Proteome Response of Meretrix Bivalves Hepatopancreas Exposed to Paralytic Shellfish Toxins Producing Dinoflagellate Gymnodinium catenatum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Animals
2.2. Cultivation of Microalgae
2.3. Experimental Design
2.4. Protein Extraction from Hepatopancreas
2.5. Protein Determination
2.6. Toxin Determination
2.7. Two-Dimensional Gel Electrophoresis and Imaging Analysis
2.8. In-Gel Digestion and MALDI-TOP Mass Spectrometry Analysis
2.9. N-Terminal Sulfonation
2.10. Statistical Analysis
3. Results and Discussion
3.1. Toxin Composition in Dinoflagellate and Bivalve
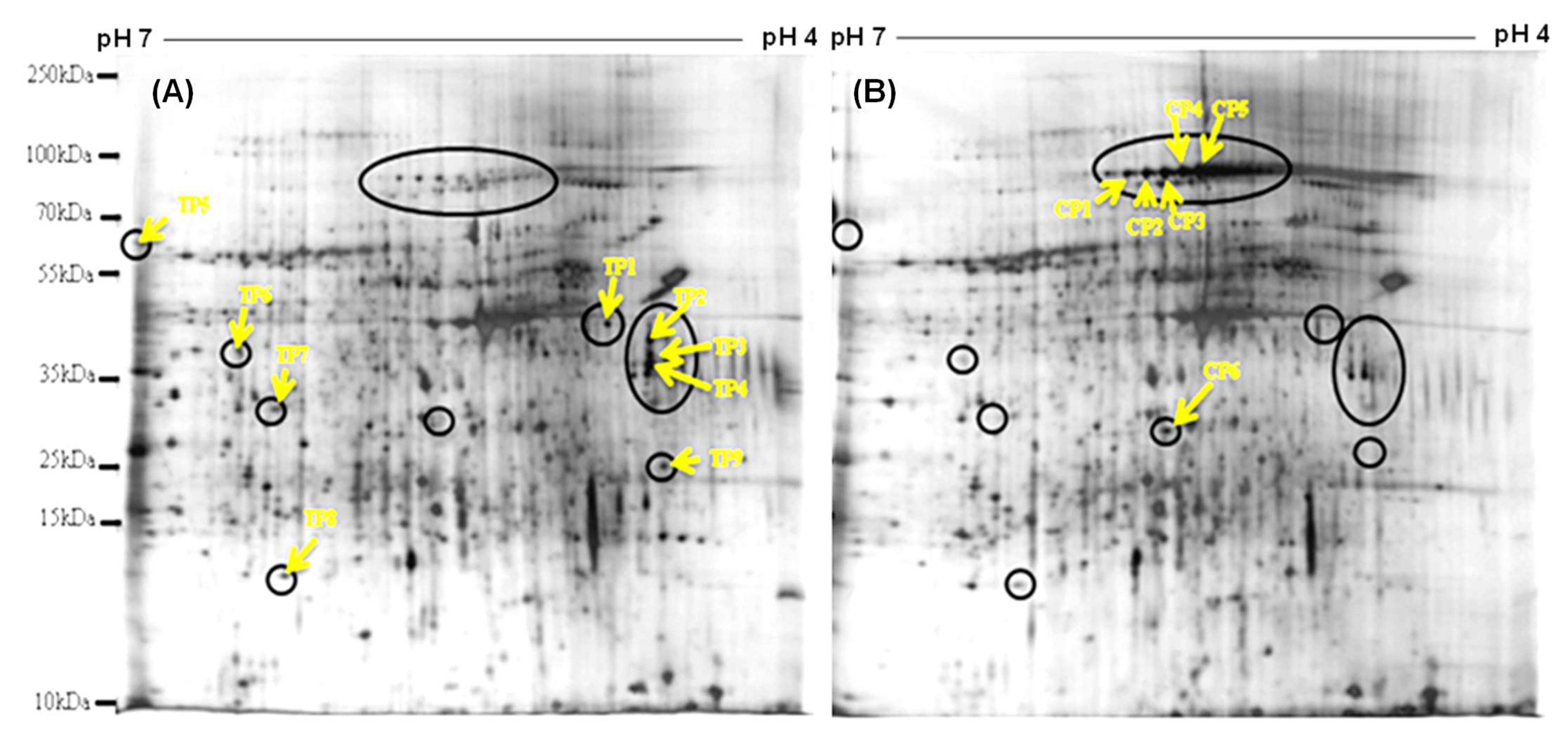
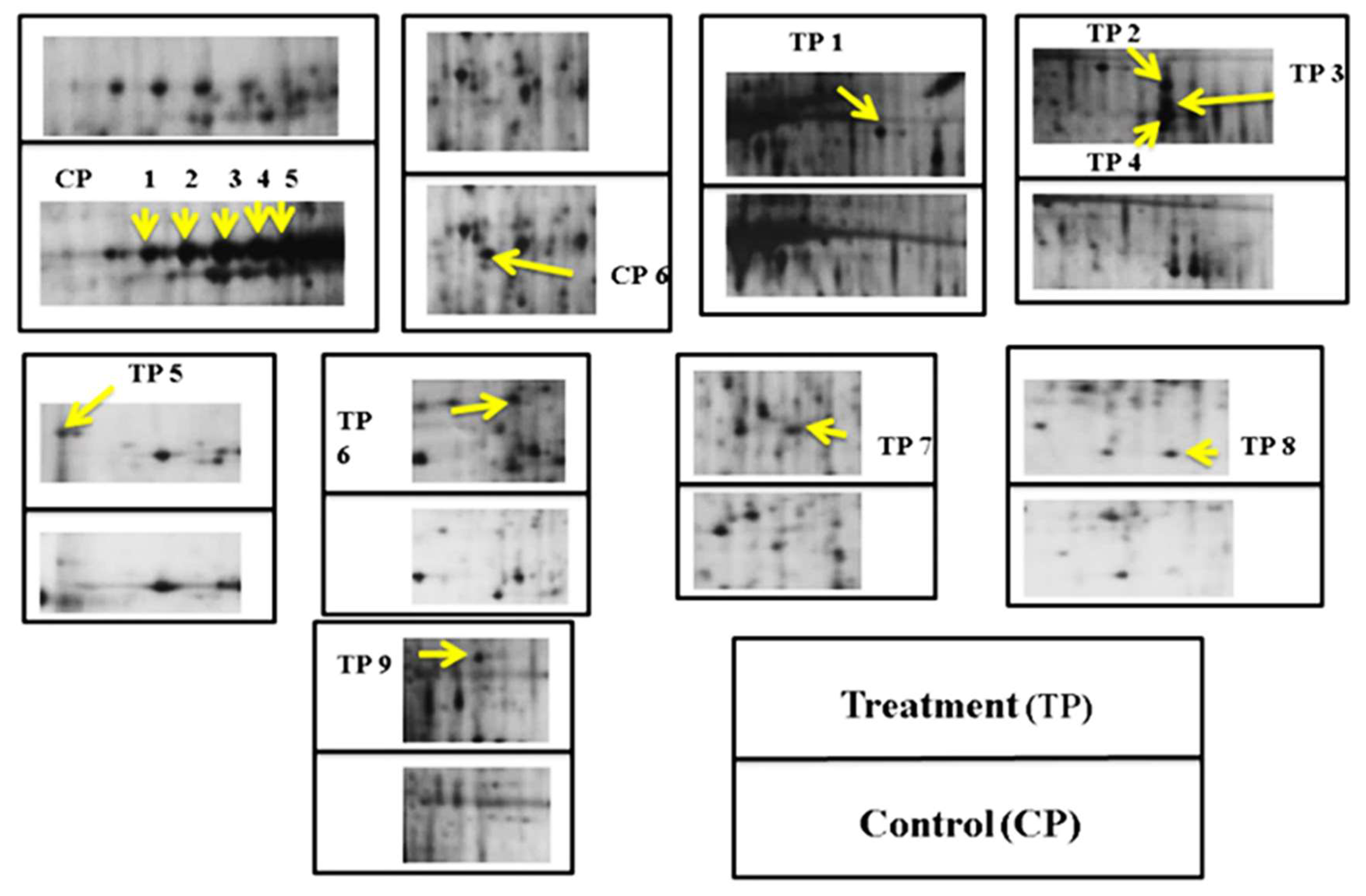
3.2. Protein Profile in the Hepatopancreas of Bivalves
3.3. Biological Functions of the Identified Proteins
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Taylor, S.L. Marine toxins of microbial origin. Food Technol. 1988, 42, 94–98. [Google Scholar]
- Yoosukh, W.; Matsukuma, A. Taxonomic study on Meretrix (Mollusca: Bivalvia) from Thailand. Phuket Mar. Biol. Cent. Spec. Publ. 2001, 25, 451–460. [Google Scholar]
- Hallegraeff, G. Harmful algal blooms: A global overview, in manual on harmful marine microalgae. Monogr. Oceanogr. Methodol 2003, 11, 25–49. [Google Scholar]
- Ho, K.C.; Hodgkiss, I.J. Red tides in sub-tropical waters: An overview of their occurrence. Asian Mar. Biol. 1991, 8, 5–23. [Google Scholar]
- Fleming, L.; Kirkpatrick, B.; Backer, L.; Walsh, C.; Nierenberg, K.; Clark, J.; Reich, A.; Hollenbeck, J.; Benson, J.; Cheng, Y.; et al. Review of Florida red tide and human health effects. Harmful Algae 2011, 10, 224–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLeroy, S. Paralytic shellfish poisoning: Seafood safety and human health perspectives. Toxicon 2010, 56, 108–122. [Google Scholar]
- Daranas, A.; Norte, M.; Fernández, J. Toxic marine microalgae. Toxicon 2001, 39, 1101–1132. [Google Scholar] [CrossRef]
- Thottumkara, A.; Parsons, W.; Bois, J. Saxitoxin. Angew. Chem. Int. Ed. 2014, 53, 5760–5784. [Google Scholar] [CrossRef] [PubMed]
- Asakawa, M.; Miyazawa, K.; Takayama, H.; Noguchi, T. Dinoflagellate Alexandrium tamarense as the source of Paralytic shellfish poison (PSP) contained in bivalves from Hiroshima Bay, Hiroshima Prefecture, Japan. Toxicon 1995, 33, 691–697. [Google Scholar] [CrossRef]
- Estrada, N.; Lagos, N.; Garcia, C.; Maeda-Martinez, A.; Ascencio, F. Effects of the toxic dinoflagellate Gymnodinium catenatum on uptake and fate of paralytic shellfish poisons in the Pacific giant lions-paw scallop Nodipecten subnodosus. Mar. Biol. 2007, 151, 1205–1214. [Google Scholar] [CrossRef]
