Liver Cancer Classification Model Using Hybrid Feature Selection Based on Class-Dependent Technique for the Central Region of Thailand
Abstract
1. Introduction
2. Background and Related Work
2.1. TNM Classification for Hepatocellular Carcinoma
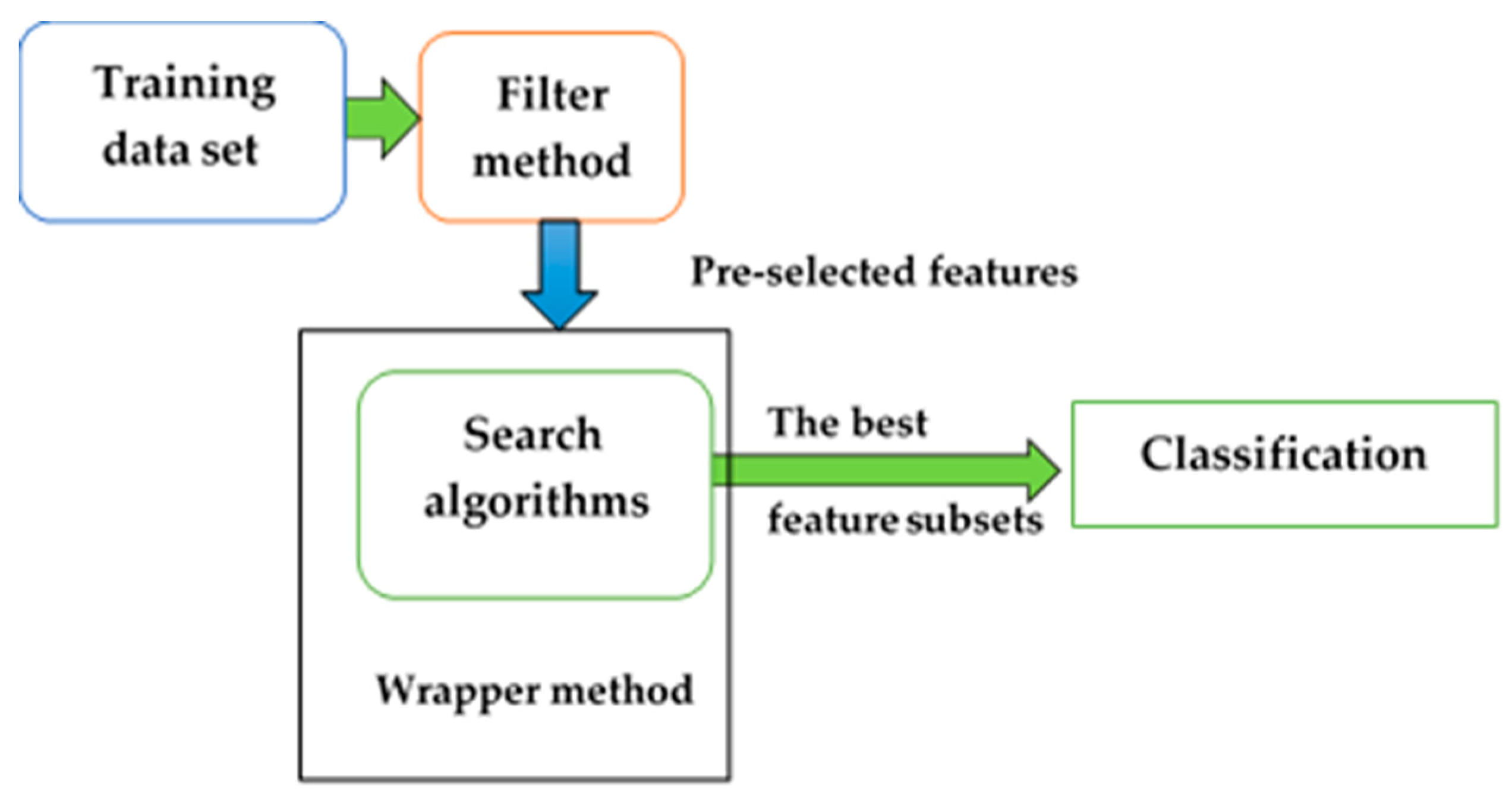
2.2. Hybrid Feature Selection Method
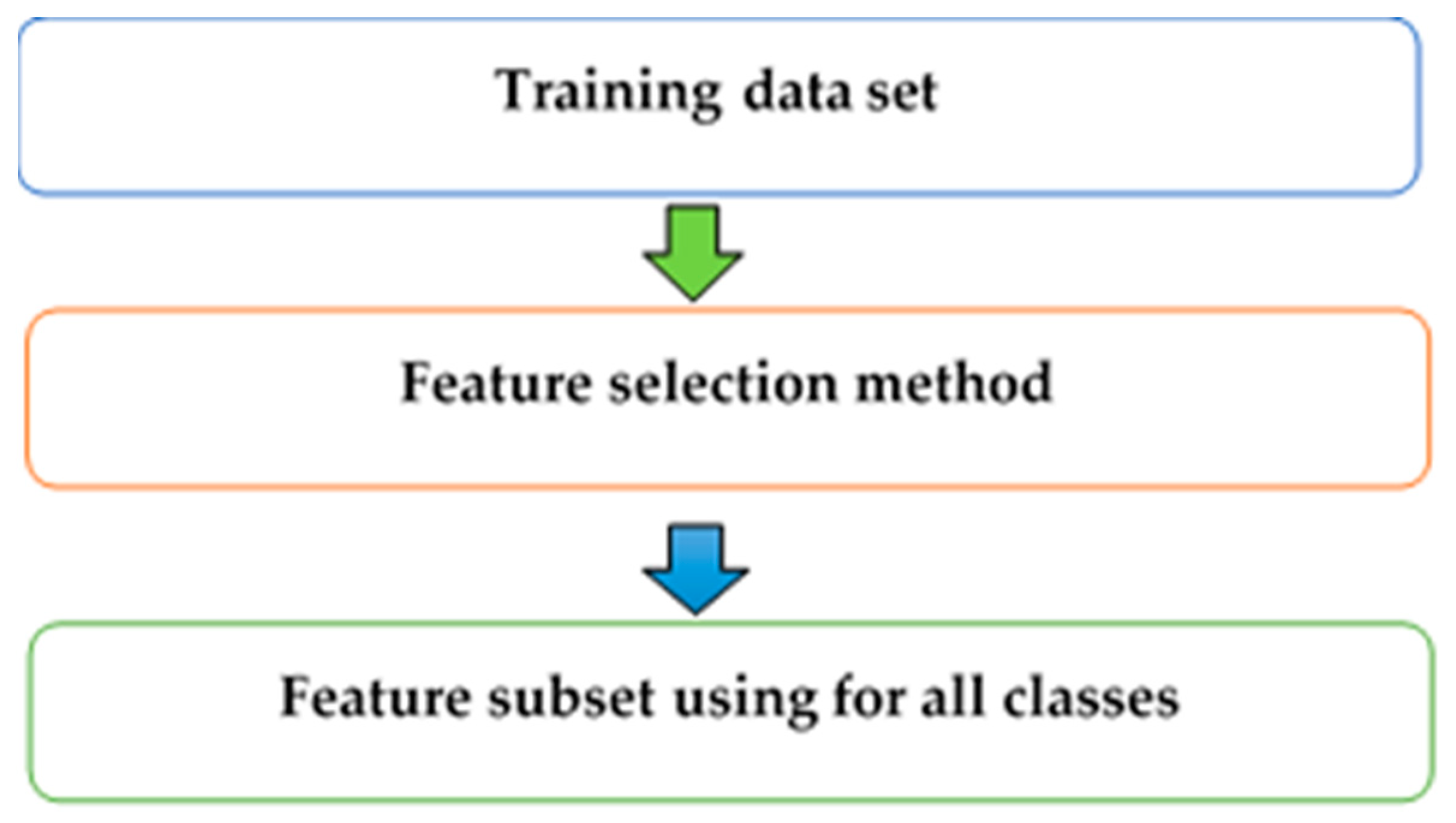
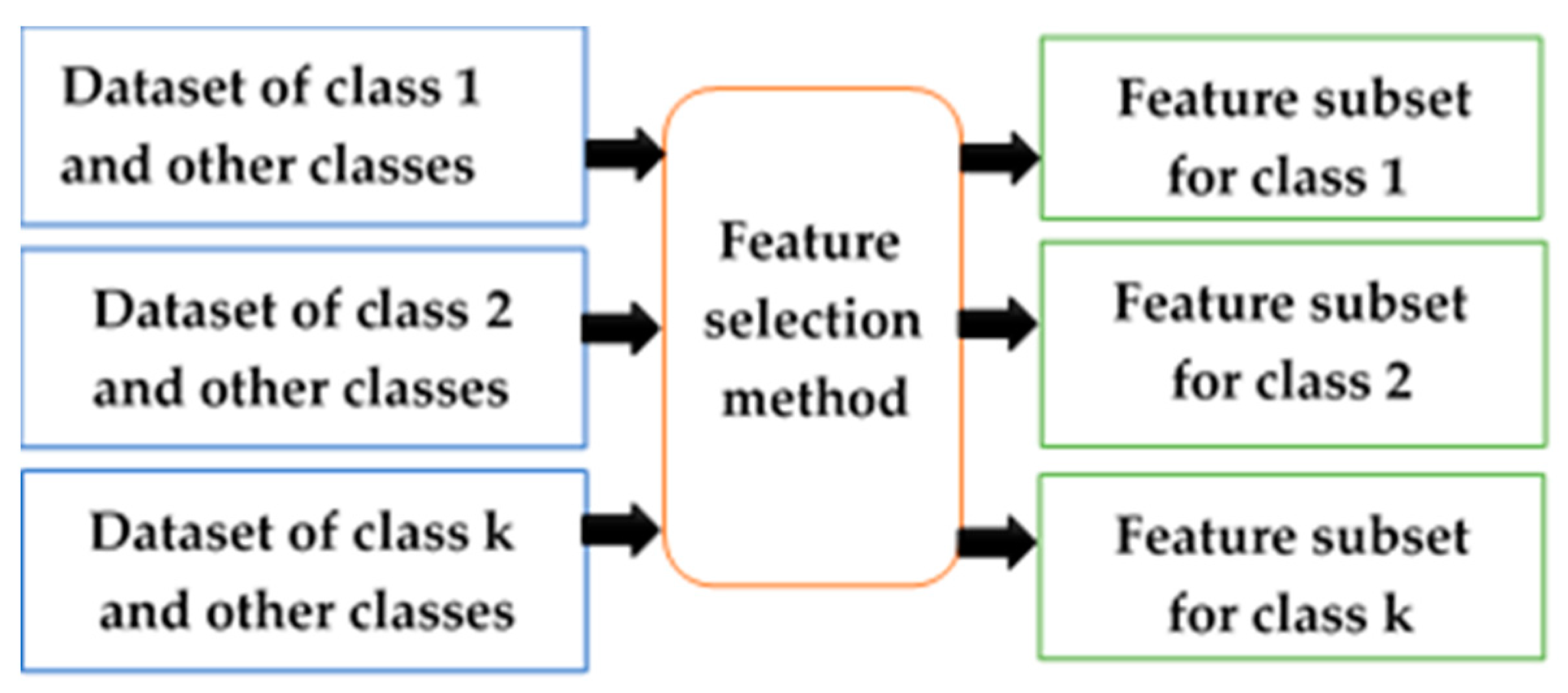
2.3. Class-Dependent and Class-Independent Techniques
2.4. Classification for Multi-Class Problem
2.5. Related Work
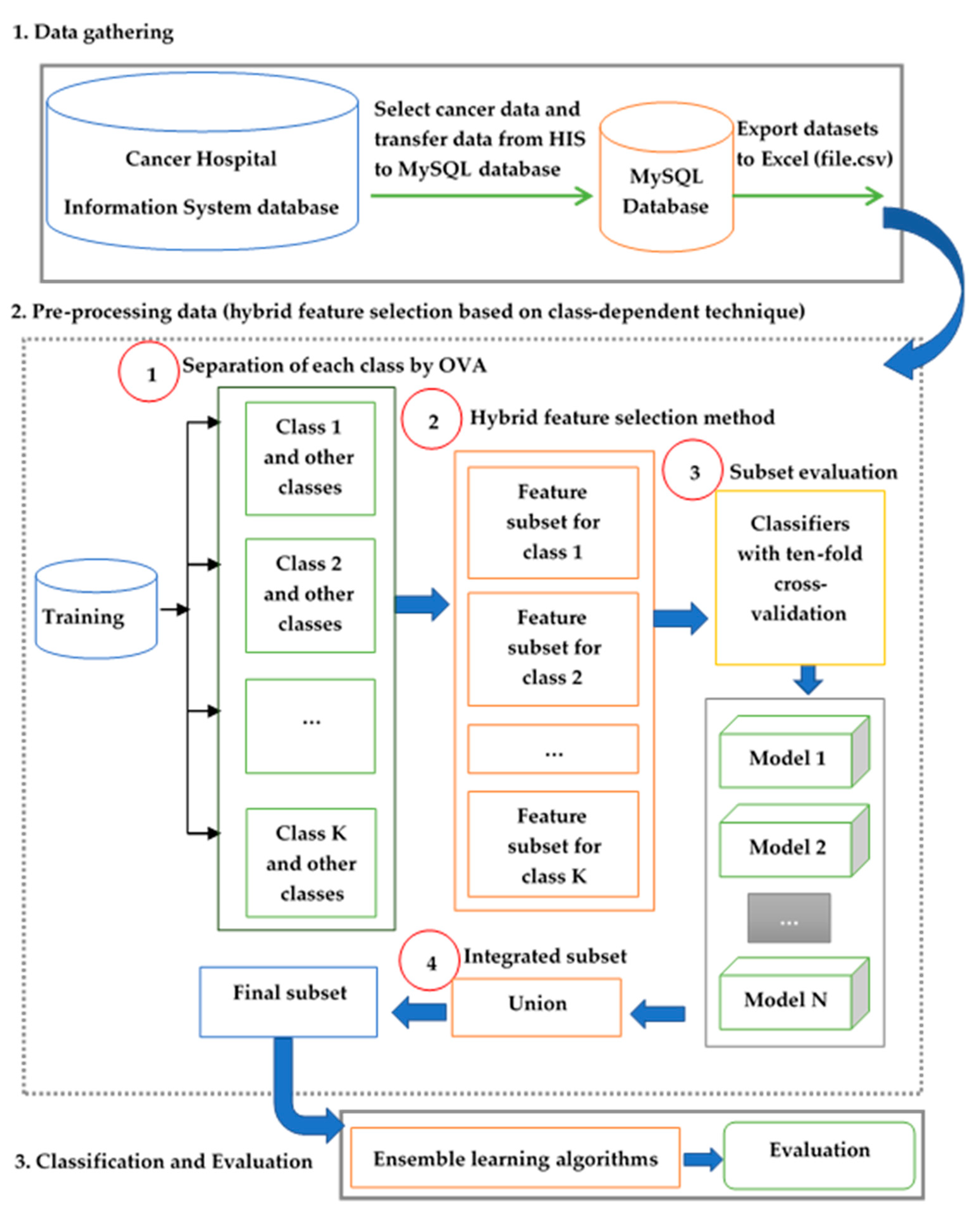
3. Materials and Methods
3.1. Data Collection
3.2. The Preprocessing Method for Class-Dependent Feature Selection
- Initialize the empty set Yk = {∅} at k = 0
