li, G.; Deng, D.; Zhang, Y.; Feng, J. Two birds with one stone: An efficient hierarchical framework for top-k and threshold-based string similarity search. In Proceedings of the 2015 IEEE 31st International Conference on Data Engineering, Seoul, Korea, 13–17 April 2015; pp. 519–530. [Google Scholar]
- McCauley, S. Approximate similarity search under edit distance using locality-sensitive hashing. arXiv 2019, arXiv:1907.01600. [Google Scholar]
- Yu, M.; Wang, J.; Li, G.; Zhang, Y.; Deng, D.; Feng, J. A unified framework for string similarity search with edit-distance constraint. VLDB J. 2017, 26, 249–274. [Google Scholar] [CrossRef]
- Pranathi, P.; Karthikeyan, C.; Charishma, D. String similarity search using edit distance and soundex algorithm. Int. J. Eng. Adv. Technol. (IJEAT) 2019, 8, 2249–8958. [Google Scholar]
- Deng, D.; Li, G.; Feng, J. A pivotal prefix based filtering algorithm for string similarity search. In Proceedings of the 2014 ACM SIGMOD International Conference on Management of Data, Snowbird, UT, USA, 22–27 June 2014; pp. 673–684. [Google Scholar]
- Matsumoto, T.; Yiu, M.L. Accelerating exact similarity search on cpu-gpu systems. In Proceedings of the 2015 IEEE International Conference on Data Mining, Atlantic City, NJ, USA, 14–17 November 2015; pp. 320–329. [Google Scholar] [CrossRef]
- Shehab, M.A.; Ghadawi, A.A.; Alawneh, L.; Al-Ayyoub, M.; Jararweh, Y. A hybrid CPU-GPU implementation to accelerate multiple pairwise protein sequence alignment. In Proceedings of the 2017 8th International Conference on Information and Communication Systems (ICICS), Irbid, Jordan, 4–6 April 2017; pp. 12–17. [Google Scholar]
- Edmiston, E.W.; Core, N.G.; Saltz, J.H.; Smith, R.M. Parallel processing of biological sequence comparison algorithms. Int. J. Parallel Program. 1988, 17, 259–275. [Google Scholar] [CrossRef]
- Zhong, C.; Chen, G.L. Parallel algorithms for approximate string matching on PRAM and LARPBS. J. Softw. 2004, 15, 159–169. [Google Scholar]
- Man, D.; Nakano, K.; Ito, Y. The approximate string matching on the hierarchical memory machine, with performance evaluation. In Proceedings of the 2013 IEEE 7th International Symposium on Embedded Multicore SoCs, Tokyo, Japan, 26–28 September 2013; pp. 79–84. [Google Scholar]
- Zhang, J.; Lan, H.; Chan, Y.; Shang, Y.; Schmidt, B.; Liu, W. BGSA: A bit-parallel global sequence alignment toolkit for multi-core and many-core architectures. Bioinformatics 2019, 35, 2306–2308. [Google Scholar] [CrossRef] [PubMed]
- Myers, G. A fast bit-vector algorithm for approximate string matching based on dynamic programming. J. ACM (JACM) 1999, 46, 395–415. [Google Scholar] [CrossRef]
- Hyyrö, H. A bit-vector algorithm for computing Levenshtein and Damerau edit distances. Nord. J. Comput. 2003, 10, 29–39. [Google Scholar]
- Xu, K.; Cui, W.; Hu, Y.; Guo, L. Bit-parallel multiple approximate string matching based on GPU. Procedia Comput. Sci. 2013, 17, 523–529. [Google Scholar] [CrossRef]
- Lin, C.H.; Wang, G.H.; Huang, C.C. Hierarchical parallelism of bit-parallel algorithm for approximate string matching on GPUs. In Proceedings of the 2014 IEEE Symposium on Computer Applications and Communications, Weihai, China, 26–27 July 2014; pp. 76–81. [Google Scholar]
- Sadiq, M.U.; Yousaf, M.M. Distributed Algorithm for Parallel Edit Distance Computation. Comput. Inform. 2020, 39, 757–779. [Google Scholar] [CrossRef]
- Sadiq, M.U.; Yousaf, M.M.; Aslam, L.; Aleem, M.; Sarwar, S.; Jaffry, S.W. NvPD: Novel parallel edit distance algorithm, correctness, and performance evaluation. Clust. Comput. 2020, 23, 879–894. [Google Scholar] [CrossRef]
- Yousaf, M.M.; Sadiq, M.A.; Aslam, L.; Ul Qounain, W.; Sarwar, S. A novel parallel algorithm for edit distance computation. Mehran Univ. Res. J. Eng. Technol. 2018, 37, 223–232. [Google Scholar] [CrossRef]
- The National Center for Biotechnology Information. 1988. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 20 March 2022).