- Zhang, F.; Xu, X.; Li, T.; Liu, Z. Shellfish toxins targeting voltage-gated sodium channels. Mar. Drugs 2013, 11, 4698–4723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bricelj, V.; Shumway, S. Paralytic shellfish toxins in bivalve molluscs: Occurrence, transfer kinetics, and biotransformation. Rev. Fish Sci. 1998, 6, 315–383. [Google Scholar] [CrossRef]
- Bricelj, V.M.; Lee, J.H.; Cembella, A.D.; Anderson, D.M. Uptake kinetics of paralytic shellfish toxins from the dinoflagellate Alexandrium fundyense in the mussel Mytilus edulis. Mar. Ecol. Progs. Ser. 1990, 63, 177–188. [Google Scholar] [CrossRef]
- Xie, W.; Liu, X.; Yang, X.; Zhang, C.; Bian, Z. Accumulation and depuration of paralytic shellfish poisoning toxins in the oyster Ostrea rivularis Gould—Chitosan facilitates the toxin depuration. Food Control 2013, 30, 446–452. [Google Scholar] [CrossRef]
- Li, S.-C.; Hsieh, D. Feeding and absorption of the toxic dinoflagellate Alexandrium tamarense by two marine bivalves from the South China Sea. Mar. Biol. 2001, 139, 617–624. [Google Scholar] [CrossRef]
- Mohamad, S.; Takatani, T.; Yamaguchi, Y.; Sagara, T.; Noguchi, T.; Arakawa, O. Accumulation and elimination profiles of paralytic shellfish poison in the short-necked clam tapes japonica fed with the toxic dinoflagellate Gymnodinium catenatum. Shokuhin Eiseigaku Zasshi J. Food Hyg. Soc. Jpn. 2007, 48, 13–18. [Google Scholar]
- Band-Schmidt, C.; Bustillos, J.; Gárate-Lizárraga, I.; Lechuga-Devéze, C.; Erler, K.; Luckas, B. Paralytic shellfish toxin profile in strains of the dinoflagellate Gymnodinium catenatum Graham and the scallop Argopecten ventricosus G.B. Sowerby II from Bahía Concepción, Gulf of California, Mexico. Harmful Algae 2005, 4, 21–31. [Google Scholar] [CrossRef]
- Mohamad, S.; Yamaguchi, Y.; Sagara, T.; Takatani, T.; Arakawa, O.; Noguchi, T. Accumulation and depuration profiles of PSP toxins in the short-necked clam Tapes japonica fed with the toxic dinoflagellate Alexandrium catenella. Toxicon 2006, 48, 323–330. [Google Scholar]
- Sassolas, A.; Hayat, A.; Catanante, G.; Marty, J. Detection of the marine toxin okadaic acid: Assessing seafood safety. Talanta 2013, 105, 306–316. [Google Scholar] [CrossRef]
- Cao, J.; Jiang, Z.; Bing, Y.; Wang, Q.; Xu, J.; Aifeng, L. Evaluation of mouse bioassay results in an inter-laboratory comparison for paralytic shellfish poisoning toxins. Chin. J. Oceanol. Limnol. 2011, 29, 912–916. [Google Scholar] [CrossRef]
- Oshima, Y. Postcolumn derivation liquid chromatographic method for paralytic shellfish toxins. J. AOAC Int. 1995, 88, 1714–1732. [Google Scholar]
- Ronzitti, G.; Milandri, A.; Scortichini, G.; Poletti, R.; Rossini, G. Protein markers of algal toxin contamination in shellfish. Toxicon 2008, 52, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Campos, A.; Puerto, M.; Prieto, A.; Cameán, A.; Almeida, A.; Coelho, A.; Vasconcelos, V. Protein Extraction and Two-dimensional gel electrophoresis of proteins in the marine mussel Mytilus galloprovincialis: An important tool for protein expression studies, food quality and safety assessment. J. Sci. Food Agric. 2013, 93, 1779–1787. [Google Scholar] [CrossRef] [PubMed]
