1. Introduction
Lack of sufficient annotated data often limits the applicability of deep learning (DL) models to real life problems. However, efficient transfer learning (TL) strategies help to utilize valuable knowledge learned with sufficient data in one task (source task) and transfer it to the task of interest (target task).
In this work we focus on a generic named entity recognition (NER) system that uses the representation learning capability of deep neural networks. NER refers to a subtask of information extraction in which entity mentions in an unstructured text are semantically labeled into pre-defined categories (e.g., in the sentence “Benjamin Franklin is known for inventing the lightening rod”, “Benjamin Franklin” has to be labeled with the tag Person). In this paper NER is evaluated with two main use cases—extraction of entity names from financial documents and from biomedical documents.
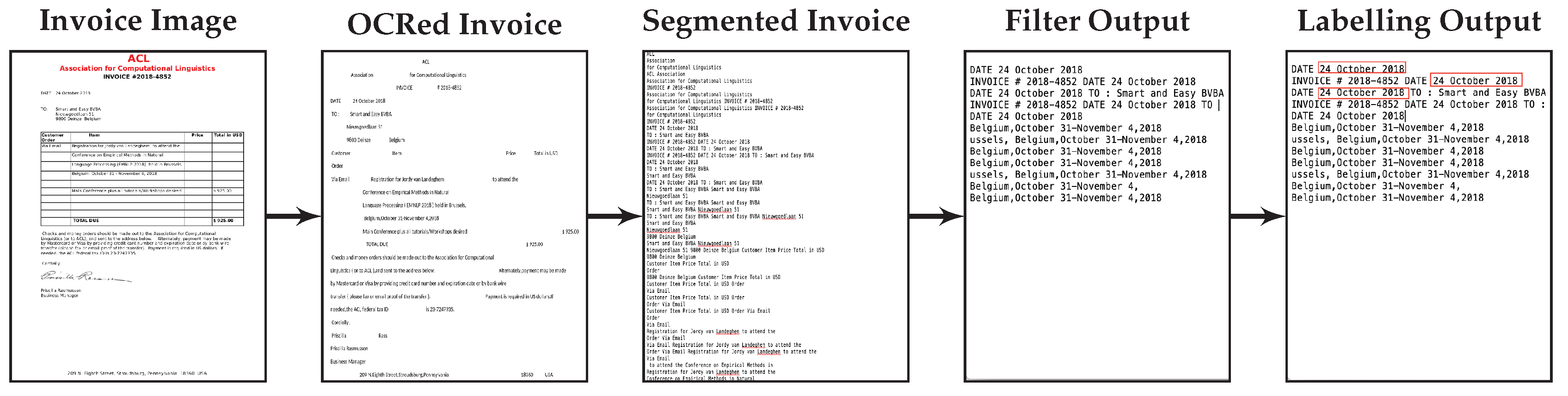
The aim of the first use case is to develop a generic NER deep learning system that is capable of recognizing entities in business documents including invoices, business forms and emails. Some examples of important named entities used in the financial documents are invoice sender name, invoice number, invoice date, International Bank Account Number (IBAN), amount payable and structured communication. The overall goal of this use case is to research and test prototypes for a self-learning SaaS platform for simplification of data-intensive customer interactions in industries such as energy, telecommunications, banking or insurance. The format of business documents from different service providers varies widely with respect to the information contained within it, the locations where it occurs, the formatting used, terminology used, and variations in context.
The growing biomedical literature requires efficient methods of text mining and information extraction tools that can extract structured information from the text for further analysis. The second use case has as objective the development and application of a generic NER system that recognizes biomedical entities like genes, proteins, chemicals, diseases and the like from biomedical text sequences.
In both cases we want to port a model learned in one domain to a target domain that is characterized by a limited number of labeled training examples. Multiple additional challenges apply. The difficulty posed by the documents in the financial domain is the noisy non-segmented OCRed text which has to be used as the training data. The documents in the biomedical domain are characterized by spelling variants of the mentioned entities.
