Homeostatic Functions of Tecrem, a CD46-Like Regulatory Protein of Complement Activation, on Epithelial Cells in Carp Fish
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Detection of Tecrem Protein by ELISA and Immunoprecipitation
2.3. Expression of Recombinant SCR1-2 and SCR3-4 Domains of cTecrem
2.4. Preparation of Rabbit Polyclonal Anti-cTecrem Domains
2.5. Flow Cytometric Analysis of Tecrem Expression on KF-1 Cells
2.6. Western Blotting Analysis of Tecrem Expression on KF-1 and CFS Cells
2.7. Adhesion Assays of KF-1 Cells
2.8. Wound Healing Assay
2.9. Immunohistochemistry Assay
3. Results
3.1. Preparation of Recombinant SCR 1-2 and SCR 3-4 and Its Polyclonal Antibodies
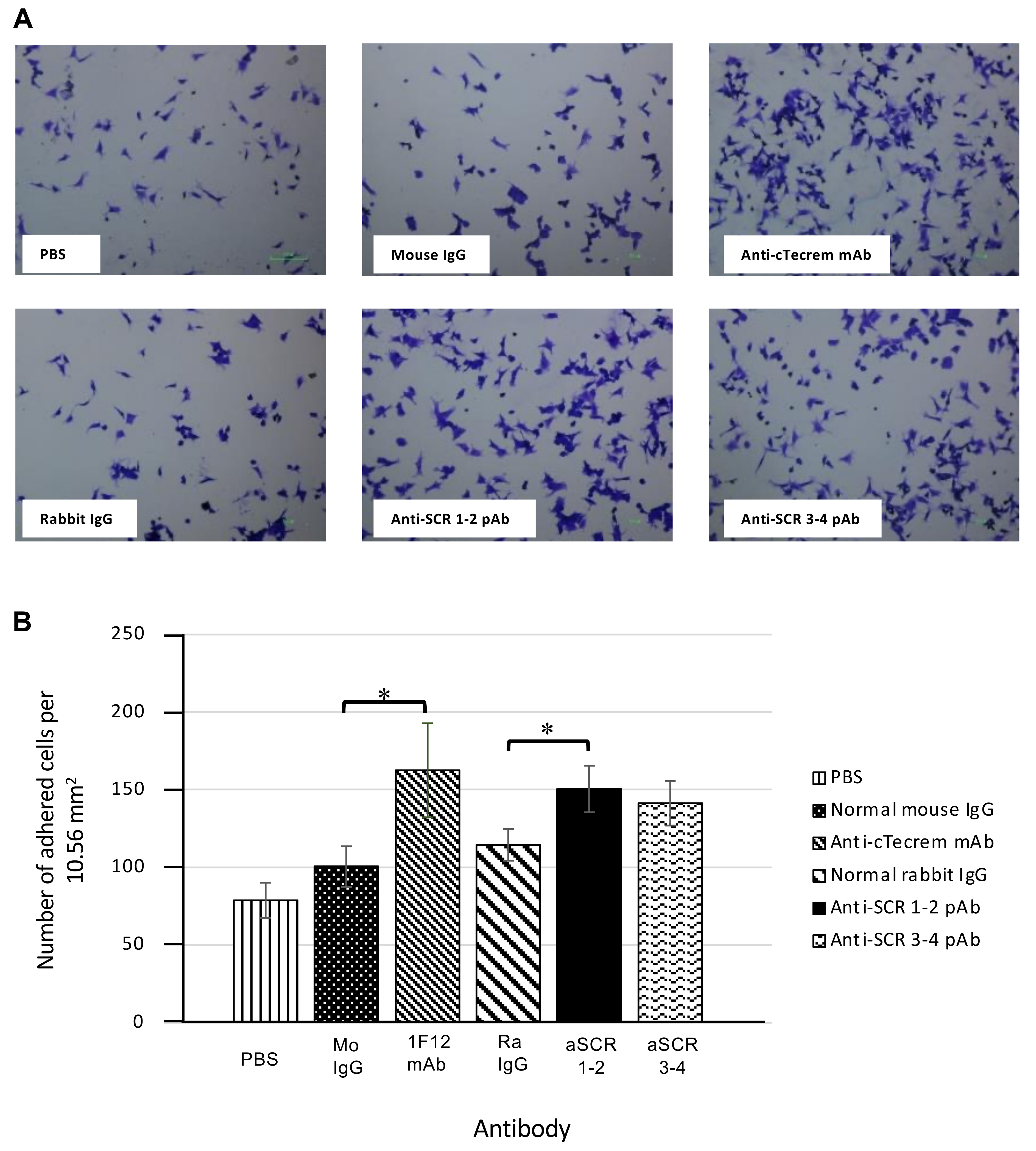
3.2. Activation of cTecrem Results in An Increase in the Adhesion of KF-1 Cells on to the Cell Culture Plate
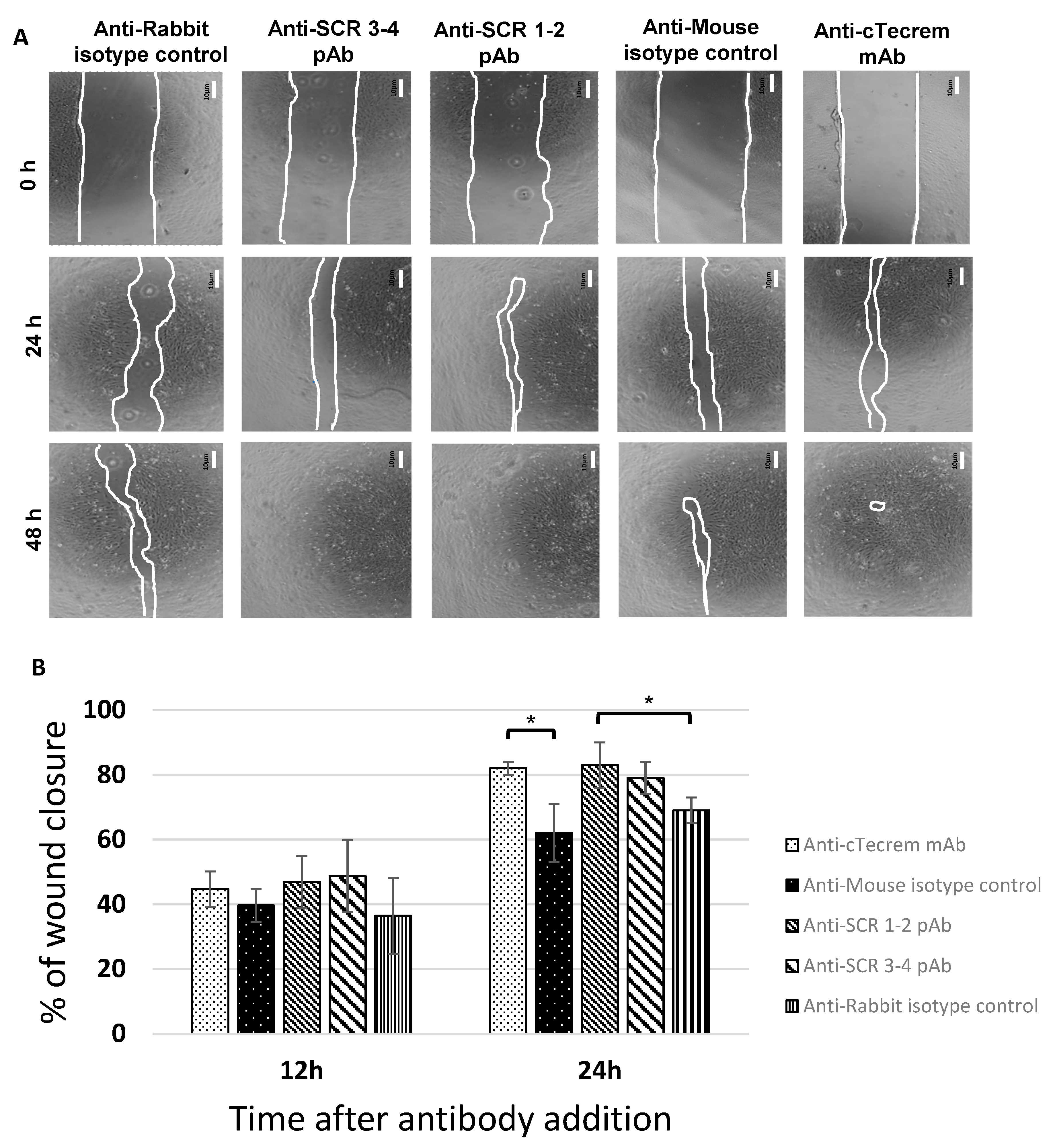
3.3. Tecrem Activation Promotes Epithelial Cell Growth as well as Wound Healing