- Select the next best feature x to add to Yk with most significant cost reduction
- Update Yk+1 = Yk + x; k = k + 1
- Repeat step 2
3.3. Classifiers
3.4. Evaluation Model with Ensemble Learning Methods
3.4.1. Heterogeneous Classifiers Use the Same Data over Diversified Learning Algorithms
3.4.2. Homogeneous Classifiers Use Different Data Sets with The Same Algorithm
- Bagging (bootstrap aggregating) is an ensemble method that creates different training sets by sampling with replacement from the original dataset on training individual classifiers [50,52]. It provides diversity in subsets that might reduce the variance of datasets and improve results of classification algorithms [53]. In this work, different training sets were obtained by resampling using bagging with a DT as the base classifier.
- Boosting is an approach to machine learning that provides a highly accurate prediction rule by combining many relatively weak and inaccurate rules. The AdaBoost (adaptive boosting) algorithm was the first practical boosting algorithm [51]. AdaBoost is an algorithm for constructing a strong classifier as a linear combination of weak classifiers. It reduces the bias of multi-class datasets and it is applicable for building ensembles that empirically improve generalization performance [48,53]. In this work, the AdaBoost approach with DT as the base classifier is used to train datasets.
3.5. Performance Measures for Classification
- CI = Point estimate ± Margin of error.