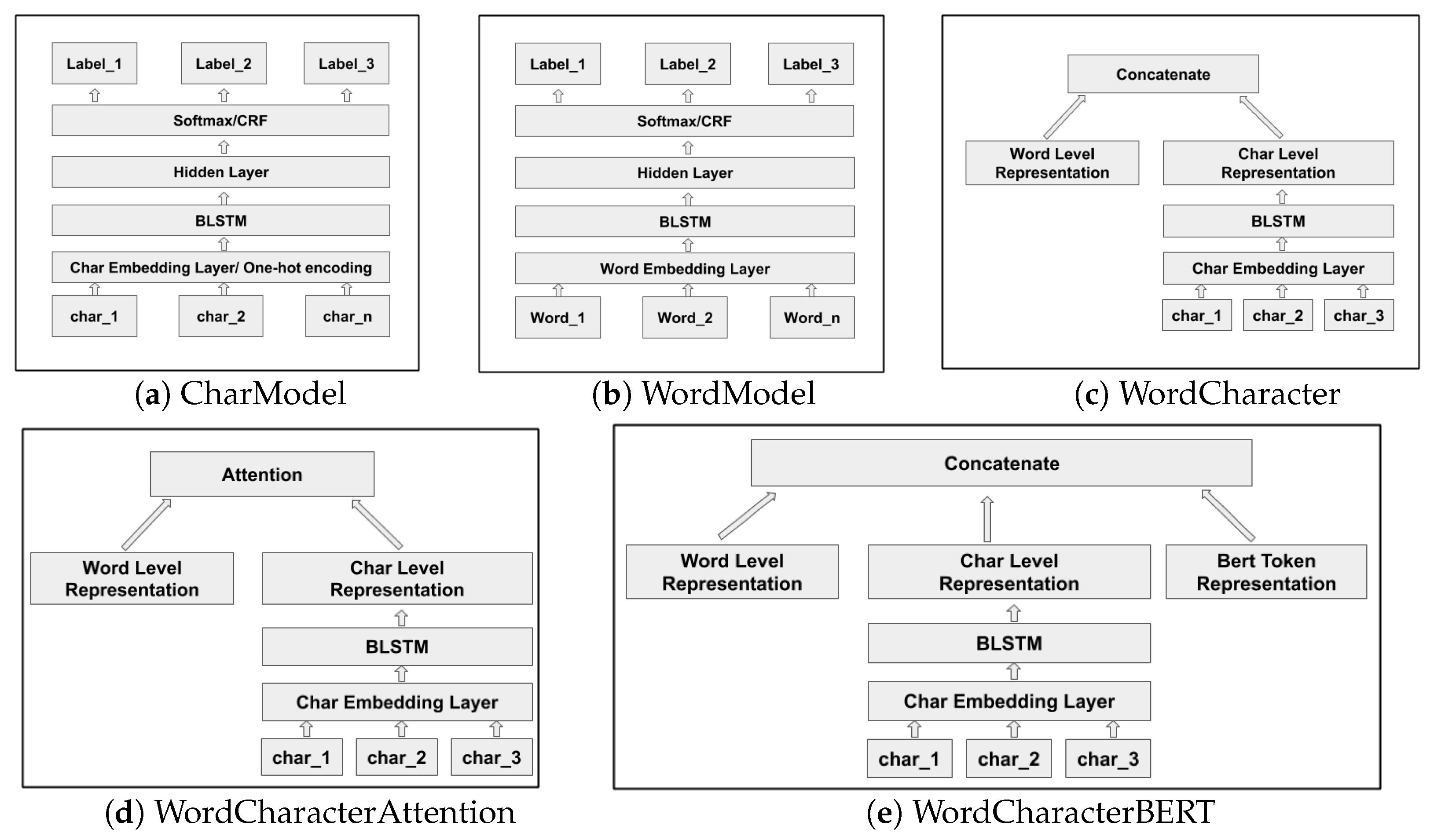
Neural network based deep learning models have become popular for processing unstructured (e.g., language) and semi-structured data. Deep learning is concerned with automatically learning the representations of the inputs where patterns at different levels of detail can be recognized and represented, a process which is referred to as representation learning. In particular, recurrent neural networks (RNNs) are capable of learning contextualized representations of text sequences. Information extraction from text using deep learning avoids the need for humans to formally specify knowledge, and even avoids the need for feature engineering. Most state-of-the-art named entity recognition (NER) systems use word level and character level representations (embeddings) to perform named entity extraction and classification (dos Santos and Guimarães [
1]).