- Zheng, Y.; Zhang, L.; Xie, X.; Ma, W.Y. Mining Interesting Locations and Travel Sequences from GPS Trajectories. In Proceedings of the 18th International Conference on World Wide Web, New York, NY, USA, 20–24 April 2009; pp. 791–800. [Google Scholar] [CrossRef]
- Python Geo-hash Library. 2011. Available online: https://pypi.org/project/python-geohash/ (accessed on 22 March 2022).
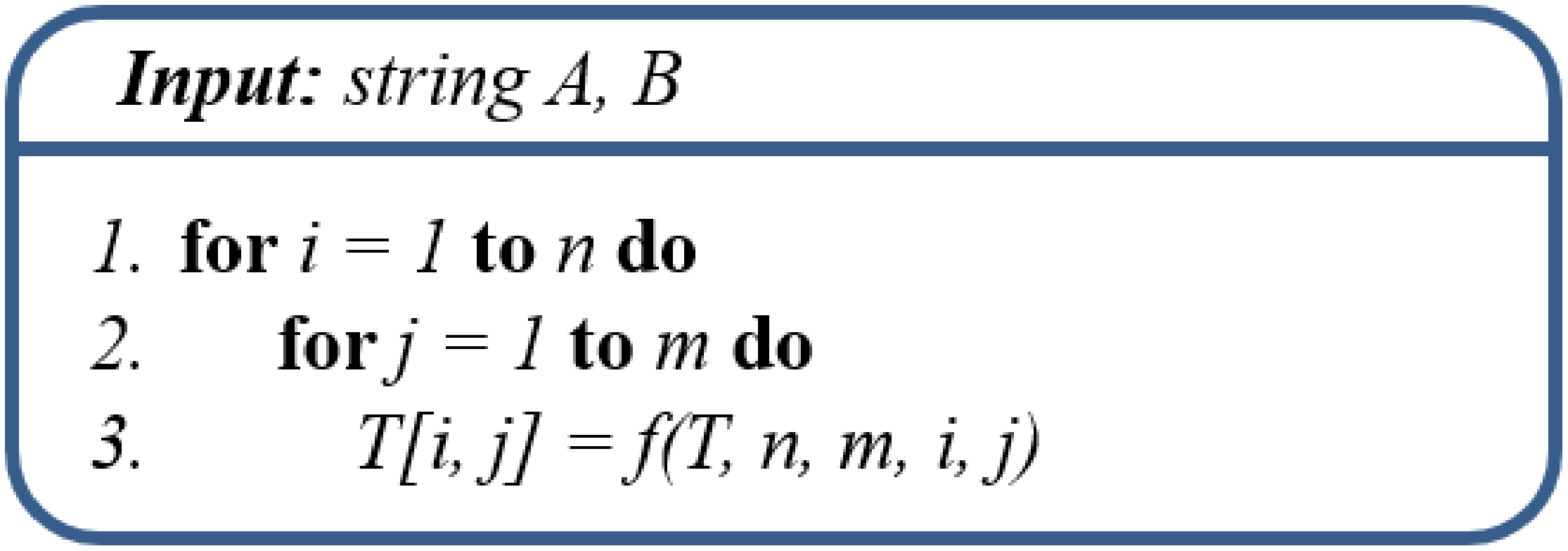



| Dataset | Query Sequence | Communication Time | Computation Time |
|---|---|---|---|
| Genome | gbgss201 | 17,539 | 13,082 |
| gbpln103 | 1771 | 1723 | |
| GeoLife | 001 | 34,689 | 29,341 |
| 006 | 10,729 | 8809 |
| Machine Name | Execution Time (s) | |
|---|---|---|
| Inter-Task Parallelism | Hybrid Parallelism | |
| WS111 | 46,455.8 | 1704.76 |
| WS112 | 35,823.9 | 2519 |
| WS113 | 39,810.7 | 619.281 |
| WS114 | 42,213.1 | 2081.14 |
| WS115 | 43,971.6 | 2329.82 |
| Total Execution time | 46,456 | 2519 |
| Standard Deviation | 3637 | 673 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khalid, M.; Yousaf, M.M.; Sadiq, M.U. Toward Efficient Similarity Search under Edit Distance on Hybrid Architectures. Information 2022, 13, 452. https://doi.org/10.3390/info13100452
Khalid M, Yousaf MM, Sadiq MU. Toward Efficient Similarity Search under Edit Distance on Hybrid Architectures. Information. 2022; 13(10):452. https://doi.org/10.3390/info13100452
Chicago/Turabian StyleKhalid, Madiha, Muhammad Murtaza Yousaf, and Muhammad Umair Sadiq. 2022. "Toward Efficient Similarity Search under Edit Distance on Hybrid Architectures" Information 13, no. 10: 452. https://doi.org/10.3390/info13100452
APA StyleKhalid, M., Yousaf, M. M., & Sadiq, M. U. (2022). Toward Efficient Similarity Search under Edit Distance on Hybrid Architectures. Information, 13(10), 452. https://doi.org/10.3390/info13100452