- MOE = Critical value (z) × Standard error of point estimate
4. Results
4.1. Performance of IGSFS-CD Approach Using Ensemble Learning Method On Lc_Dataset-1
4.2. Performance of the IGSFS-CD Approach by the Ensemble Learning Method on LC_dataset-II
4.3. Selected Features by the Igsfs-Cd Approach
4.4. Estimation of Performance Measures
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Deerasamee, S.; Martin, N.; Sontipong, S.; Sriamporn, S.; Sriplung, H.; Srivatanakul, P.; Vatanasapt, V.; Parkin, D. Cancer registration in Thailand. Asian Pac. J. Cancer Prev. 2001, 2, 79–84. [Google Scholar]
- Wongphan, T.; Bundhamcharoen, K. Health-Related Quality of Life as Measured by EQ-5D and TFLIC-2 in Liver Cancer Patients. Siriraj Med. J. 2018, 70, 406–412. [Google Scholar]
- Kitiyakara, T. Advances in biomarkers for HCC. Thai J. Hepatol. 2018, 1, 29–32. [Google Scholar] [CrossRef]
- Intaraprasong, P. Review New therapy including combination. Thai J. Hepatol. 2018, 1, 33–36. [Google Scholar] [CrossRef]
- Fujiwara, N.; Friedman, S.L.; Goossens, N.; Hoshida, Y. Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J. Hepatol. 2018, 68, 526–549. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, S.; Kelley, R.K.; Venook, A.P. A review of hepatocellular carcinoma (HCC) staging systems. Chin. Clin. Oncol. 2013, 2, 33. [Google Scholar] [PubMed]
- Clark, H.P.; Carson, W.F.; Kavanagh, P.V.; Ho, C.P.; Shen, P.; Zagoria, R.J. Staging and Current Treatment of Hepatocellular Carcinoma. Radiographics 2005, 25, S3–S23. [Google Scholar] [CrossRef] [PubMed]
- Sutha, K.; Tamilselvi, J.J. A Review of Feature Selection Algorithms for Data mining Techniques. Int. J. Comput. Sci. Eng. 2015, 7, 63–67. [Google Scholar]
- Bolón-Canedo, V.; Sánchez-Marono, N.; Alonso-Betanzos, A.; Benítez, J.M.; Herrera, F. A review of microarray datasets and applied feature selection methods. Inf. Sci. 2014, 282, 111–135. [Google Scholar] [CrossRef]
- Chandrashekar, G.; Sahin, F. A survey on feature selection methods. Comput. Electr. Eng. 2014, 40, 16–28. [Google Scholar] [CrossRef]
- Kotsiantis, S.B. Feature selection for machine learning classification problems: A recent overview. Artif. Intell. Rev. 2011, 42, 157–176. [Google Scholar] [CrossRef]
- Liu, J.; Ranka, S.; Kahveci, T. Classification and Feature Selection Algorithms for Multi-class CGH data. Bioinformatics 2008, 24, i86–i95. [Google Scholar] [CrossRef] [PubMed]
- Abeel, T.; Helleputte, T.; Van de Peer, Y.; Dupont, P.; Saeys, Y. Robust Biomarker Identification for Cancer Diagnosis Using Ensemble Feature Selection Methods. Bioinformatics 2009, 26, 392–398. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Osuna, R. Pattern Analysis for Machine Olfaction: A Review. IEEE Sens. J. 2002, 2, 189–202. [Google Scholar] [CrossRef]
- Bertolazzi, P.; Felici, G.; Festa, P.; Fiscon, G.; Weitschek, E. Integer programming models for feature selection: New extensions and a randomized solution algorithm. Eur. J. Oper. Res. 2016, 250, 389–399. [Google Scholar] [CrossRef]
- Gnana, D.A.A.; Appavu, S.; Leavline, E.J. Literature Review on Feature Selection Methods for High-dimensional Data. Int. J. Comput. Appl. 2016, 136, 9–17. [Google Scholar]
- Hsu, H.H.; Hsieh, C.W.; Lu, M.D. Hybrid Feature Selection by Combining Filters and Wrappers. Expert Syst. Appl. 2011, 38, 8144–8150. [Google Scholar] [CrossRef]
- Panthong, R.; Srivihok, A. Wrapper Feature Subset Selection for Dimension Reduction Based on Ensemble Learning Algorithm. In Proceedings of the 3rd Information Systems International Conference Procedia Computer Science (ISICO2015), Surabaya, Indonesia, 2–4 November 2015; pp. 162–169. [Google Scholar]
- Zhou, W.; Dickerson, J.A. A Novel Class Dependent Feature Selection Method for Cancer Biomarker Discovery. Comput. Med. Biol. 2014, 47, 66–75. [Google Scholar] [CrossRef]
- Bailey, A. Class-Dependent Features and Multicategory Classification. Ph.D. Thesis, Department of Electronics and Computer Science, University of Southampton, Southampton, UK, 7 February 2001. [Google Scholar]
- Cateni, S.; Colla, V.; Vannucci, M. A Hybrid Feature Selection Method for Classification Purposes. In Proceedings of the 8th IEEE Modelling Symposium (EMS) European, Pisa, Italy, 21–23 October 2014; pp. 39–44. [Google Scholar]
- Naqvi, G. A Hybrid Filter-Wrapper Approach for Feature Selection. Master’s Thesis, Dept. Technology, Orebro University, Orebro, Sweden, 2012. [Google Scholar]
- Oh, I.S.; Lee, J.S.; Suen, C.Y. Analysis of class separation and combination of class-dependent features for handwriting recognition. IEEE Trans. Pattern Anal. Mach. Intell. 1999, 21, 1089–1094. [Google Scholar]
- Oh, I.S.; Lee, J.S.; Suen, C.Y. Using class separation for feature analysis and combination of class-dependent features. In Proceedings of the 14th International Conference Pattern Recognit, Washington, DC, USA, 16–20 August 1998; pp. 453–455. [Google Scholar]
- Mehra, N.; Gupta, S. Survey on Multiclass Classification Methods. Int. J. Comput. Sci. Inf. Technol. 2013, 4, 572–576. [Google Scholar]
- Prachuabsupakij, W.; Soonthornphisaj, N. A New Classification for Multiclass Imbalanced Datasets Based on Clustering Approach. In Proceedings of the 26th Annual Conference the Japanese Society for Artificial Intelligence, Yamaguchi, Japan, 12–15 June 2012; pp. 1–10. [Google Scholar]
- Liu, H.; Jiang, H.; Zheng, R. The Hybrid Feature Selection Algorithm Based on Maximum Minimum Backward Selection Search Strategy for Liver Tissue Pathological Image Classification. Comput. Math. Methods Med. 2016, 2016, 7369137. [Google Scholar] [CrossRef] [PubMed]