- Campos, A.; Tedesco, S.; Vasconcelos, V.; Cristobal, S. Proteomic research in bivalves: Towards the identification of molecular markers of aquatic pollution. J. Proteom. 2012, 75, 4346–4359. [Google Scholar] [CrossRef] [PubMed]
- López-Pedrouso, M.; Varela, Z.; Franco, D.; Fernández, J.A.; Aboal, J.R. Can proteomics contribute to biomonitoring of aquatic pollution? A critical review. Environ. Pollut. 2020, 267, 115473. [Google Scholar] [CrossRef]
- Puerto, M.; Campos, A.; Prieto, A.; Cameán, A.; de Almeida, A.M.; Coelho, A.V.; Vasconcelos, V. Differential protein expression in two bivalve species; Mytilus galloprovincialis and Corbicula fluminea; exposed to Cylindrospermopsis raciborskii cells. Aquat. Toxicol. 2011, 101, 109–116. [Google Scholar] [CrossRef]
- Gomes, T.; Chora, S.; Guerreiro Pereira, C.; Cardoso, C.; Bebianno, M. Proteomic response of mussels Mytilus galloprovincialis exposed to CuO NPs and Cu2+: An exploratory biomarker discovery. Aquat. Toxicol. 2014, 155, 327–336. [Google Scholar] [CrossRef]
- Corporeau, C.; Tamayo, D.; Pernet, F.; Quéré, C.; Madec, S. Proteomic signatures of the oyster metabolic response to herpesvirus OsHV-1 μVar infection. J. Proteom. 2014, 109, 176–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, C.; Wu, H.; Wei, L.; Zhao, J.; Wang, Q.; Lu, H. Responses of Mytilus galloprovincialis to bacterial challenges by metabolomics and proteomics. Fish Shellfish Immunol. 2013, 35, 489–498. [Google Scholar] [CrossRef]
- Fernández-Boo, S.; Villalba, A.; Cao, A. Protein expression profiling in haemocytes and plasma of the Manila clam Ruditapes philippinarum in response to infection with Perkinsus olseni. J. Fish Dis. 2016, 39, 1369–1385. [Google Scholar] [CrossRef]
- Gu, H.; Wu, Y.; Lu, S.; Lu, D.; Tang, Y.Z.; Qi, Y. Emerging harmful algal bloom species over the last four decades in China. Harmful Algae 2021, 102059. [Google Scholar] [CrossRef]
- Zhang, C.; Lim, P.T.; Li, X.; Gu, H.; Li, X.; Anderson, D. Wind-driven development and transport of Gymnodinium catenatum blooms along the coast of Fujian, China. Reg. Stud. Mar. Sci. 2020, 39, 101397. [Google Scholar] [CrossRef] [PubMed]
- Sierra-Beltrán, A.P.; Lluch-Cota, D.B.; Lluch-Cota, S.E.; Cortés-Altamirano, R.; Cortés-Lara, M.C.; Castillo-Chávez, M.; Carrillo, L.; Pacas, L.; Víquez, R.; García-Hansen, I. Spatial-temporal dynamics of red tide precursor organisms at the Pacific coast of North and Central America. Rev. Biol. Trop. 2004, 52 (Suppl. 1), 99–107. [Google Scholar]
