A Deep-Learning-Based Framework for Automated Diagnosis of COVID-19 Using X-ray Images
Abstract
1. Introduction
2. Literature Review
3. Data Set Description
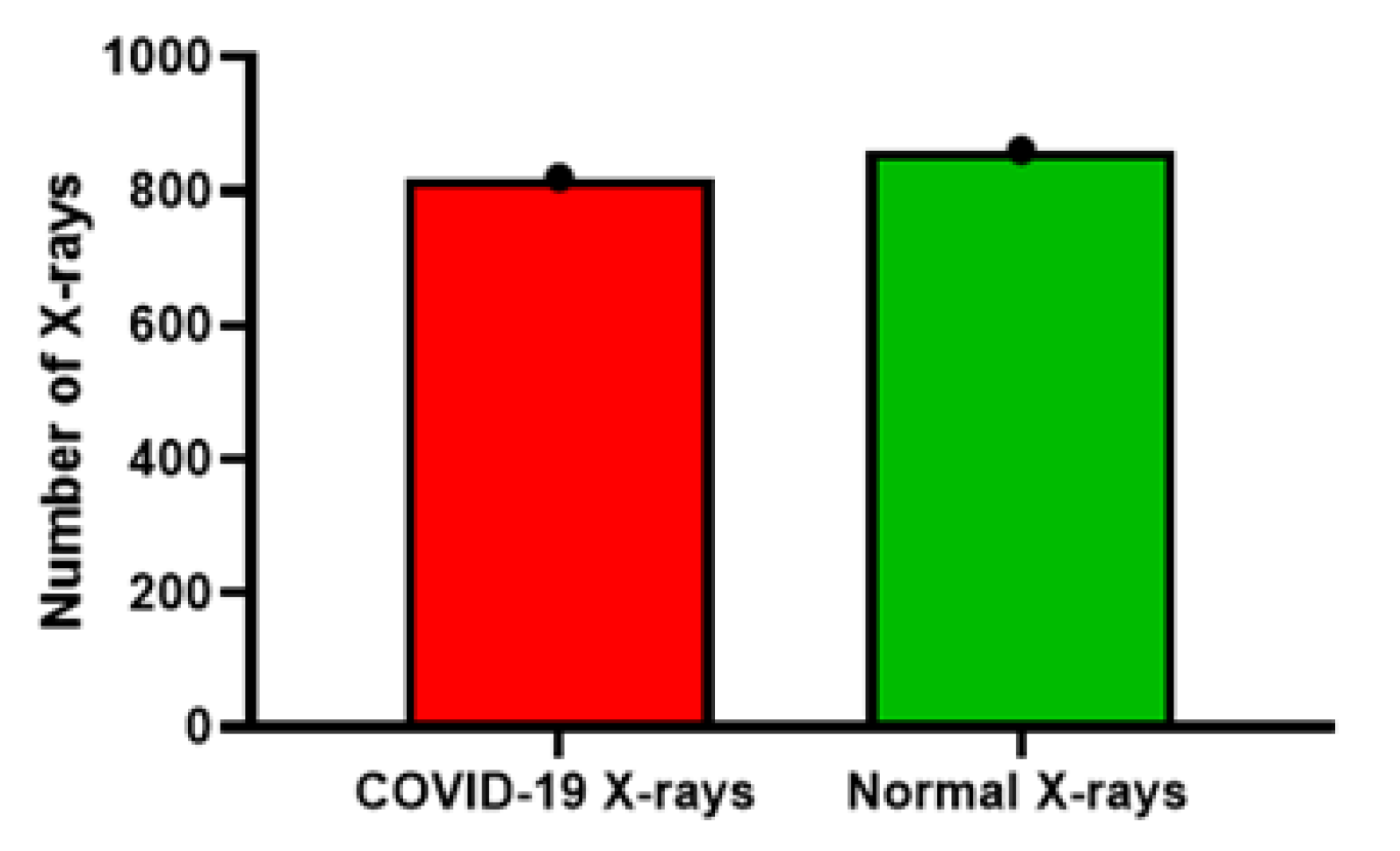
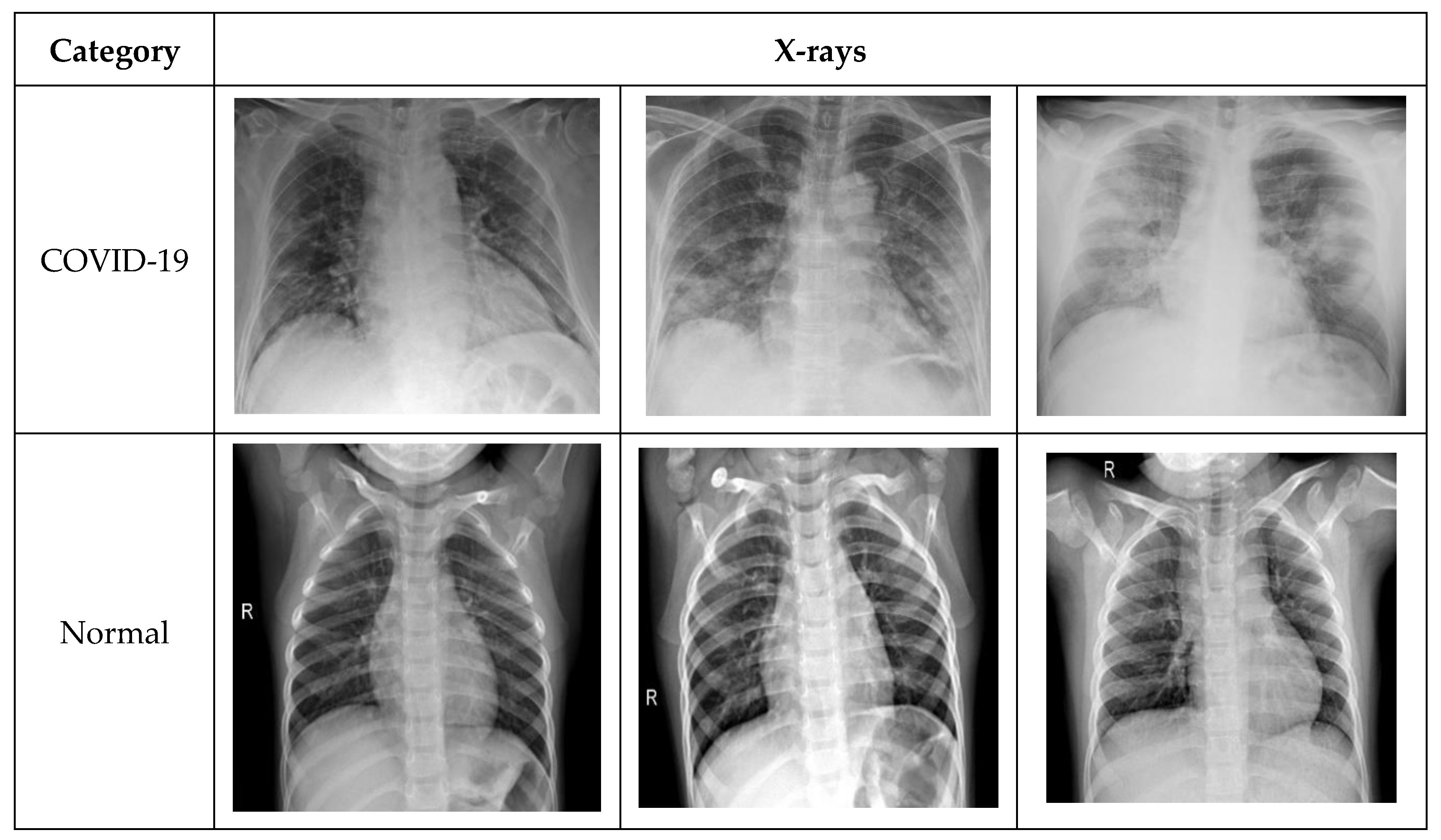
- COVID-19 X-ray image database collected by Cohen et al. [30] consists of a total of 660 images; some of the images in the data set were CT-scan, and some were nonfrontal chest X-rays. CT-scan and nonfrontal X-rays of non-COVID-19 patients X-rays were removed. Moreover, the images tagged with pneumonia were also removed from the data set. The selected frontal chest X-ray of positive COVID-19 patients from Cohen’s data set was 390 X-rays.
- Furthermore, 25 X-ray images of COVID-19 patients were selected from the COVID-19 chest X-ray data initiative [33] data set. The original data set consisted of 55 X-rays. Some of the images in the data set were not clear and were not considered in our experiments.
- Additionally, 180 X-ray images of COVID-19 were also selected from the Actualmed COVID-19 chest X-ray data initiative [35]. Originally, the data set consisted of 237 scans.
- Finally, the X-ray images of both the normal and COVID-19 categories were selected from the COVID-19 radiography database [36]. The data set contained 1057 X-ray images (219 COVID-19, 1341 normal, and 1345 viral pneumonia). In our study, we selected 195 X-rays for COVID-19 and 862 images for the normal category.
4. Methodology
4.1. Data PreProcessing and Augmentation
4.2. Deep Neural Networks and Transfer-Learning