- Gunasundari, S.; Janakiraman, S. A Hybrid PSO-SFS-SBS Algorithm in Feature Selection for Liver Cancer Data. In Power Electronics and Renewable Energy Systems; Kamalakannan, C., Suresh, L.P., Dash, S.S., Panigrahi, B.K., Eds.; Springer: New Delhi, India, 2015; Volume 326, pp. 1369–1376. [Google Scholar]
- Hassan, A.; Abou-Taleb, A.S.; Mohamed, O.A.; Hassan, A.A. Hybrid Feature Selection approach of ensemble multiple Filter methods and wrapper method for Improving the Classification Accuracy of Microarray Data Set. Int. J. Comput. Sci. Inf. Technol. Secur. 2013, 3, 185–190. [Google Scholar]
- Ding, J.; Fu, L. A Hybrid Feature Selection Algorithm Based on Information Gain and Sequential Forward Floating Search. J. Intell. Comput. 2018, 9, 93–101. [Google Scholar] [CrossRef]
- Chuang, L.Y.; Ke, C.H.; Yang, C.H. A Hybrid Both Filter and Wrapper Feature Selection Method for Microarray Classification. arXiv 2016, arXiv:1612.08669. [Google Scholar]
- Zhou, N.; Wang, L. Processing Bio-medical Data with Class-Dependent Feature Selection. In Advances in Neural Networks Computational Intelligence for ICT; Bassis, S., Esposito, A., Morabito, F.C., Pasero, E., Eds.; Springer: Cham, Switzerland, 2016; Volume 54, pp. 303–310. [Google Scholar]
- Zhou, N.; Wang, L. A Novel Support Vector Machine with Class-dependent Features for Biomedical Data. In Proceedings of the 2006 IEEE International Conference on Systems, Man and Cybernetics, Taipei, Taiwan, 8–11 October 2006; pp. 1666–1670. [Google Scholar]
- Wang, L.; Zhou, N.; Chu, F. A General Wrapper Approach to Selection of Class-Dependent Features. IEEE Trans. Neural Netw. 2008, 19, 1267–1278. [Google Scholar] [CrossRef]
- Azhagusundari, B.; Thanamani, A.S. Feature Selection based on Information Gain. Int. J. Innov. Technol. Explor. Eng. 2013, 2, 18–21. [Google Scholar]
- Karaca, T. Functıonal Health Patterns Model—A Case Study. Case Stud. J. 2016, 5, 14–22. [Google Scholar]
- Yilmaz, F.T.; Sabanciogullari, S.; Aldemir, K. The opinions of nursing students regarding the nursing process and their levels of proficiency in Turkey. J. Caring Sci. 2015, 4, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Weitschek, E.; Felici, G.; Bertolazzi, P. Clinical data mining: Problems, pitfalls and solutions. In Proceedings of the 24th International Workshop on Database and Expert Systems Applications (DEXA 2013), Los Alamitos, CA, USA, 26–30 August 2013; Wagner, R.R., Tjoa, A.M., Morvan, F., Eds.; IEEE: Prague, Czech Republic, 26 August 2013; pp. 90–94. [Google Scholar]
- Yang, C.H.; Chuang, L.Y.; Yang, C.H. IG-GA: A Hybrid Filter/Wrapper Method for Feature Selection of Microarray Data. J. Med. Biol. Eng. 2010, 30, 23–28. [Google Scholar]
- Zhang, T. Adaptive Forward-Backward Greedy Algorithm for Learning Sparse Representations. IEEE Trans. Inf. Theory. 2011, 57, 4689–4708. [Google Scholar] [CrossRef]
- RapidMiner Studio. Available online: https://rapidminer. com/products/studio (accessed on 9 May 2016).
- Danjuma, K.J. Performance Evaluation of Machine Learning Algorithms in Post-operative Life Expectancy in the Lung Cancer. arXiv 2015, arXiv:1504.04646. [Google Scholar]
- Farid, D.M.; Zhang, L.; Rahman, C.M.; Hossain, M.A.; Strachan, R. Hybrid decision tree and Naïve Bayes classifiers for multiclass classification tasks. Expert Syst. Appl. 2014, 41, 1937–1946. [Google Scholar] [CrossRef]
- Tan, F. Improving Feature Selection Techniques for Machine Learning. Ph.D. Thesis, Department of Computer Science, Georgia State University, Atlanta, GA, USA, 27 November 2007. [Google Scholar]
- Gavrilov, V. Benefits of Decision Trees in Solving Predictive Analytics Problems. Available online: http://www.prognoz.com/blog/platform/benefits-of-decision-trees-in-solving-predictive-analytics-problems/ (accessed on 20 July 2017).
- Guo, G.; Wang, H.; Bell, D.; Bi, Y.; Greer, K. KNN Model-Based Approach in Classification. In Proceedings of the OTM Confederated International Conferences, On the Move to Meaningful Internet System, Catania, Italy, 3–7 November 2003; pp. 986–996. [Google Scholar]
- Bin Basir, M.A.; Binti Ahmad, F. Attribute Selection-based Ensemble Method for Dataset Classification. Int. J. Comput. Sci. Electron. Eng. 2016, 4, 70–74. [Google Scholar]
- Polikar, R. Ensemble Learning. Available online: http://www. scholarpedia.org/article/Ensemble_learning (accessed on 23 April 2017).
- Ruta, D.; Gabrys, B. Classifier selection for majority voting. Inf. Fusion 2005, 6, 63–81. [Google Scholar] [CrossRef]
- Breiman, L. Bagging predictors. Mach. Learn. 1996, 24, 123–140. [Google Scholar] [CrossRef]
- Freund, Y.; Schapire, R.E. A decision-theoretic generalization of on-line learning and an application to boosting. J. Comput. Syst. Sci. 1977, 55, 119–139. [Google Scholar] [CrossRef]
- Ozcift, A.; Gulten, A. A Robust Multi-Class Feature Selection Strategy Based on Rotation Forest Ensemble Algorithm for Diagnosis. J. Med. Syst. 2012, 36, 941–949. [Google Scholar] [CrossRef] [PubMed]
- Saeys, Y.; Abeel, T.; Peer, Y.V. Robust Feature Selection Using Ensemble Feature Selection Techniques. In Proceedings of the Joint European Conference on Machine Learning and Knowledge Discovery in Databases (ECML PKDD 2008), Antwerp, Belgium, 15–19 September 2008; pp. 313–325. [Google Scholar]
- Shams, R. Micro- and Macro-average of Precision, Recall and F-Score. Available online: http://rushdishams.blogspot.com/2011/08/micro-and-macro-average-of-precision.html (accessed on 15 June 2017).