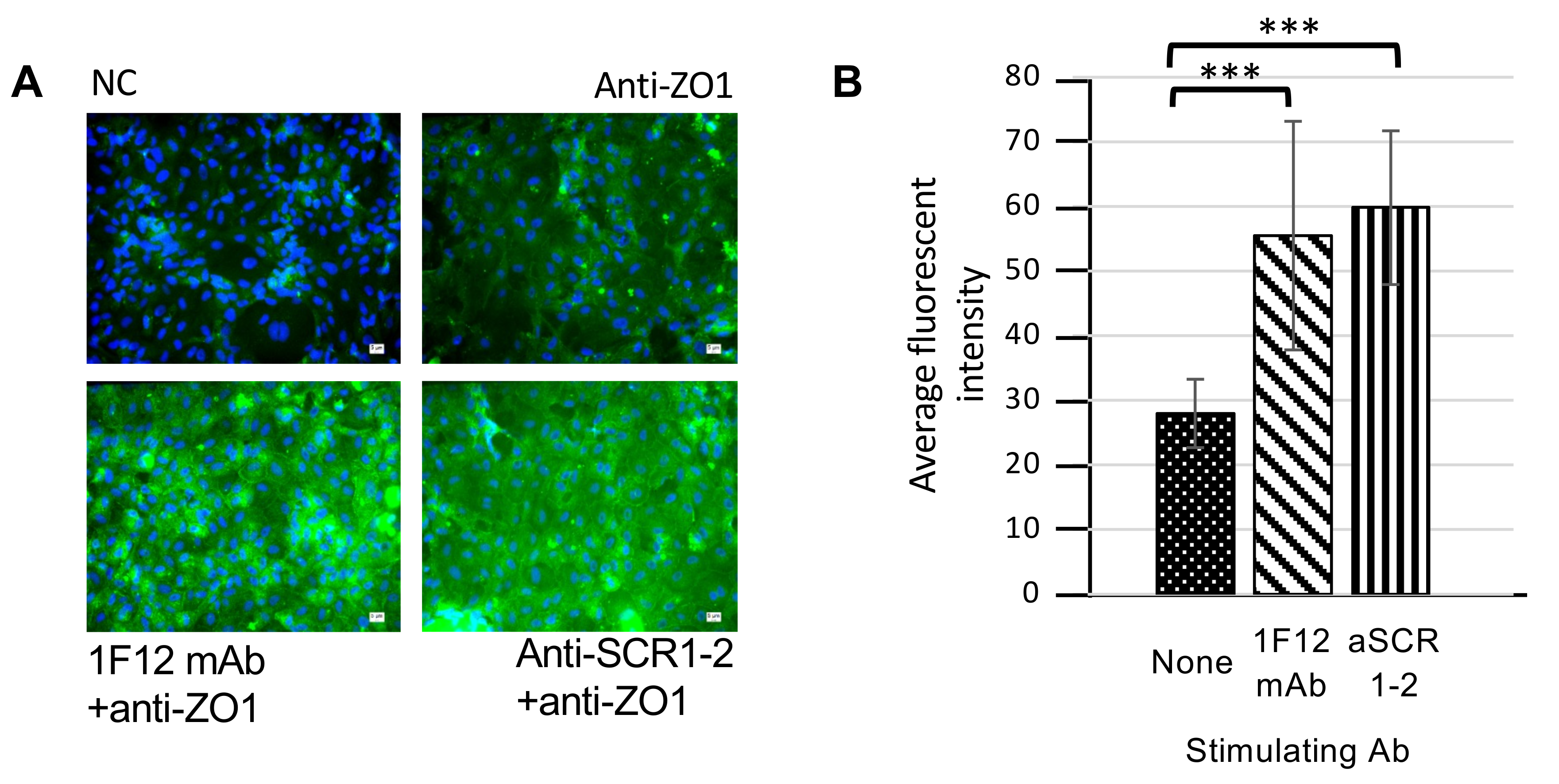
3.4. Enhancement of Tight Junction Protein Expression by cTecrem Stimulation in CFS Cells
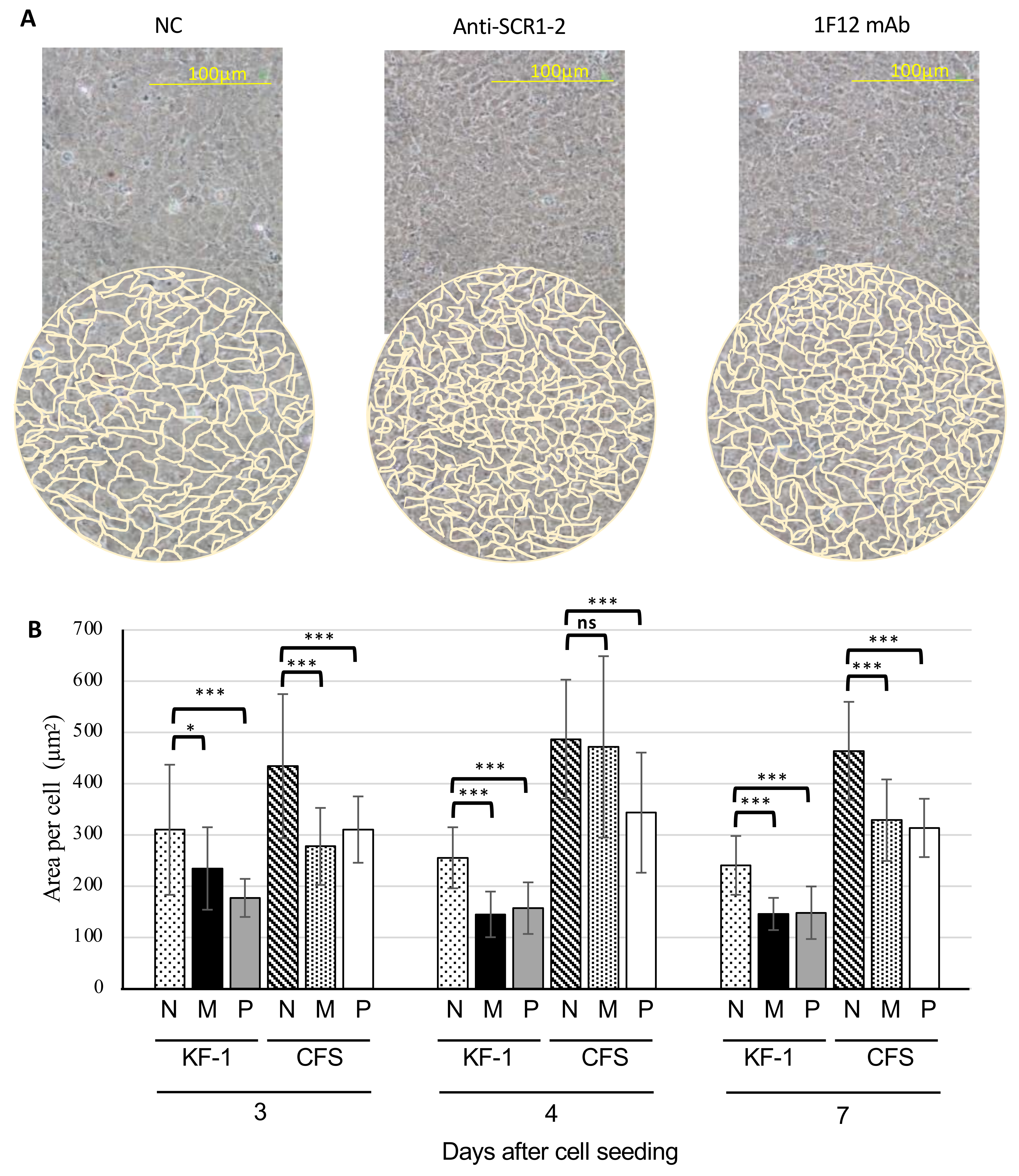
3.5. Morphological Changes in the cTecrem-Activated KF-1 and CFS Cell Lines
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Parra, D.; Reyes-Lopez, F.E.; Tort, L. Mucosal immunity and B cells in teleosts: Effect of vaccination and stress. Front. Immunol. 2015, 6, 354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merle, N.S.; Church, S.E.; Fremeaux-Bacchi, V.; Roumenina, L.T. Complement system part I – molecular mechanisms of activation and regulation. Front. Immunol. 2015, 6, 262. [Google Scholar] [CrossRef] [Green Version]
- Nakao, M.; Tsujikura, M.; Ichiki, S.; Vo, T.K.; Somamoto, T. The complement system in teleost fish: Progress of post-homolog-hunting researches. Dev. Comp. Immunol. 2011, 35, 1296–1308. [Google Scholar] [CrossRef]