- DenseNet121: The dense convolutional neural network (DenseNet) is a feed forward fully connected neural network. Each layer in DenseNet consists of a feature map. The feature map of each layer serves as an input to the next layer. Among the advantages of the DenseNet is that it requires less parameters. The number of filters or feature maps used in DenseNet is 12. Traditional convolutional neural networks consisting of L layers contain L number of connections, while in DenseNet, the number of direct connections is [60]. The dense connectivity of the model circumvents the need for redundant learning. In addition to this, DenseNet decreases the chance of model overfitting due to the small size training data set by applying regularization.
- ResNet50: ResNet, also known as the deep residual network, was initially proposed in 2015 with the motivation of a “identity shortcut connection”. It is also among the pretrained models using ImageNet. ResNet skips one or more layers and handles the gradient vanishing issue. Among the key advantages of ResNet is easier optimization. Moreover, the accuracy of the model can be enhanced with the increase in the depth of the model [60]. ResNet model skips one, two, or more layers and is directly connected to any layer, not necessarily the adjacent layer, using a ReLu nonlinear activation function. ResNet uses the forward and backward propagation method.
- VGG: VGG, also known as a very deep convolutional network, was first introduced in 2014. VGG is an advanced version of AlexNet with an increased number of layers. The increase in the number of layers increases the generalization of the model [61]. The benefit of VGG is the use of only 3 × 3 convolutional filters. The only difference between VGG16 and VGG19 is the number of layers. However, the convolutional neural network is used for analyzing the object of the image. We used both models for COVID-19 X-ray.