A word or character is mapped to a continuous vector representation (embedding) that captures the context of the word and character respectively. While word based models need accurate token-level segmentation, character level models have the ability to perform accurate labelling of tokens or character units without the need for a-priori word segmentation (Kann and Schütze [
2]).
The aim is that the extraction system can be trained with few manually annotated training data, especially when a trained model needs to be ported to a novel domain (e.g., another provider of documents to be analyzed, other entity classes to be recognized).
We witness a rising interest in language models to improve NER in a transfer learning setting. The current most successful language model is BERT (Devlin et al. [
3]). We integrate BERT representations to analyse its impact when using it along with separate character and word representations.
The research questions posed in the paper are:
How to train models (e.g., character based neural networks) with small sized semi-structured labeled datasets (e.g., contracts and invoices)?
What is the impact of transfer learning when a NER model that is trained on a source dataset labeled with one type of entities is ported to a target dataset to be labeled with another but related entity type and only few labeled documents in the target domain are available?
What is the impact of using the BERT language model along with character and word character level representations?
The major contributions presented in the paper are: (a) Development of an efficient pipeline for entity extraction from financial documents that is efficient at runtime and easily parameterizable, and which includes efficient segmentation, filtering of entity elements and finally labelling and extraction; (b) Effective document segmentation by splitting a document text into meaningful and contextual sub-segments; (c) Easy porting from models trained on one task to another task using transfer learning; and (d) Comparison of the impact of integrating the BERT language model with the character and word level representations. The impact of using limited training data in the target domain has been investigated by using different percentages of target training examples.
2. Related Work
NER is a well studied task in natural language processing, yet it has mostly been studied in the frame of recognition of entity types that refer to persons, organizations, locations, or dates. Traditionally NER is seen as a sequence labeling task in which word tokens are labeled with the corresponding named entity class (often using a BIO-format referring to respectively the Beginning, Intermediate or Outside class token). There has been a lot of previous work on optimizing neural architectures for sequence labeling. Collobert et al. [
4] introduced one of the first task-independent neural sequence labeling models using a convolutional neural network (CNN). They were able to achieve good results on other sequence labelling tasks such as Part-of-Speech (POS) tagging, chunking, and semantic role labeling (SRL) without relying on hand-engineered features. Huang et al. [
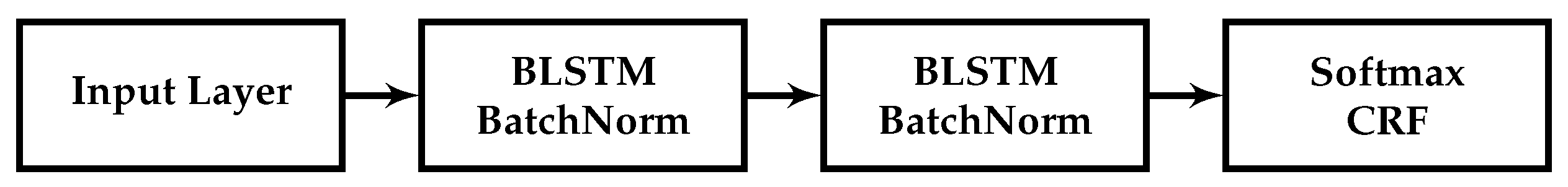
5] describe a bidirectional long short-term memory network (LSTM) model, a type of recurrent neural network (RNN), with a conditional random field (CRF) layer, which includes hand-crafted features specialised for the task of named entity recognition. The architecture of our baseline sequence labelling word model for biomedical data uses a similar bidirectional LSTM (BiLSTM) and CRF with batch normalization.
Character level models are capable of capturing morpheme patterns and hence improve the generalisation of the model on both frequent and unseen words. Kim et al. [
6] have implemented a language model with a convolutional neural network (CNN) and LSTM architecture using characters as input while the predictions are made at the word level. We use similar neural architectures when extracting named entities from the financial and biomedical documents. For the biomedical domain, we chose to implement word based models and also combinations of word and character based models firstly due to the availability of pretrained word embeddings trained on huge biomedical corpora and general Wikipedia text which considerably mitigates the problem of out-of-vocabulary (OOV) words. Character level models are semantically more void, yet morphologically more informed, and thus word based and character based models are hypothesized to perform better for biomedical domain texts.