- Xia, L.; Chen, S.; Dahms, H.U.; Ying, X.; Peng, X. Cadmium induced oxidative damage and apoptosis in the hepatopancreas of Meretrix meretrix. Ecotoxicology 2016, 25, 959–969. [Google Scholar] [CrossRef]
- Chen, L.S.; Wang, X.B.; Chen, D.Q. Research on the development of hard clam market in China mainland. J. Shanghai Ocean Univ. 2004, 283–287. [Google Scholar]
- Ho, J.S.; Zheng, G.X. Ostrincola koe (Copepoda, Myicolidae) and mass mortality of cultured hard clam (Meretrix meretrix) in China. Hydrobiologia 1994, 284, 169–173. [Google Scholar] [CrossRef]
- Li, H.; Liu, W.; Gao, X.G.; Zhu, D.; Wang, J.; Li, Y.F.; He, C.B. Identification of host-defense genes and development of microsatellite markers from ESTs of hard clam Meretrix meretrix. Mol. Biol. Rep. 2010, 38, 769–775. [Google Scholar] [CrossRef]
- Haoujar, I.; Abrini, J.; Chadli, H.; Essafi, A.; Nhhala, H.; Chebbaki, K.; Cacciola, F.; Skali-Senhaji, N. Effect of four diets based on three microalgae on the growth performance and quality of Mediterranean mussel flesh, Mytilus galloprovincialis. Int. Aquat. Res. 2020, 12, 137–145. [Google Scholar]
- Choi, N.; Yeung, L.; Siu, W.; So, I.; Jack, R.; Hsieh, D.; Wu, R.; Lam, P. Relationships between tissue concentrations of paralytic shellfish toxins and antioxidative responses of clams, Ruditapes philippinarum. Mar. Pollut. Bull. 2006, 52, 572–578. [Google Scholar] [CrossRef] [PubMed]
- Lee, F.W.F.; Lo, S.C.L. The use of Trizol reagent (phenol/guanidine isothiocyanate) for producing high quality two-dimensional gel electrophoretograms (2-DE) of dinoflagellates. J. Microbiol. Methods 2008, 73, 26–32. [Google Scholar] [CrossRef]
- Ramagli, L.; Rodriguez, L. Quantitation of Microgram Amounts of Protein in two-dimensional polyacrylamide-gel electrophoresis sample buffer. Electrophoresis 1985, 6, 559–563. [Google Scholar] [CrossRef]
- Chen, P.; Nie, S.; Mi, W.; Wang, X.C.; Liang, S.P. De Novo Sequencing of tryptic peptides sulfonated by 4-sulfophenyl isothiocyanate for unambiguous protein identification using post-source decay matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2004, 18, 191–198. [Google Scholar] [CrossRef]
- Band-Schmidt, C.J.; Bustillos-Guzmán, J.J.; Hernández-Sandoval, F.E.; Núñez-Vázquez, E.J.; López-Cortés, D.J. Effect of temperature on growth and paralytic toxin profiles in isolates of Gymnodinium catenatum (Dinophyceae) from the Pacific coast of Mexico. Toxicon 2014, 90, 199–212. [Google Scholar] [CrossRef] [PubMed]
- Park, T.G.; Kim, C.H.; Oshima, Y. Paralytic shellfish toxin profiles of different geographic populations of Gymnodinium catenatum (Dinophyceae) in Korean coastal waters. Phycol. Res. 2004, 52, 300–305. [Google Scholar] [CrossRef]