4.3. Model Evaluation
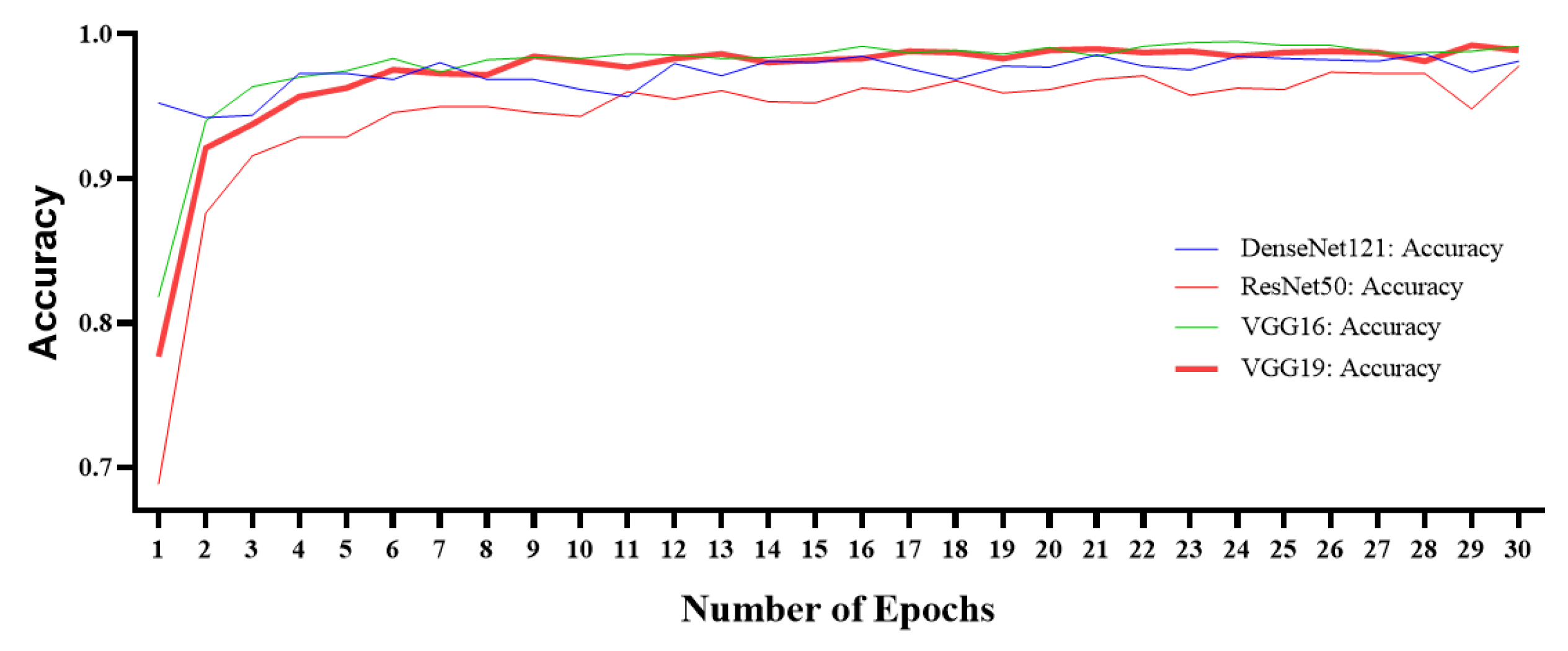
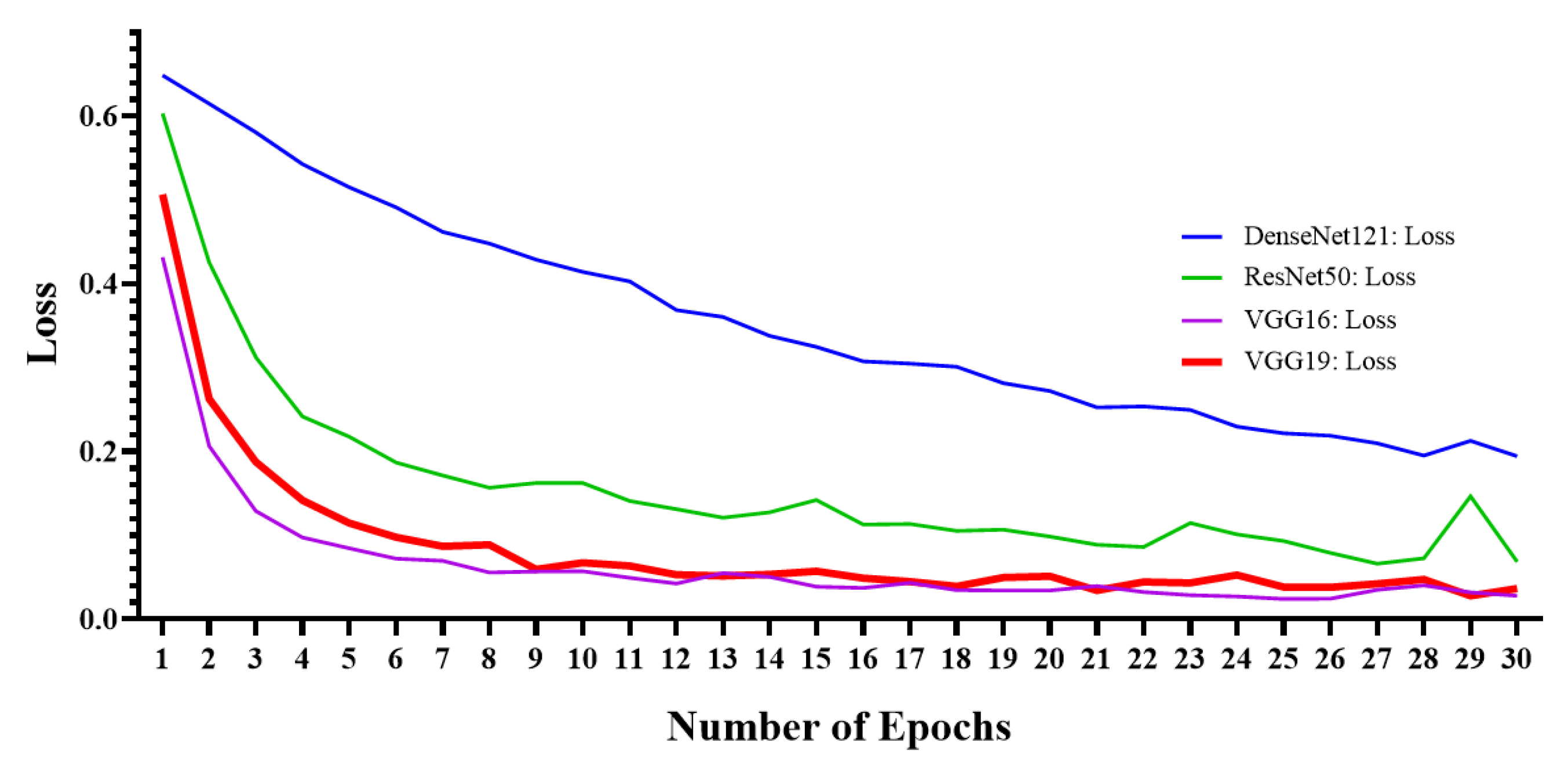
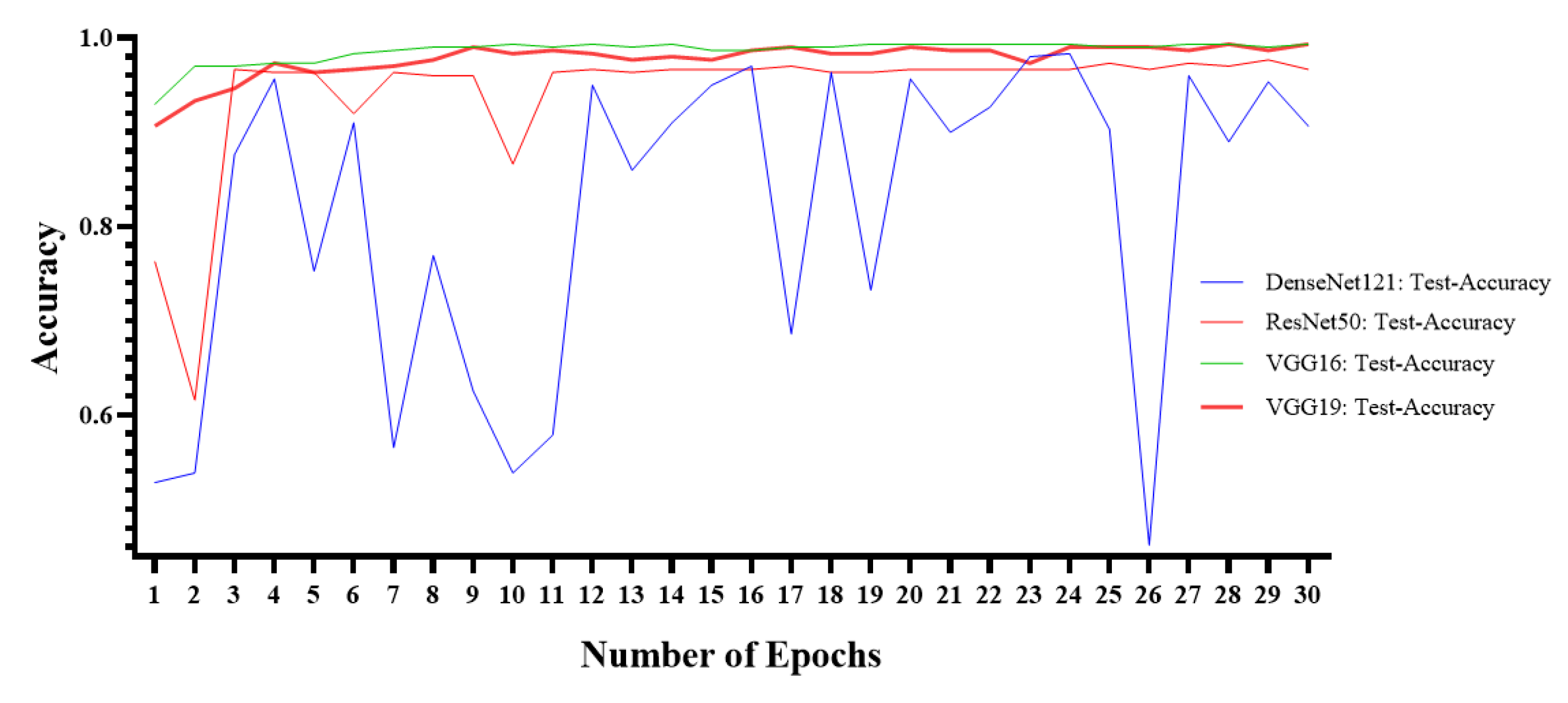
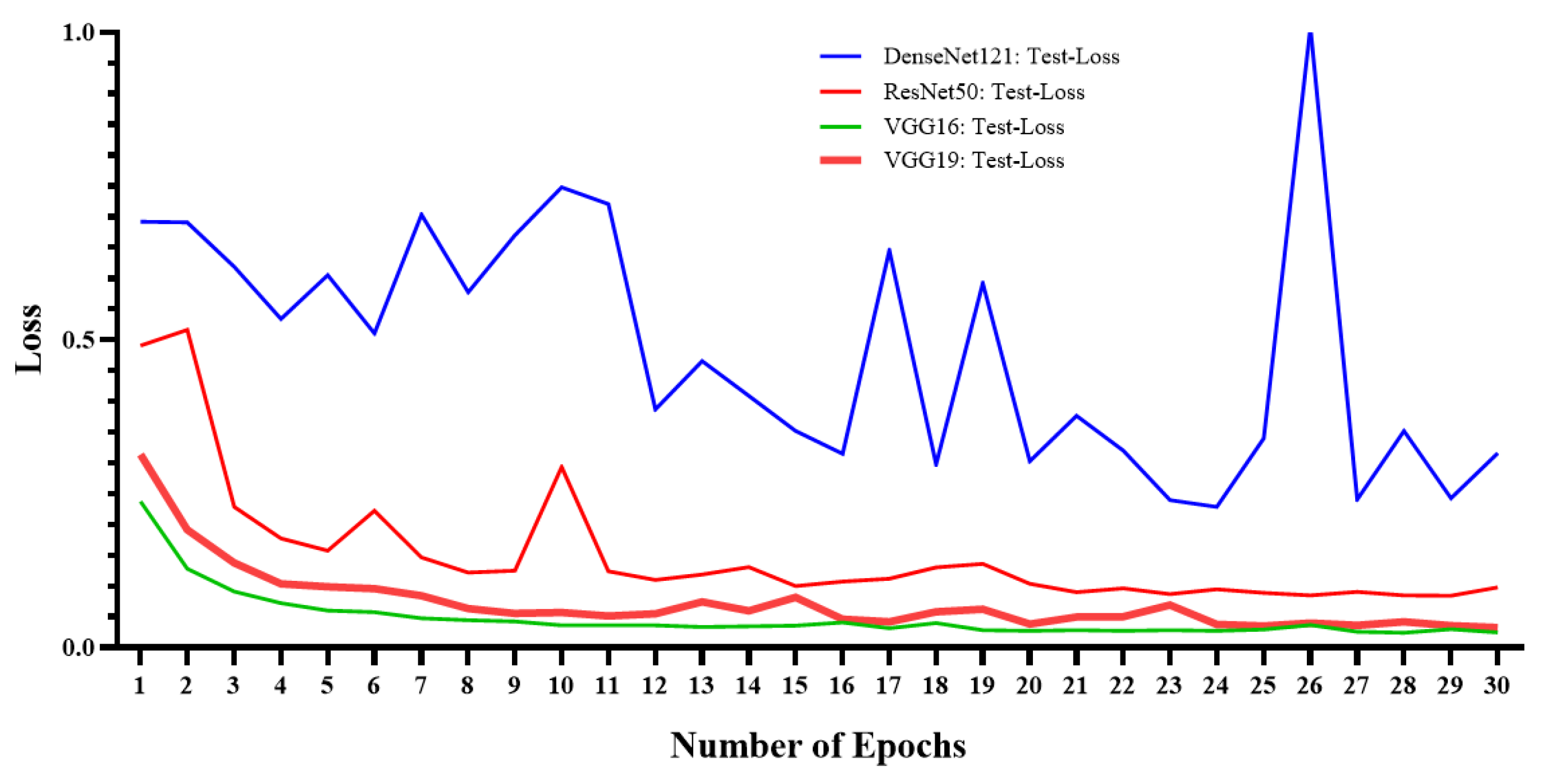
5. Experimental Results
6. Comparison with Existing Studies
- The study does not suffer from data imbalance.
- The model was trained using a large number of COVID-19 X-ray radiology images when compared to the previous studies.
- The proposed model is a fully automated diagnosis method and does not require any separate feature extraction or annotation prior to the diagnosis.
- Data augmentation was applied to increase the generalization of the proposed model.
- The model outperforms the the benchmark studies.
- The proposed system needs to be trained for other respiratory diseases. The current model only diagnoses COVID-19 and healthy individuals and is unable to diagnose other kinds of pneumonia and respiratory infections.
- The number of COVID-19 X-ray radiology images needs to be increased for better model training. The deep-learning model performance can be further enhanced with the increase in the size of the data set.
- The current study was based on the data set curated using several open-source chest X-ray images. These samples were collected from various research publications or uploaded by volunteers. Therefore, these X-ray images were not collected in rigorous manner.
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- COVID-19 Worldwide Statistics. Available online: https://www.worldometers.info/coronavirus/? (accessed on 15 June 2020).