- Liszewski, M.K.; Farries, T.C.; Lublin, D.M.; Rooney, I.A.; Atkinson, J.P. Control of the Complement System. Adv. Immunol. 1996, 61, 201–283. [Google Scholar] [CrossRef]
- Yamamoto, H.; Fara, A.F.; Dasgupta, P.; Kemper, C. CD46: The ‘multitasker’ of complement proteins. Int. J. Biochem. Cell. Biol. 2013, 45, 2808–2820. [Google Scholar] [CrossRef]
- Tsujikura, M.; Nagasawa, T.; Ichiki, S.; Nakamura, R.; Somamoto, T.; Nakao, M. A CD46-like Molecule Functional in Teleost Fish Represents an Ancestral Form of Membrane-Bound Regulators of Complement Activation. J. Immunol. 2015, 194, 262–272. [Google Scholar] [CrossRef]
- Nur, I.; Harada, H.; Tsujikura, M.; Somamoto, T.; Nakao, M. Molecular characterization and expression analysis of three membrane-bound complement regulatory protein isoforms in the ginbuna crucian carp Carassius auratus langsdorfii. Fish Shellfish. Immunol. 2013, 35, 1333–1337. [Google Scholar] [CrossRef]
- Nur, I.; Abdelkhalek, N.K.; Motobe, S.; Nakamura, R.; Tsujikura, M.; Somamoto, T.; Nakao, M. Functional analysis of membrane bound complement regulatory protein on T-cell immune response in Ginbuna Crucian Carp. Mol. Immunol. 2016, 70, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Li, H.; Zhang, S. Regulator of complement activation (RCA) group 2 gene cluster in zebrafish: Identification, expression, and evolution. Funct. Integr. Genomics 2012, 12, 367–377. [Google Scholar] [CrossRef]
- Li, M.F.; Sui, Z.H.; Sun, L. A teleost CD46 is involved in the regulation of complement activation and pathogen infection. Sci. Rep. 2017, 7, 15028. [Google Scholar] [CrossRef] [Green Version]
- West, E.E.; Kolev, M.; Kemper, C. Complement and the Regulation of T Cell Responses. Annu. Rev. Immunol. 2018, 36, 309–338. [Google Scholar] [CrossRef]
- Cardone, J.; Shouli, A.L.; Kemper, C. A novel role for CD46 in wound repair. Front. Immunol. 2011, 2, 28. [Google Scholar] [CrossRef] [Green Version]