- Albert, J. How to Build a Confusion Matrix for a Multiclass Classifier. Available online: https://stats.stackexchange.com/questions/179835/how-to-build-a-confusion-matrix-for-a-multiclass-classifier (accessed on 20 May 2018).
- Sokolova, M.; Lapalme, G. A systematic analysis of performance measures for classification tasks. Inf. Process. Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]
- Caballero, J.C.F.; Martínez, F.J.; Hervás, C.; Gutiérrez, P.A. Sensitivity Versus Accuracy in Multiclass Problems Using Memetic Pareto Evolutionary Neural Networks. IEEE Trans. Neural Netw. 2010, 21, 750–770. [Google Scholar] [CrossRef]
- Hazra, A. Using the confidence interval confidently. J. Thorac. Dis. 2017, 9, 4125–4130. [Google Scholar] [CrossRef]
| Algorithm | Abbreviation |
|---|---|
| Class-independent with filter by IG [35] | IG-CID |
| Class-independent with wrapper by SFS [10] | SFS-CID |
| Class-independent with hybrid feature selection method (IG+SFS) [30] | IGSFS-CID |
| Class-dependent with filter by IG | IG-CD |
| Class-dependent with wrapper by SFS | SFS-CD |
| Class-dependent with hybrid feature selection method (IG+SFS) | * IGSFS-CD |
| Dataset | No. of Features | No. of Instances | No. of Classes | No. of Instances Per Class | Type of Features |
|---|---|---|---|---|---|
| LC_dataset-1 | 56 | 264 | 4 | 13, 33, 44, 174 | Nominal, Numeric |
| LC_dataset-II | 117 | 79 | 4 | 8, 16, 7, 48 | Nominal, Numeric |
| Predicted Class | |||||
|---|---|---|---|---|---|
| Class 1 | Class 2 | … | Class k | ||
| True Class/Actual Class | Class1 | C11 | C12 | … | C1k |
| Class2 | C21 | C22 | … | C2k | |
| … | … | … | … | … | |
| Class k | Ck1 | Ck2 | … | Ckk | |
| Subset Evaluation with Decision Tree (DT) | Subset Evaluation with Naïve Bayes (NB) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Method | No. of Selected Features | Ensemble Classifiers | Bagging | AdaBoost | Avg * | No. of Selected Features | Ensemble Classifiers | Bagging | AdaBoost | Avg * |
| Without feature selection | 56 | 72.35 | 68.86 | 70.77 | 70.66 | 56 | 72.35 | 68.86 | 70.77 | 70.66 |
| IG-CID | 35 | 75.40 | 71.14 | 69.25 | 71.93 | 35 | 75.40 | 71.88 | 69.25 | 72.18 |
| SFS-CID | 6 | 71.98 | 75.38 | 75.77 | 74.38 | 6 | 78.36 | 76.10 | 75.36 | 76.61 |
| IGSFS-CID | 4 | 74.20 | 76.14 | 76.13 | 75.49 | 4 | 78.35 | 75.73 | 76.10 | 76.73 |
| IG-CD | 41 | 74.62 | 69.60 | 68.87 | 71.03 | 41 | 74.99 | 70.34 | 69.25 | 71.53 |
| SFS-CD | 6 | 74.94 | 78.02 | 76.88 | 76.61 | 11 | 74.22 | 72.68 | 73.40 | 73.43 |
| IGSFS-CD | 7 | 75.70 | 76.88 | 78.36 | 76.98 | 14 | 76.87 | 73.03 | 74.57 | 74.82 |
| Subset Evaluation with Decision Tree (DT) | Subset Evaluation with Naïve Bayes (NB) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | Ensemble Classifiers | Bagging | AdaBoost | Ensemble Classifiers | Bagging | AdaBoost | ||||||
| Snµ | SnM | Snµ | SnM | Snµ | SnM | Snµ | SnM | Snµ | SnM | Snµ | SnM | |
| Without feature selection | 0.7235 | 0.4814 | 0.6894 | 0.4348 | 0.7083 | 0.4322 | 0.7235 | 0.4814 | 0.6894 | 0.4348 | 0.7083 | 0.4322 |
| IG-CID | 0.7538 | 0.5774 | 0.7121 | 0.4458 | 0.6932 | 0.4340 | 0.7538 | 0.5774 | 0.7197 | 0.4486 | 0.6932 | 0.4340 |
| SFS-CID | 0.7197 | 0.5832 | 0.7538 | 0.5209 | 0.7576 | 0.5382 | 0.7841 | 0.5977 | 0.7614 | 0.5469 | 0.7538 | 0.5391 |
| IGSFS-CID | 0.7424 | 0.5600 | 0.7614 | 0.5458 | 0.7614 | 0.5475 | 0.7841 | 0.6290 | 0.7576 | 0.5393 | 0.7614 | 0.5659 |
| IG-CD | 0.7462 | 0.5487 | 0.6970 | 0.4376 | 0.6894 | 0.4032 | 0.7500 | 0.5502 | 0.7045 | 0.4405 | 0.6932 | 0.4046 |
| SFS-CD | 0.7500 | 0.5886 | 0.7803 | 0.6056 | 0.7689 | 0.5725 | 0.7424 | 0.5500 | 0.7273 | 0.4913 | 0.7348 | 0.4889 |
| IGSFS-CD | 0.7576 | 0.6043 | 0.7689 | 0.5555 | 0.7841 | 0.6047 | 0.7689 | 0.5855 | 0.7311 | 0.4699 | 0.7462 | 0.4858 |