- Holmes, M.J.; Bolch, C.J.; Green, D.H.; Cembella, A.D.; Teo, S.L.M. Singapore isolates of the dinoflagellate Gymnodinium catenatum (Dinophyceae) produce a unique profile of paralytic shellfish poisoning toxins 1. J. Phycol. 2002, 38, 96–106. [Google Scholar] [CrossRef]
- Seok Jin, O.; Matsuyama, Y.; Yoon, Y.H.; Miyamura, K.; Choi, C.G.; Yang, H.S.; Kang, I.J. Comparative analysis of paralytic shellfish toxin content and profile produced by dinoflagellate Gymnodinium catenatum isolated from Inokushi Bay, Japan. J. Fac. Agric. Kyushu 2010, 55, 47–54. [Google Scholar]
- Band-Schmidt, C.; Bustillos-Guzmán, J.; Morquecho, L.; Gárate-Lizárraga, I.; Alonso-Rodríguez, R.; Reyes-Salinas, A.; Erler, K.; Luckas, B. Variations of psp toxin profiles during different growth phases in Gymnodinium catenatum (dinophyceae) strains isolated from three locations in the Gulf of California, Mexico. J. Phycol. 2006, 42, 757–768. [Google Scholar] [CrossRef]
- Bustillos-Guzman, J. Variations in growth and toxicity in Gymnodinium catenatum Graham from the Gulf of California under different ratios of nitrogen and phosphorus. Ciencias Marinas 2012, 38, 101–117. [Google Scholar] [CrossRef]
- Oshima, Y.; Blackburn, S.I.; Hallegraeff, G. Comparative study on paralytic shellfish toxin profiles of the dinoflagellate Gymnodinium catenatum from three different countries. Mar. Biol. 1993, 116, 471–476. [Google Scholar] [CrossRef]
- Kwong, R.; Lam, P.; Yu, P. The uptake, distribution and elimination of paralytic shellfish toxins in mussels and fish exposed to toxic dinoflagellates. Aquat. Toxicol. 2006, 80, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Wekell, J.C.; Hurst, J.; Lefebvre, K. The origin of the regulatory limits for PSP and ASP toxins in shellfish. J. Shellfish Res. 2004, 23, 927–930. [Google Scholar]
- Reqia, S.; Amanhir, R.; Taleb, H.; Vale, P.; Blaghen, M.; Loutfi, M. Comparative study on differential accumulation of PSP toxins between cockle (Acanthocardia tuberculatum) and sweet clam (Callista chione). Toxicon 2005, 46, 612–618. [Google Scholar]
- Yasmeen, S. Changes in hepatopancreas of the bivalve molluscs Lamellidens marginalis exposed to acute toxicity of cadmium in summer. World J. Fish Mar. Sci. 2013, 5, 437–440. [Google Scholar]
- Qiu, J.; Ma, F.; Fan, H.; Aifeng, L. Effects of feeding Alexandrium tamarense, a paralytic shellfish toxin producer, on antioxidant enzymes in scallops (Patinopecten yessoensis) and mussels (Mytilus galloprovincialis). Aquaculture 2013, 396, 76–81. [Google Scholar] [CrossRef]
- Rőszer, T. The invertebrate midintestinal gland (“hepatopancreas”) is an evolutionary forerunner in the integration of immunity and metabolism. Cell Tissue Res. 2014, 358, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Braga, A.C.; Pereira, V.; Marçal, R.; Marques, A.; Guilherme, S.; Costa, P.R.; Pacheco, M. DNA damage and oxidative stress responses of mussels Mytilus galloprovincialis to paralytic shellfish toxins under warming and acidification conditions—Elucidation on the organ-specificity. Aquat. Toxicol. 2020, 228, 105619. [Google Scholar] [CrossRef] [PubMed]