- Lv, H.; Zhou, T.; Dong, C.; Kong, S.; Chen, L.; Pu, F.; Li, X.; Xu, P. Genome-wide identification, evolution, and mRNA expression of complement genes in common carp (Cyprinus carpio). Fish Shellfish. Immunol. 2020, 96, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, S.F.; Buchmann, K.; Nielsen, M.E. Complement expression in common carp (Cyprinus carpio L.) during infection with Ichthyophthirius multifiliis. Dev. Comp. Immunol. 2007, 31, 576–586. [Google Scholar] [CrossRef]
- Brinchmann, M.F. Immune relevant molecules identified in the skin mucus of fish using -omics technologies. Mol. Biosyst. 2016, 12, 2056–2063. [Google Scholar] [CrossRef] [Green Version]
- Balda, M.S.; Matter, K. The tight junction protein ZO-1 and an interacting transcription factor regulate ErbB-2 expression. EMBO J. 2000, 19, 2024–2033. [Google Scholar] [CrossRef]
- Anderson, J.M.; Itallie, C.M.V. Physiology and Function of the Tight Junction. Cold Spring Harb. Perspect. Biol. 2009, 1, a002584. [Google Scholar] [CrossRef]
- Balda, M.S.; Matter, K. Review-Tight junctions and the regulation of gene expression. Biochim. Biophys. Acta 2009, 1788, 761–767. [Google Scholar] [CrossRef] [Green Version]
- Tsukita, S.; Furuse, M.; Itoh, M. Structural and signalling molecules come together at tight junctions. Curr. Opin. Cell Biol. 1999, 11, 628–633. [Google Scholar] [CrossRef]
- Georgiadis, A.; Tschernutter, M.; Bainbridge, J.W.B.; Balaggan, K.S.; Mowat, F.; West, E.L.; Munro, P.M.G.; Thrasher, A.J.; Matter, K.; Balda, M.S.; et al. The Tight Junction Associated Signalling Proteins ZO-1 and ZONAB Regulate Retinal Pigment Epithelium Homeostasis in Mice. PLoS ONE 2010, 5, e15730. [Google Scholar] [CrossRef] [Green Version]