| Subset Evaluation with Decision Tree (DT) | Subset Evaluation with Naïve Bayes (NB) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | Ensemble Classifiers | Bagging | AdaBoost | Ensemble Classifiers | Bagging | AdaBoost | ||||||
| Spµ | SpM | Spµ | SpM | Spµ | SpM | Spµ | SpM | Spµ | SpM | Spµ | SpM | |
| Without feature selection | 0.8870 | 0.8120 | 0.8694 | 0.8313 | 0.8793 | 0.7980 | 0.8870 | 0.8120 | 0.8694 | 0.8313 | 0.8793 | 0.7980 |
| IG-CID | 0.9018 | 0.8542 | 0.8813 | 0.8164 | 0.8714 | 0.7915 | 0.9018 | 0.8542 | 0.8851 | 0.8194 | 0.8714 | 0.7915 |
| SFS-CID | 0.8851 | 0.8551 | 0.9018 | 0.8371 | 0.9036 | 0.8436 | 0.9159 | 0.8754 | 0.9054 | 0.8530 | 0.9018 | 0.8470 |
| IGSFS-CID | 0.8963 | 0.8407 | 0.9054 | 0.8439 | 0.9054 | 0.8435 | 0.9159 | 0.8766 | 0.9036 | 0.8502 | 0.9054 | 0.8528 |
| IG-CD | 0.8982 | 0.8468 | 0.8734 | 0.8250 | 0.8694 | 0.8194 | 0.9000 | 0.8483 | 0.8774 | 0.8111 | 0.8714 | 0.7895 |
| SFS-CD | 0.9000 | 0.8469 | 0.9142 | 0.8615 | 0.9090 | 0.8494 | 0.8963 | 0.8457 | 0.8889 | 0.8341 | 0.8926 | 0.8245 |
| IGSFS-CD | 0.9036 | 0.8545 | 0.9090 | 0.8517 | 0.9159 | 0.8609 | 0.9090 | 0.8603 | 0.8908 | 0.8370 | 0.8982 | 0.8302 |
| Subset Evaluation with Decision Tree (DT). | Subset Evaluation with Naïve Bayes (NB) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Method | No. of Selected Features | Ensemble Classifiers | Bagging | AdaBoost | Avg * | No. of Selected Features | Ensemble Classifiers | Bagging | AdaBoost | Avg * |
| Without feature selection | 117 | 75.71 | 67.32 | 73.39 | 72.14 | 117 | 75.71 | 67.32 | 73.39 | 72.14 |
| IG-CID | 40 | 82.32 | 75.00 | 72.14 | 76.49 | 30 | 80.71 | 72.50 | 70.89 | 74.70 |
| SFS-CID | 5 | 81.07 | 72.32 | 77.32 | 76.90 | 7 | 83.57 | 65.89 | 73.39 | 74.28 |
| IGSFS-CID | 5 | 81.07 | 72.32 | 77.32 | 76.90 | 7 | 84.64 | 73.39 | 77.14 | 78.39 |
| IG-CD | 102 | 80.71 | 68.57 | 74.64 | 74.64 | 102 | 79.46 | 68.57 | 74.82 | 74.28 |
| SFS-CD | 6 | 83.39 | 75.89 | 78.39 | 79.22 | 14 | 82.14 | 72.32 | 69.64 | 74.70 |
| IGSFS-CD | 6 | 83.39 | 75.89 | 78.39 | 79.22 | 14 | 84.82 | 69.82 | 74.82 | 76.49 |
| Subset Evaluation with Decision Tree (DT) | Subset Evaluation with Naïve Bayes (NB) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | Ensemble Classifiers | Bagging | AdaBoost | Ensemble Classifiers | Bagging | AdaBoost | ||||||
| Snµ | SnM | Snµ | SnM | Snµ | SnM | Snµ | SnM | Snµ | SnM | Snµ | SnM | |
| Without feature selection | 0.7595 | 0.5149 | 0.6709 | 0.3906 | 0.7342 | 0.4837 | 0.7595 | 0.5149 | 0.6709 | 0.3906 | 0.7342 | 0.4837 |
| IG-CID | 0.8228 | 0.6808 | 0.7468 | 0.5097 | 0.7215 | 0.4740 | 0.8101 | 0.6087 | 0.7215 | 0.4732 | 0.7089 | 0.4531 |
| SFS-CID | 0.8101 | 0.7016 | 0.7215 | 0.4889 | 0.7722 | 0.5781 | 0.8354 | 0.6615 | 0.6582 | 0.3594 | 0.7342 | 0.5156 |
| IGSFS-CID | 0.8101 | 0.7016 | 0.7215 | 0.4889 | 0.7722 | 0.5781 | 0.8481 | 0.6712 | 0.7342 | 0.4531 | 0.7722 | 0.5469 |
| IG-CD | 0.8101 | 0.6087 | 0.6835 | 0.4219 | 0.7468 | 0.5253 | 0.7975 | 0.5930 | 0.6835 | 0.4115 | 0.7468 | 0.5305 |
| SFS-CD | 0.8354 | 0.6458 | 0.7595 | 0.5261 | 0.7848 | 0.6250 | 0.8228 | 0.6198 | 0.7215 | 0.4948 | 0.6962 | 0.4271 |
| IGSFS-CD | 0.8354 | 0.6458 | 0.7595 | 0.5261 | 0.7848 | 0.6250 | 0.8481 | 0.7173 | 0.6962 | 0.4375 | 0.7468 | 0.5469 |
| Subset Evaluation with Decision Tree (DT) | Subset Evaluation with Naïve Bayes (NB) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | Ensemble Classifiers | Bagging | AdaBoost | Ensemble Classifiers | Bagging | AdaBoost | ||||||
| Spµ | SpM | Spµ | SpM | Spµ | SpM | Spµ | SpM | Spµ | SpM | Spµ | SpM | |
| Without feature selection | 0.9045 | 0.8479 | 0.8595 | 0.7695 | 0.8923 | 0.8299 | 0.9045 | 0.8479 | 0.8595 | 0.7695 | 0.8923 | 0.8299 |
| IG-CID | 0.9330 | 0.9125 | 0.8985 | 0.8436 | 0.8860 | 0.8254 | 0.9275 | 0.8938 | 0.8860 | 0.8291 | 0.8796 | 0.8197 |