Cao and Rei [
7] proposed a method for learning both word embedding and morphological segmentation with a bidirectional RNN over characters. Ling et al. [
8] have proposed a neural architecture that replaces word embeddings with dynamically constructed character based representations. Lample et al. [
9] describe a model where the character level representation is combined with a word embedding through concatenation. Miyamoto and Cho [
10] have also described a related method for the task of language modelling, combining character and word embeddings using gating.
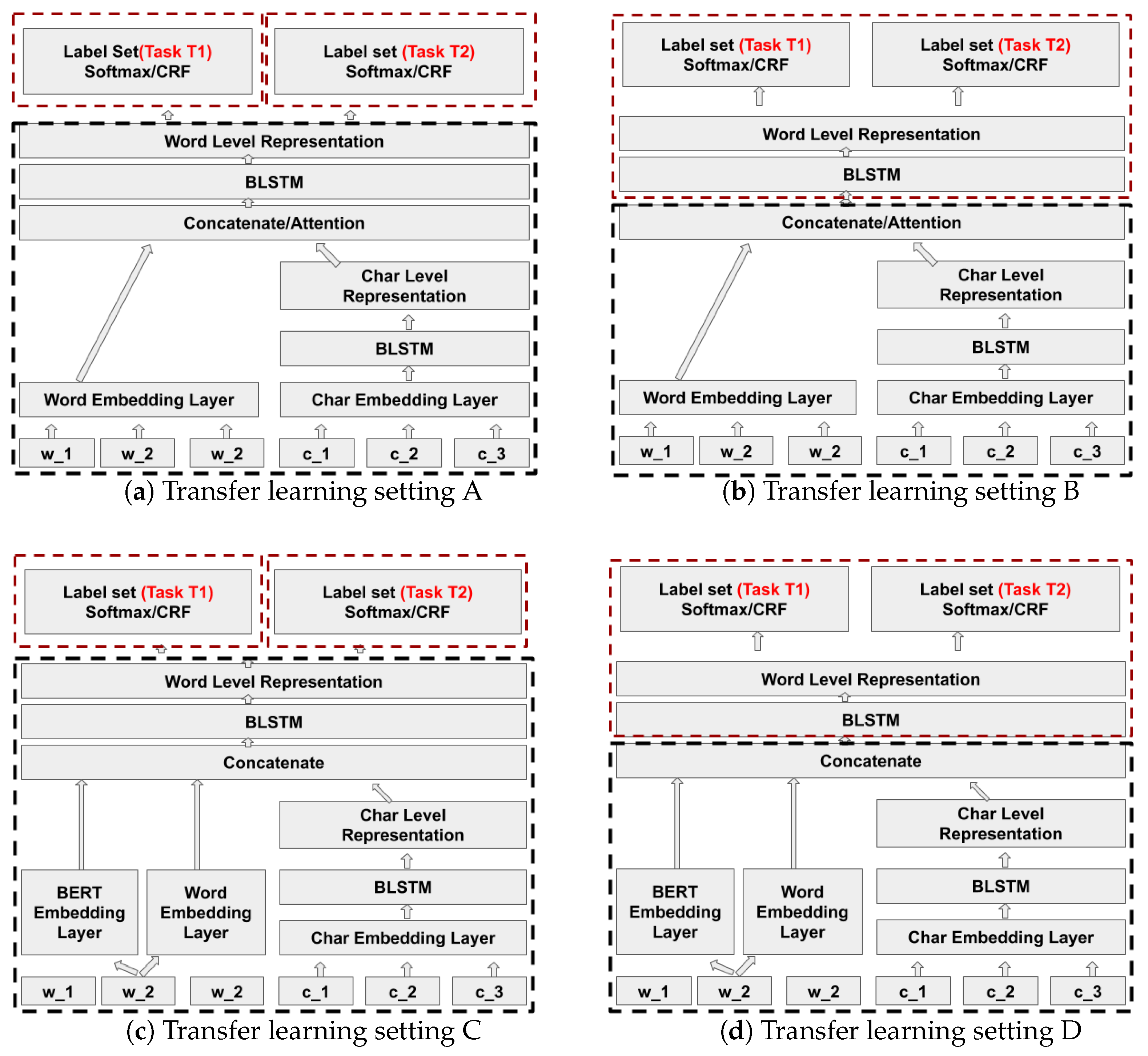
Transfer learning is a machine learning technique where a model pre-trained for one task is reused for a second related task that has less labeled training data. Transfer learning in general improves the generalisation of the model. In this paper we apply transfer learning in two different use cases, that is, NER in financial and biomedical documents. We study how a neural model for NER of one entity type which is structurally or contextually similar to some other entity type can avoid the need for training the target model from scratch. For example, we transfer knowledge from a trained model that has learned to properly extract the entity invoice date to the extraction of a related entity expiration date. This is a case of learning a new task where the character pattern of the new entity type is similar but the context in which this entity occurs is different.