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- West, C.P.; Montori, V.M.; Sampathkumar, P. COVID-19 Testing: The Threat of False-Negative Results. Mayo Clin. Proc. 2020, 95, 1127–1129. [Google Scholar] [CrossRef]
- Guyatt, G.; Rennie, D.; Maureen, O.M.; Cook, D.J. Users’ Guides to the Medical Literature: A Manual for Evidence-Based Clinical Practice, 3rd ed.; McGraw-Hill Medical: New York, NY, USA, 2015. [Google Scholar]
- Yoon, S.H.; Lee, K.H.; Kim, J.Y.; Lee, Y.K.; Ko, H.; Kim, K.H.; Park, C.M.; Kim, Y.-H. Chest Radiographic and CT Findings of the 2019 Novel Coronavirus Disease (COVID-19): Analysis of Nine Patients Treated in Korea. Korean J. Radiol. 2020, 21, 494–500. [Google Scholar] [CrossRef]
- Xie, X. Chest CT for Typical Covid-19 pneumonia. Radiology 2020, 21, 494–500. [Google Scholar]
- Fang, Y.; Zhang, H.; Xie, J.; Lin, M.; Ying, L.; Pang, P.; Ji, W. Sensitivity of Chest CT for COVID-19: Comparison to RT-PCR. Radiology 2020, 296, E115–E117. [Google Scholar] [CrossRef]
- Chan, J.F.-W.; Yuan, S.; Kok, K.-H.; To, K.K.-W.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C.C.-Y.; Poon, R.W.-S.; et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef]
- Ai, T.; Yang, Z.; Hou, H.; Zhan, C.; Chen, C.; Lv, W.; Tao, Q.; Sun, Z.; Xia, L. Correlation of Chest CT and RT-PCR Testing in Coronavirus Disease 2019 (COVID-19) in China: A Report of 1014 Cases. Radiology 2020, 296, E32–E40. [Google Scholar]
- Guan, W.-J.; Ni, Z.-Y.; Hu, Y.; Liang, W.-H.; Ou, C.-Q.; He, J.-X.; Liu, L.; Shan, H.; Lei, C.-L.; Hui, D.S.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Latif, S.; Usman, M.; Manzoor, S.; Iqbal, W.; Qadir, J.; Tyson, G.; Castro, I.; Razi, A.; Boulos, M.N.K.; Weller, A.; et al. Leveraging Data Science to Combat COVID-19: A Comprehensive Review. TechRxiv 2020, 1–19. [Google Scholar] [CrossRef]
- Wynants, L.; Van Calster, B.; Collins, G.S.; Riley, R.D.; Heinze, G.; Schuit, E.; Bonten, M.M.J.; Dahly, D.L.; Damen, J.A.A.; Debray, T.P.; et al. Prediction models for diagnosis and prognosis of covid-19: Systematic review and critical appraisal. BMJ 2020, 369, m1328. [Google Scholar] [CrossRef]
- Swapnarekha, H.; Behera, H.S.; Nayak, J.; Naik, B.; Hanumanthu, S.R. Role of intelligent computing in COVID-19 prognosis: A state-of-the-art review. Chaos Solitons Fractals 2020, 138, 109947. [Google Scholar] [CrossRef] [PubMed]
- Hesamian, M.H.; Jia, W.; He, X.; Kennedy, P. Deep Learning Techniques for Medical Image Segmentation: Achievements and Challenges. J. Digit. Imaging 2019, 32, 582–596. [Google Scholar] [CrossRef] [PubMed]
- Balyen, L.; Peto, T. Promising Artificial Intelligence-Machine Learning-Deep Learning Algorithms in Ophthalmology. Asia Pacific J. Ophthalmol. 2019, 8, 264–272. [Google Scholar] [CrossRef]
- Connor, C.W. Artificial Intelligence and Machine Learning in Anesthesiology. Anesthesiology 2019, 131, 1346–1359. [Google Scholar] [CrossRef]
- Galbusera, F.; Casaroli, G.; Bassani, T. Artificial intelligence and machine learning in spine research. JOR Spine 2019, 2, e1044. [Google Scholar] [CrossRef]
- Gjoreski, M.; Gradišek, A.; Budna, B.; Gams, M.Z.; Poglajen, G. Machine Learning and End-to-End Deep Learning for the Detection of Chronic Heart Failure from Heart Sounds. IEEE Access 2020, 8, 20313–20324. [Google Scholar] [CrossRef]
- Kumar, A.; Fulham, M.; Feng, D.; Kim, J. Co-Learning Feature Fusion Maps From PET-CT Images of Lung Cancer. IEEE Trans. Med. Imaging 2020, 39, 204–217. [Google Scholar] [CrossRef]
- Podnar, S.; Kukar, M.; Gunčar, G.; Notar, M.; Gošnjak, N.; Notar, M. Diagnosing brain tumours by routine blood tests using machine learning. Sci. Rep. 2019, 9, 14481–14487. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Celik, Y.; Talo, M.; Yildirim, O.; Karabatak, M.; Acharya, U.R. Automated invasive ductal carcinoma detection based using deep transfer learning with whole-slide images. Pattern Recognit. Lett. 2020, 133, 232–239. [Google Scholar] [CrossRef]
- Deng, L. Deep Learning: Methods and Applications. Deep Learn. Methods Appl. 2014, 7, 197–387. [Google Scholar] [CrossRef]
- Gaál, G.; Maga, B.; Lukács, A. Attention U-Net Based Adversarial Architectures for Chest X-ray Lung Segmentation. arXiv 2020, arXiv:2003.10304. [Google Scholar]
- Souza, J.C.; Diniz, J.O.B.; Ferreira, J.L.; Da Silva, G.L.F.; Silva, A.C.; De Paiva, A.C. An automatic method for lung segmentation and reconstruction in chest X-ray using deep neural networks. Comput. Methods Programs Biomed. 2019, 177, 285–296. [Google Scholar] [CrossRef]
- Rajpurkar, P.; Irvin, J.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.; Shpanskaya, K.; et al. CheXNet: Radiologist-Level Pneumonia Detection on Chest X-rays with Deep Learning. arXiv 2017, arXiv:1711.05225. [Google Scholar]
- Lakhani, P.; Sundaram, B. Deep Learning at Chest Radiography: Automated Classification of Pulmonary Tuberculosis by Using Convolutional Neural Networks. Radiology 2017, 284, 574–582. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Liu, F.; Li, Z. A Review of Automatically Diagnosing COVID-19 based on Scanning Image. arXiv 2020, arXiv:2006.05245. [Google Scholar]
- Hemdan, E.E.D.; Shouman, M.A.; Karar, M.E. COVIDX-Net: A Framework of Deep Learning Classifiers to Diagnose COVID-19 in X-ray Images. arXiv 2020, arXiv:2003.11055. [Google Scholar]
- Cohen, J.P.; Morrison, P.; Dao, L. COVID-19 Image Data Collection. arXiv 2020, arXiv:2003.11597v1. [Google Scholar]
- Detecting COVID-19 in X-ray Images with Keras, TensorFlow, and Deep Learning. Available online: https://www.pyimagesearch.com/2020/03/16/detecting-covid-19-in-x-ray-images-with-keras-tensorflow-and-deep-learning/ (accessed on 16 June 2020).