- Martins, J.C.; Leão, P.N.; Vasconcelos, V. Differential protein expression in Corbicula fluminea upon exposure to a Microcystis aeruginosa toxic strain. Toxicon 2009, 53, 409–416. [Google Scholar] [CrossRef]
- Huang, L.; Zou, Y.; Weng, H.W.; Li, H.Y.; Liu, J.S.; Yang, W.D. Proteomic profile in Perna viridis after exposed to Prorocentrum lima, a dinoflagellate producing DSP toxins. Environ. Pollut. 2015, 196, 350–357. [Google Scholar] [CrossRef]
- Wu, H.; Ji, C.; Wei, L.; Zhao, J.; Lu, H. Proteomic and metabolomic responses in hepatopancreas of Mytilus galloprovincialis challenged by Micrococcus luteus and Vibrio anguillarum. J. Proteom. 2013, 94, 54–67. [Google Scholar] [CrossRef]
- Bourret, R. Receiver domain structure and function in response regulator proteins. Curr. Opin. Microbiol. 2010, 13, 142–149. [Google Scholar] [CrossRef] [Green Version]
- Wolanin, P.; Thomason, P.; Stock, J. Histidine protein kinases: Key signal transducers outside the animal kingdom. Genome Biol. 2002, 3, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roetzer, A.; Klopf, E.; Gratz, N.; Marcet-Houben, M.; Hiller, E.; Rupp, S.; Gabaldón, T.; Kovarik, P.; Schüller, C. Regulation of Candida glabrata oxidative stress resistance is adapted to host environment. FEBS Lett. 2011, 585, 319–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jack, D.L.; Yang, N.M.; Saier, M.H., Jr. The drug/metabolite transporter superfamily. Eur. J. Biochem. 2001, 268, 3620–3639. [Google Scholar] [CrossRef]
- Pazos, A.J.; Ventoso, P.; Martínez-Escauriaza, R.; Pérez-Parallé, M.L.; Blanco, J.; Triviño, J.C.; Sánchez, J.L. Transcriptional response after exposure to domoic acid-producing Pseudo-nitzschia in the digestive gland of the mussel Mytilus galloprovincialis. Toxicon 2017, 140, 60–71. [Google Scholar] [CrossRef]
- Mat, A.; Klopp, C.; Payton, L.; Jeziorski, C.; Chalopin, M.; Amzil, Z.; Tran, D.; Wikfors, G.; Hégaret, H.; Soudant, P.; et al. Oyster transcriptome response to Alexandrium exposure is related to saxitoxin load and characterized by disrupted digestion, energy balance, and calcium and sodium signaling. Aquat. Toxicol. 2018, 199, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Xun, X.; Cheng, J.; Wang, J.; Li, Y.; Li, X.; Moli, L.; Lou, J.; Kong, Y.; Bao, Z.; Hu, X. Solute carriers in scallop genome: Gene expansion and expression regulation after exposure to toxic dinoflagellate. Chemosphere 2019, 241, 124968. [Google Scholar] [CrossRef]