Model based transfer uses the similarity and relatedness between the source task and the target task by modifying the model architectures, training algorithms, or feature representations. For example, Ando and Zhang [
11] propose a transfer learning framework that shares structural parameters across multiple tasks, and improve the performance on various tasks including NER. Collobert et al. [
4] present a task-independent CNN and employ joint training to transfer knowledge from NER and POS tagging to sentence chunking. Peng and Dredze [
12] study transfer learning between NER and word segmentation in Chinese based on RNN architectures. We have exploited model based transfer learning in our models to improve the target task models by investigating different levels of freezing and fine-tuning for the target architecture based on Yang et al. [
13], Rodriguez et al. [
14] and Lee et al. [
15].
We witness an interest in language models to improve NER as a way of coping with limited labeled data. The current most successful language model is BERT (Devlin et al. [
3]). BERT makes use of a Transformer architecture, which integrates an attention mechanism that learns contextual relations between words (or sub-words) in a text.
BioBERT (Lee et al. [
16]) provides a pre-trained biomedical language model trained with the BERT architecture that can be used in various biomedical text mining tasks such as NER. Because of its success in state-of-the-art models we integrate embeddings obtained with the BERT language model in our NER models for biomedical texts.
Author Contributions
Conceptualization, S.F., J.V.L. and M.-F.M.; data curation, S.F. and J.V.L.; methodology, S.F.; investigation, S.F. and J.V.L.; writing—original draft preparation, S.F.; writing—review and editing, S.F., J.V.L. and M.-F.M.; visualization, S.F.; supervision, project administration and funding acquisition, M.-F.M.
Funding
The research in collaboration with the company Contract.fit was sponsored by VLAIO (Flanders Innovation & Entrepreneurship) under contract number HBC.2017.0264, and another part was financed by the SBO project ACCUMULATE (IWT-SBO-Nr. 150056).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dos Santos, C.N.; Guimarães, V. Boosting named entity recognition with neural character embeddings. In Proceedings of the Fifth Named Entity Workshop (NEWS@ACL 2015), Beijing, China, 31 July 2015; pp. 25–33. [Google Scholar]
- Kann, K.; Schütze, H. Single-model encoder-decoder with explicit morphological representation for reinflection. In Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (ACL), Berlin, Germany, 7–12 August 2016. [Google Scholar]
- Devlin, J.; Chang, M.; Lee, K.; Toutanova, K. BERT: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Collobert, R.; Weston, J.; Bottou, L.; Karlen, M.; Kavukcuoglu, K.; Kuksa, P. Natural language processing (almost) from scratch. J. Mach. Learn. Res. 2011, 12, 2493–2537. [Google Scholar]
- Huang, Z.; Xu, W.; Yu, K. Bidirectional LSTM-CRF models for sequence tagging. arXiv 2015, arXiv:1508.01991. [Google Scholar]
- Kim, Y.; Jernite, Y.; Sontag, D.; Rush, A.M. Character-aware neural language models. In Proceedings of the Thirtieth AAAI Conference on Artificial Intelligence, Phoenix, AZ, USA, 12–17 February 2016. [Google Scholar]
- Cao, K.; Rei, M. A joint model for word embedding and word morphology. arXiv 2016, arXiv:1606.02601. [Google Scholar]
- Ling, W.; Luís, T.; Marujo, L.; Astudillo, R.F.; Amir, S.; Dyer, C.; Black, A.W.; Trancoso, I. Finding function in form: Compositional character models for open vocabulary word representation. arXiv 2015, arXiv:1508.02096. [Google Scholar]