- Wang, L.; Wong, A. COVID-Net: A Tailored Deep Convolutional Neural Network Design for Detection of COVID-19 Cases from Chest X-ray Images. arXiv 2020, arXiv:2003.09871. [Google Scholar]
- COVID-19 Chest X-ray Dataset Initiative. Available online: https://github.com/agchung/Figure1-COVID-chestxray-dataset (accessed on 16 June 2020).
- RSNA Pneumonia Detection Challenge. Available online: https://www.kaggle.com/c/rsna-pneumonia-detection-challenge/data (accessed on 16 June 2020).
- Actualmed-COVID-chestxray-dataset. Available online: https://github.com/agchung/Actualmed-COVID-chestxray-dataset (accessed on 16 June 2020).
- COVID-19 Radiography Database. Available online: https://www.kaggle.com/tawsifurrahman/covid19-radiography-database (accessed on 16 June 2020).
- Apostolopoulos, I.D.; Mpesiana, T.A. Covid-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020, 43, 635–640. [Google Scholar] [CrossRef]
- Radiopaedia. Available online: https://radiopaedia.org/ (accessed on 16 June 2020).
- Italian Society of Medical and Interventional Radiology (SIRM). Available online: https://www.sirm.org/en/italian-society-of-medical-and-interventional-radiology/ (accessed on 16 June 2020).
- Kumar, P.; Kumari, S. Detection of coronavirus Disease (COVID-19) based on Deep Features. Preprints 2020. [Google Scholar] [CrossRef]
- Chest X-ray Images (Pneumonia). Available online: https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia (accessed on 16 June 2020).
- Narin, A.; Kaya, C.; Pamuk, Z. Automatic Detection of Coronavirus Disease (COVID-19) Using X-ray Images and Deep Convolutional Neural Networks. arXiv 2020, arXiv:2003.10849. [Google Scholar]
- Ozturk, T.; Talo, M.; Yildirim, E.A.; Baloglu, U.B.; Yildirim, O.; Acharya, U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Boil. Med. 2020, 121, 103792. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. ChestX-ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 3462–3471. [Google Scholar]
- Minaee, S.; Kafieh, R.; Sonka, M.; Yazdani, S.; Soufi, G.J. Deep-COVID: Predicting COVID-19 from chest X-ray images using deep transfer learning. Med. Image Anal. 2020, 65, 101794. [Google Scholar] [CrossRef] [PubMed]
- Irvin, J.; Rajpurkar, P.; Ko, M.; Yu, Y.; Ciurea-Ilcus, S.; Chute, C.; Marklund, H.; Haghgoo, B.; Ball, R.; Shpanskaya, K.; et al. CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison. Proc. AAAI Conf. Artif. Intell. 2019, 33, 590–597. [Google Scholar] [CrossRef]
- Afshar, P.; Heidarian, S.; Naderkhani, F.; Oikonomou, A.; Plataniotis, K.N.; Mohammadi, A. COVID-CAPS: A Capsule Network-based Framework for Identification of COVID-19 cases from X-ray Images. arXiv 2020, arXiv:2004.02696. [Google Scholar]
- Ucar, F.; Korkmaz, D. COVIDiagnosis-Net: Deep Bayes-SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images. Med. Hypotheses 2020, 140, 109761. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.; Hafeez, A. COVID-ResNet: A Deep Learning Framework for Screening of COVID19 from Radiographs. arXiv 2020, arXiv:2003.14395. [Google Scholar]
- Oh, Y.; Park, S.; Ye, J.C. Deep Learning COVID-19 Features on CXR using Limited Training Data Sets. IEEE Trans. Med. Imaging 2020, 39, 2688–2700. [Google Scholar] [CrossRef]
- Shiraishi, J.; Katsuragawa, S.; Ikezoe, J.; Matsumoto, T.; Kobayashi, T.; Komatsu, K.-I.; Matsui, M.; Fujita, H.; Kodera, Y.; Doi, K. Development of a Digital Image Database for Chest Radiographs With and Without a Lung Nodule. Am. J. Roentgenol. 2000, 174, 71–74. [Google Scholar] [CrossRef]
- Van Ginneken, B.; Stegmann, M.B.; Loog, M. Segmentation of anatomical structures in chest radiographs using supervised methods: A comparative study on a public database. Med. Image Anal. 2006, 10, 19–40. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, S.; Candemir, S.; Antani, S.; Wáng, Y.-X.J.; Lu, P.-X.; Thoma, G. Two public chest X-ray datasets for computer-aided screening of pulmonary diseases. Quant. Imaging Med. Surg. 2014, 4, 475–477. [Google Scholar] [PubMed]
- CoronaHack—Chest X-ray-Dataset. Available online: https://www.kaggle.com/praveengovi/coronahack-chest-xraydataset (accessed on 17 June 2020).