- Hermann, A.; Cox, J. Sarcoplasmic calcium-binding protein. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1995, 111, 337–345. [Google Scholar] [CrossRef]
- Truebano, M.; Burns, G.; Thorne, M.A.S.; Hillyard, G.; Peck, L.S.; Skibinski, D.O.F.; Clark, M.S. Transcriptional response to heat stress in the Antarctic bivalve Laternula elliptica. J. Exp. Mar. Biol. Ecol. 2010, 391, 65–72. [Google Scholar] [CrossRef]
- Toescu, E.C. Hypoxia sensing and pathways of cytosolic Ca2+ increases. Cell Calcium 2004, 36, 187–199. [Google Scholar] [CrossRef]
- Nelson, T.; McEachron, D.; Freedman, W.; Yang, W.-P. Cold acclimation increases gene transcription of two calcium transport molecules, calcium transporting ATPase and parvalbumin beta, in carassius auratus lateral musculature. J. Therm. Biol. 2003, 28, 227–234. [Google Scholar] [CrossRef]
- Lian, S.; Zhao, L.; Xun, X.; Lou, J.; Moli, L.; Li, X.; Wang, S.; Zhang, L.; Hu, X.; Bao, Z. Genome-wide identification and characterization of SODs in zhikong scallop reveals gene expansion and regulation divergence after toxic dinoflagellate exposure. Mar. Drugs 2019, 17, 700. [Google Scholar] [CrossRef] [Green Version]
- Vitolo, M. Decomposition of hydrogen peroxide by catalase. World J. Pharm. Pharm. Sci. 2021, 10, 47. [Google Scholar]
- Garcia Lagunas, N.; Romero-Geraldo, R.; Hernandez Saavedra, N. Genomics study of the exposure effect of Gymnodinium catenatum, a paralyzing toxin producer, on Crassostrea gigas’ defense system and detoxification genes. PLoS ONE 2013, 8, e72323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, J.; Liu, Y.; Yang, Y.; Zhang, H.; Li, Z.; Yang, Q.; Zhang, S.; Zhang, Q.; Liu, X. Molecular characterization and functional analysis of the ultraspiracle (USP) in the oriental fruit moth Grapholita molesta (Lepidoptera: Olethreutidae). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2015, 190, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Grøsvik, B.E.; Jonsson, H.; Rodríguez-Ortega, M.J.; Roepstorff, P.; Goksøyr, A. CYP1A-immunopositive proteins in bivalves identified as cytoskeletal and major vault proteins. Aquat. Toxicol. 2006, 79, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Ortega, M.J.; Grøsvik, B.E.; Rodríguez-Ariza, A.; Goksøyr, A.; López-Barea, J. Changes in protein expression profiles in bivalve molluscs (Chamaelea gallina) exposed to four model environmental pollutants. Proteomics 2003, 3, 1535–1543. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.S.; Li, H.; Song, B.; Liu, X. Polo-like kinase 1 phosphorylation of G2 and S-phase-expressed 1 protein is essential for p53 inactivation during G2 checkpoint recovery. EMBO Rep. 2010, 11, 626–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lockwood, B.L.; Somero, G.N. Transcriptomic responses to salinity stress in invasive and native blue mussels (genus Mytilus). Mol. Ecol. 2011, 20, 517–529. [Google Scholar] [CrossRef]