- Lample, G.; Ballesteros, M.; Subramanian, S.; Kawakami, K.; Dyer, C. Neural architectures for named entity recognition. arXiv 2016, arXiv:1603.01360. [Google Scholar]
- Miyamoto, Y.; Cho, K. Gated word-character recurrent language model. In Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing, EMNLP 2016, Austin, TX, USA, 1–4 November 2016; pp. 1992–1997. [Google Scholar]
- Ando, R.K.; Zhang, T. A framework for learning predictive structures from multiple tasks and unlabeled data. J. Mach. Learn. Res. 2005, 6, 1817–1853. [Google Scholar]
- Peng, N.; Dredze, M. Multi-task multi-domain representation learning for sequence tagging. arXiv 2016, arXiv:1608.02689. [Google Scholar]
- Yang, Z.; Salakhutdinov, R.; Cohen, W.W. Transfer learning for sequence tagging with hierarchical recurrent networks. arXiv 2017, arXiv:1703.06345. [Google Scholar]
- Rodriguez, J.D.; Caldwell, A.; Liu, A. Transfer learning for entity recognition of novel classes. In Proceedings of the 27th International Conference on Computational Linguistics, Santa Fe, NM, USA, 20–26 August 2018; pp. 1974–1985. [Google Scholar]
- Lee, J.Y.; Dernoncourt, F.; Szolovits, P. Transfer learning for named-entity recognition with neural networks. arXiv 2017, arXiv:1705.06273. [Google Scholar]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. arXiv 2019, arXiv:1901.08746. [Google Scholar]
- Lafferty, J.D.; McCallum, A.; Pereira, F.C.N. Conditional Random Fields: Probabilistic models for segmenting and labeling sequence data. In Proceedings of the Eighteenth International Conference on Machine Learning (ICML), Williamstown, MA, USA, 28 June–1 July 2001; pp. 282–289. [Google Scholar]
- Sakharov, A.; Sakharov, T. The Viterbi algorithm for subsets of stochastic context-free languages. Inf. Process. Lett. 2018, 135, 68–72. [Google Scholar] [CrossRef]
- Rei, M.; Crichton, G.K.O.; Pyysalo, S. Attending to characters in neural sequence labeling models. In Proceedings of the 26th International Conference on Computational Linguistics (COLING), Osaka, Japan, 11–16 December 2016; pp. 309–318. [Google Scholar]
- Howard, J.; Ruder, S. Universal Language Model fine-tuning for text classification. In Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics, ACL, Melbourne, Australia, 15–20 July 2018; pp. 328–339. [Google Scholar]
- Erhan, D.; Manzagol, P.; Bengio, Y.; Bengio, S.; Vincent, P. The difficulty of training deep architectures and the effect of unsupervised pre-training. In Proceedings of the Twelfth International Conference on Artificial Intelligence and Statistics (AISTATS), Clearwater, FL, USA, 16–18 April 2009; pp. 153–160. [Google Scholar]
- Giorgi, J.M.; Bader, G.D. Transfer learning for biomedical named entity recognition with neural networks. Bioinformatics 2018, 34, 4087–4094. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.; Tanabe, L.K.; nee Ando, R.J.; Kuo, C.J.; Chung, I.F.; Hsu, C.N.; Lin, Y.S.; Klinger, R.; Friedrich, C.M.; Ganchev, K.; et al. Overview of BioCreative II gene mention recognition. Genome Biol. 2008, 9, S2. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.D.; Ohta, T.; Pyysalo, S.; Kano, Y.; Tsujii, J. Extracting bio-molecular events from literature: The BIONLP’09 shared task. Comput. Intell. 2011, 27, 513–540. [Google Scholar] [CrossRef]
- Mikolov, T.; Chen, K.; Corrado, G.; Dean, J. Efficient estimation of word representations in vector space. arXiv 2013, arXiv:1301.3781. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).