| SFS-CID | 0.9275 | 0.9098 | 0.8860 | 0.8292 | 0.9104 | 0.8674 | 0.9384 | 0.9164 | 0.8525 | 0.7681 | 0.8923 | 0.8249 |
| IGSFS-CID | 0.9275 | 0.9098 | 0.8860 | 0.8292 | 0.9104 | 0.8674 | 0.9437 | 0.9210 | 0.8923 | 0.8424 | 0.9104 | 0.8736 |
| IG-CD | 0.9275 | 0.8832 | 0.8663 | 0.7733 | 0.8985 | 0.8454 | 0.9220 | 0.8745 | 0.8663 | 0.7978 | 0.8985 | 0.8390 |
| SFS-CD | 0.9384 | 0.9260 | 0.9045 | 0.8812 | 0.9163 | 0.9022 | 0.9330 | 0.9003 | 0.8860 | 0.8337 | 0.8730 | 0.8031 |
| IGSFS-CD | 0.9384 | 0.9260 | 0.9045 | 0.8812 | 0.9163 | 0.9022 | 0.9437 | 0.9273 | 0.8730 | 0.8101 | 0.8985 | 0.8480 |
| IGSFS-CD with DT | IGSFS-CD with NB | ||||
|---|---|---|---|---|---|
| Class | Selected Features | No. of Features | Class | Selected Features | No. of Features |
| LC_dataset-1 | LC_dataset-1 | ||||
| Class1 | T | 1 | Class1 | T, LATER, PROC1, IM001, IM008 | 5 |
| Class2 | AGE | 1 | Class2 | SDX9 | 1 |
| Class3 | BASIC | 1 | Class3 | MOR, PROC2, PDX2, SDX6, BC005 | 5 |
| Class4 | M, EXT, BC006, BC009 | 4 | Class4 | T, EXT, MOR, GRADE, PROC2, IM008, BC006 | 7 |
| LC_dataset-II | LC_dataset-II | ||||
| Class1 | T | 1 | Class1 | T, IM008 | 2 |
| Class2 | AGE | 1 | Class2 | BASIC, CHEM, MET, N, BC009, BC015 | 6 |
| Class3 | AGE | 1 | Class3 | T | 1 |
| Class4 | EXT, SURG, IM011, 60_list | 4 | Class4 | T, EXT, BASIC, LATER, RADI, MET, PDX2, SDX3, 57_list | 9 |
| Method | Accuracy (95%CI) | Sensitivity (95%CI) | Specificity (95%CI) |
|---|---|---|---|
| Without feature selection | 70.66 (69.66; 71.66) | 0.5783 (0.4983; 0.6583) | 0.8462 (0.8262; 0.8662) |
| IG-CID | 72.05 (70.05; 74.05) | 0.6036 (0.5236; 0.6836) | 0.8533 (0.8333; 0.8733) |
| SFS-CID | 75.49 (73.49; 77.49) | 0.6547 (0.5947; 0.7147) | 0.8771 (0.8571; 0.8971) |
| IGSFS-CID | 76.11 (75.11; 77.11) | 0.6630 (0.6030; 0.7230) | 0.8783 (0.8583; 0.8983) |
| IG-CD | 71.28 (69.28; 73.28) | 0.5888 (0.5088; 0.6688) | 0.8525 (0.8325; 0.8725) |
| SFS-CD | 75.02 (73.02; 77.02) | 0.6501 (0.5901; 0.7101) | 0.8719 (0.8519; 0.8919) |
| IGSFS-CD | 75.90 (74.90; 76.09) | 0.6552 (0.5852; 0.7252) | 0.8768 (0.8568; 0.8968) |
| Method | Accuracy (95%CI) | Sensitivity (95%CI) | Specificity (95%CI) |
|---|---|---|---|
| Without feature selection | 72.14 (69.14; 75.14) | 0.5923 (0.5123; 0.6723) | 0.8506 (0.8260; 0.8806) |
| IG-CID | 75.59 (71.59; 79.59) | 0.6443 (0.5643; 0.7243) | 0.8779 (0.8579; 0.8979) |
| SFS-CID | 75.59 (70.59; 80.59) | 0.6531 (0.5731; 0.7331) | 0.8769 (0.8469; 0.9096) |
| IGSFS-CID | 77.65 (73.65; 81.65) | 0.6748 (0.6048; 0.7448) | 0.8928 (0.8728; 0.9128) |
| IG-CD | 74.46 (70.46; 78.46) | 0.6299 (0.5499; 0.7099) | 0.8660 (0.8360; 0.8960) |
| SFS-CD | 76.96 (72.63; 81.29) | 0.6632 (0.5932; 0.7332) | 0.8915 (0.8715; 0.9115) |
| IGSFS-CD | 77.86 (73.86; 81.86) | 0.6808 (0.6108; 0.7508) | 0.8974 (0.8774; 0.9174) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panthong, R.; Srivihok, A. Liver Cancer Classification Model Using Hybrid Feature Selection Based on Class-Dependent Technique for the Central Region of Thailand. Information 2019, 10, 187. https://doi.org/10.3390/info10060187
Panthong R, Srivihok A. Liver Cancer Classification Model Using Hybrid Feature Selection Based on Class-Dependent Technique for the Central Region of Thailand. Information. 2019; 10(6):187. https://doi.org/10.3390/info10060187
Chicago/Turabian StylePanthong, Rattanawadee, and Anongnart Srivihok. 2019. "Liver Cancer Classification Model Using Hybrid Feature Selection Based on Class-Dependent Technique for the Central Region of Thailand" Information 10, no. 6: 187. https://doi.org/10.3390/info10060187
APA StylePanthong, R., & Srivihok, A. (2019). Liver Cancer Classification Model Using Hybrid Feature Selection Based on Class-Dependent Technique for the Central Region of Thailand. Information, 10(6), 187. https://doi.org/10.3390/info10060187