- Chouhan, V.; Singh, S.K.; Khamparia, A.; Gupta, N.; Tiwari, P.; Moreira, C.; Damasevicius, R.; De Albuquerque, V.H.C. A Novel Transfer Learning Based Approach for Pneumonia Detection in Chest X-ray Images. Appl. Sci. 2020, 10, 559. [Google Scholar] [CrossRef]
- Rahman, T.; Chowdhury, M.E.H.; Khandakar, A.; Islam, K.R.; Islam, K.F.; Mahbub, Z.B.; Kadir, M.A.; Kashem, S.; Rahman, T. Transfer Learning with Deep Convolutional Neural Network (CNN) for Pneumonia Detection Using Chest X-ray. Appl. Sci. 2020, 10, 3233. [Google Scholar] [CrossRef]
- Pan, S.J.; Yang, Q. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 2009, 22, 1345–1359. [Google Scholar] [CrossRef]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015, San Diego, CA, USA, 7–9 May 2015; pp. 1–14. [Google Scholar]
- Hossin, M.; Sulaiman, M.N. A Review on Evaluation Metrics for Data Classification Evaluations. Int. J. Data Min. Knowl. Manag. Process. 2015, 5, 1–11. [Google Scholar] [CrossRef]
| Split | COVID-19 | Normal |
|---|---|---|
| Training | 630 | 642 |
| Validation | 60 | 60 |
| Testing | 100 | 100 |
| Measures | ResNet50 | VGG16 | VGG19 | DenseNet121 |
|---|---|---|---|---|
| Accuracy (ACC) | 97% | 99.33% | 99.33% | 96.66% |
| Sensitivity (SEN) | 98.48% | 99.28% | 100.00% | 99.23% |
| Specificity (SPE) | 95.21% | 99.38% | 98.77% | 94.67% |
| False Negative Rate (FNR) | 1.52% | 0.72% | 0.00% | 0.77% |
| False Positive Rate (FPR) | 4.79% | 0.62% | 1.23% | 5.33% |
| Positive Predicted Value (PPV) | 94.20% | 99.28% | 98.55% | 93.48% |
| F1 Score (F1) | 96.30% | 99.28% | 99.27% | 96.27% |
| Study | Number of Samples | Technique | ACC | SEN | SPE | |
|---|---|---|---|---|---|---|
| Hamdan et al. [29] | 50 (25 healthy and 25 COVID-19) | COVIDX-Net | 90% | - | - | |
| Wang et al. [32] | 13,975 (normal, pneumonia, and COVID-19) | Tailored CNN (COVID-Net) | 93.3% | - | - | |
| Apostolopoulos et al. [37] | 1427 (224 COVID-19, 700 pneumonia, 504 normal) | VGG19, MobileNet, Inception, Xception, Inception ResNetV2 | 98.75% | 99.1% | - | |
| Kumar et al. [40] | 50 (25 healthy and 25 COVID-19) | ResNet50 + SVM | 95.38% | - | - | |
| Ali et al. [42] | 100 (50 normal and 50 COVID-19) | ResNet50 | 98% | - | - | |
| Ozturk et al. [43] | 625 (125 COVID-19, 500 no-findings) | DarkNet | 98.08% | - | - | |
| Minaee et al. [45] | (100 COVID-19, 5000 Non-COVID-19) | Deep-COVID | - | 97.5% | 90% | |
| Ucar et al. [48] | 2839 (1203 normal, 1591 pneumonia and 45 COVID-19) | COVIDiagnosis-Net | 98.30% | - | - | |
| Farooq et al. [49] | 2813 (1203 normal, 931 bacterial pneumonia, 660 viral pneumonia, 19 COVID-19 cases) | COVID-ResNet | 96.23% | - | - | |
| Oh et al. [50] | 502 (191 normal, 54 bacterial, 57 tuberculosis, 20 viral, 180 COVID-19) | ResNet18 | 88.90% | - | 96.4% | |
| Proposed Study | VGG16, VGG19 | 99.38% | 100% | 99.33% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, I.U.; Aslam, N. A Deep-Learning-Based Framework for Automated Diagnosis of COVID-19 Using X-ray Images. Information 2020, 11, 419. https://doi.org/10.3390/info11090419
Khan IU, Aslam N. A Deep-Learning-Based Framework for Automated Diagnosis of COVID-19 Using X-ray Images. Information. 2020; 11(9):419. https://doi.org/10.3390/info11090419
Chicago/Turabian StyleKhan, Irfan Ullah, and Nida Aslam. 2020. "A Deep-Learning-Based Framework for Automated Diagnosis of COVID-19 Using X-ray Images" Information 11, no. 9: 419. https://doi.org/10.3390/info11090419
APA StyleKhan, I. U., & Aslam, N. (2020). A Deep-Learning-Based Framework for Automated Diagnosis of COVID-19 Using X-ray Images. Information, 11(9), 419. https://doi.org/10.3390/info11090419