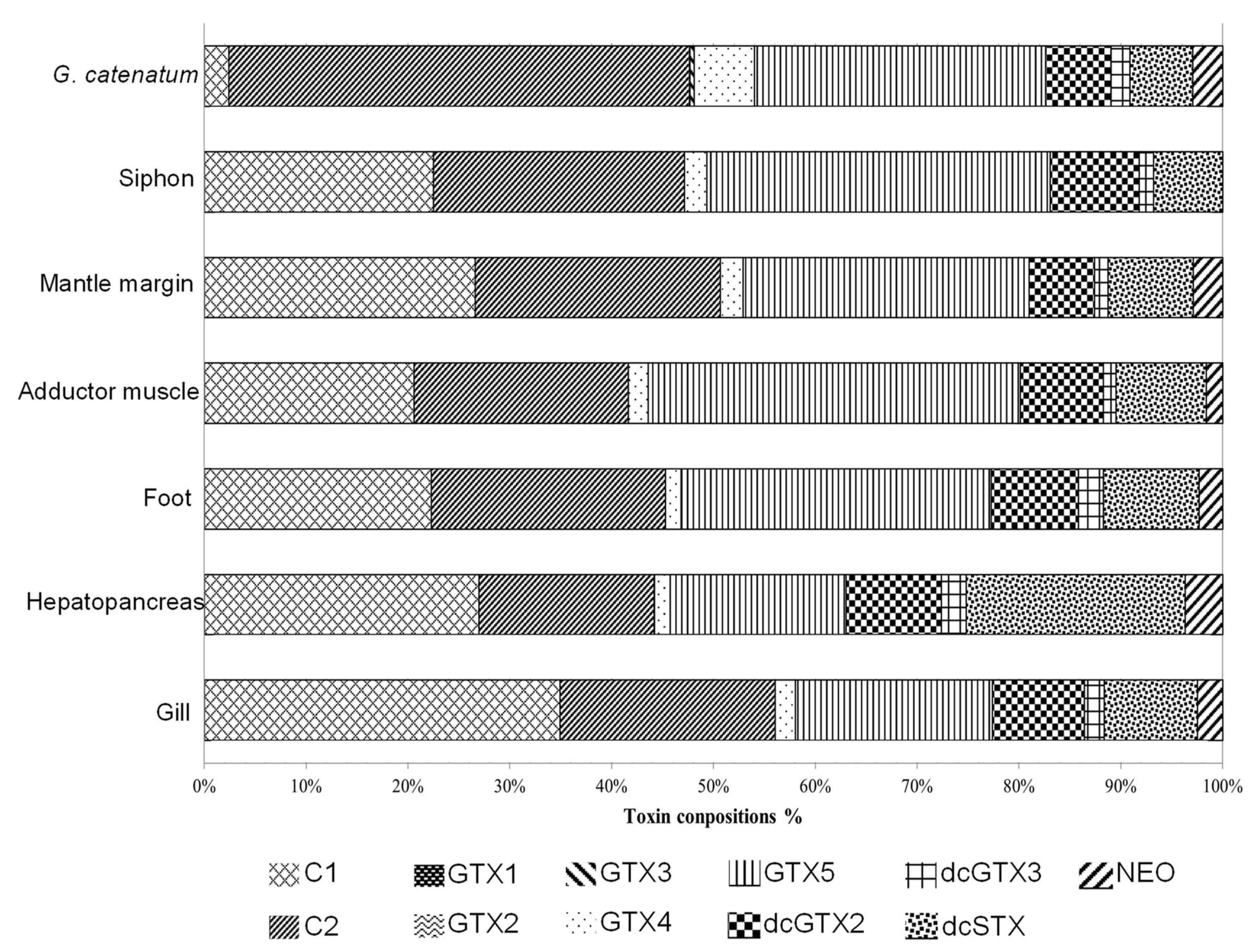
| Organ | Toxin Content (μg STX Equivalents Per Gram) | Relative Abundant (% Molar) |
|---|---|---|
| Gill | 0.74 ± 0.14 a | 1.82 |
| Hepatopancreas | 38.71 ± 3.63 b | 95.27 |
| Foot | 0.73 ± 0.10 a | 1.79 |
| Adductor muscle | 0.212 ± 0.05 a | 0.52 |
| Mantle margin | 0.107 ± 0.06 a | 0.26 |
| Sighon | 0.131 ± 0.03 a | 0.32 |
| Spot | pI | MW (kDa) | Fold Changes | Protein Name | Source | Amino Acid Sequence |
|---|---|---|---|---|---|---|
| TP1 | 4.8 | 46 | 4.2 | Response regulator receiver domain-containing protein | De novo peptide sequencing 1634.9 m/z | FAI/LAKAVNHHNWAR |
| TP2 | 4.6 | 42 | 4.9 | Major facilitator superfamily transporters | De novo peptide sequencing 1154.3 m/z | AI/LFSFWI/LDR |
| TP3 | 4.6 | 40 | 5.3 | Sarcoplasmic calcium-binding protein | De novo peptide sequencing 842.2 m/z | VATVSI/LPR |
| TP5 | 7.0 | 60 | 2.3 | Catalase | PMF | -- |
| TP6 | 6.5 | 18 | 3.2 | Protein ultraspiracle homolog | PMF | -- |
| TP7 | 6.3 | 34 | 3.0 | Paramyosin | De novo peptide sequencing 1839.8 m/z | DI/LEI/LAVI/LSHESAEASI/LR |
| TP8 | 6.1 | 20 | 3.8 | Mg superoxide dismutase | De novo peptide sequencing 1646.6 m/z | AI/LFKI/LANWEEVGNR |
| TP9 | 4.5 | 29 | 3.3 | G2 and S phase-expressed protein 1 | PMF | -- |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chan, K.-K.; Tam, N.F.-Y.; Ng, C.; Kwok, C.S.-N.; Xu, S.J.-L.; Sze, E.T.-P.; Lee, F.W.-F. Proteome Response of Meretrix Bivalves Hepatopancreas Exposed to Paralytic Shellfish Toxins Producing Dinoflagellate Gymnodinium catenatum. J. Mar. Sci. Eng. 2021, 9, 1039. https://doi.org/10.3390/jmse9091039
Chan K-K, Tam NF-Y, Ng C, Kwok CS-N, Xu SJ-L, Sze ET-P, Lee FW-F. Proteome Response of Meretrix Bivalves Hepatopancreas Exposed to Paralytic Shellfish Toxins Producing Dinoflagellate Gymnodinium catenatum. Journal of Marine Science and Engineering. 2021; 9(9):1039. https://doi.org/10.3390/jmse9091039
Chicago/Turabian StyleChan, Kin-Ka, Nora Fung-Yee Tam, Christie Ng, Celia Sze-Nga Kwok, Steven Jing-Liang Xu, Eric Tung-Po Sze, and Fred Wang-Fat Lee. 2021. "Proteome Response of Meretrix Bivalves Hepatopancreas Exposed to Paralytic Shellfish Toxins Producing Dinoflagellate Gymnodinium catenatum" Journal of Marine Science and Engineering 9, no. 9: 1039. https://doi.org/10.3390/jmse9091039
APA StyleChan, K.-K., Tam, N. F.-Y., Ng, C., Kwok, C. S.-N., Xu, S. J.-L., Sze, E. T.-P., & Lee, F. W.-F. (2021). Proteome Response of Meretrix Bivalves Hepatopancreas Exposed to Paralytic Shellfish Toxins Producing Dinoflagellate Gymnodinium catenatum. Journal of Marine Science and Engineering, 9(9), 1039. https://doi.org/10.3390/jmse9091039