- Mariscal, L.G.; Lechuga, S.; Garay, E. Role of tig.ht junctions in cell proliferation and cancer. Prog. Histochem. Cytochem. 2007, 42, 1–57. [Google Scholar] [CrossRef]
- Mariscal, L.G.; Tapia, R.; Chamorro, D. Crosstalk of tight junction components with signaling pathways. Biochim. Biophys. Acta 2008, 1778, 729–756. [Google Scholar] [CrossRef] [Green Version]
- Kolosov, D.; Chasiotis, H.; Kelly, S.P. Tight junction protein gene expression patterns and changes in transcript abundance during development of model fish gill epithelia. J. Exp. Biol. 2014, 217, 1667–1681. [Google Scholar] [CrossRef] [Green Version]
- Balda, M.S.; Garrett, M.S.; Matter, K. The ZO-1–associated Y-box factor ZONAB regulates epithelial cell proliferation and cell density. J. Cell. Biol. 2003, 160, 423–432. [Google Scholar] [CrossRef]
- Freshney, R.I. Culture of Animal Cells: A Manual of Basic Technique and Specialized Applications. Biol. Cult. Cells 2010, 11–23. [Google Scholar] [CrossRef]
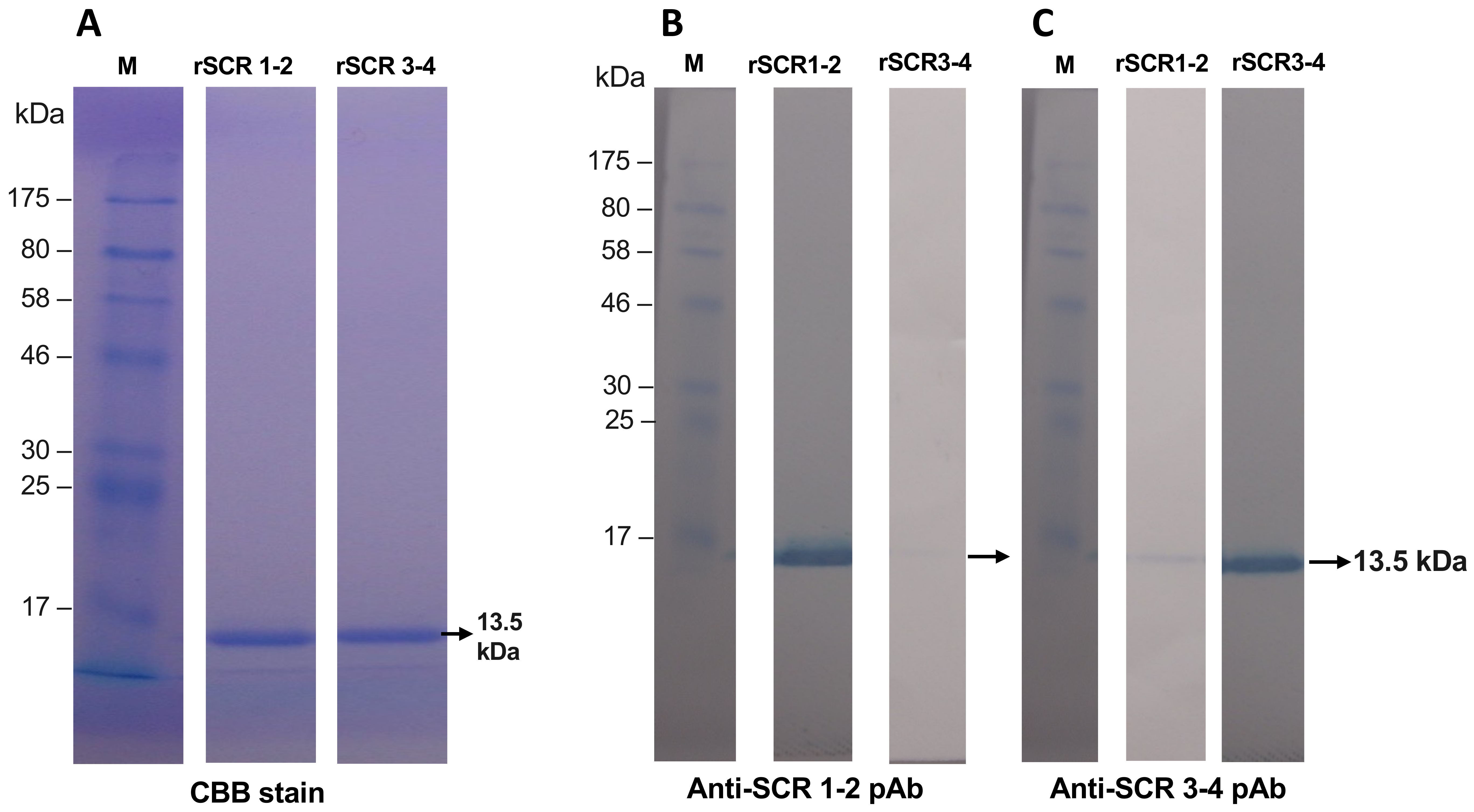
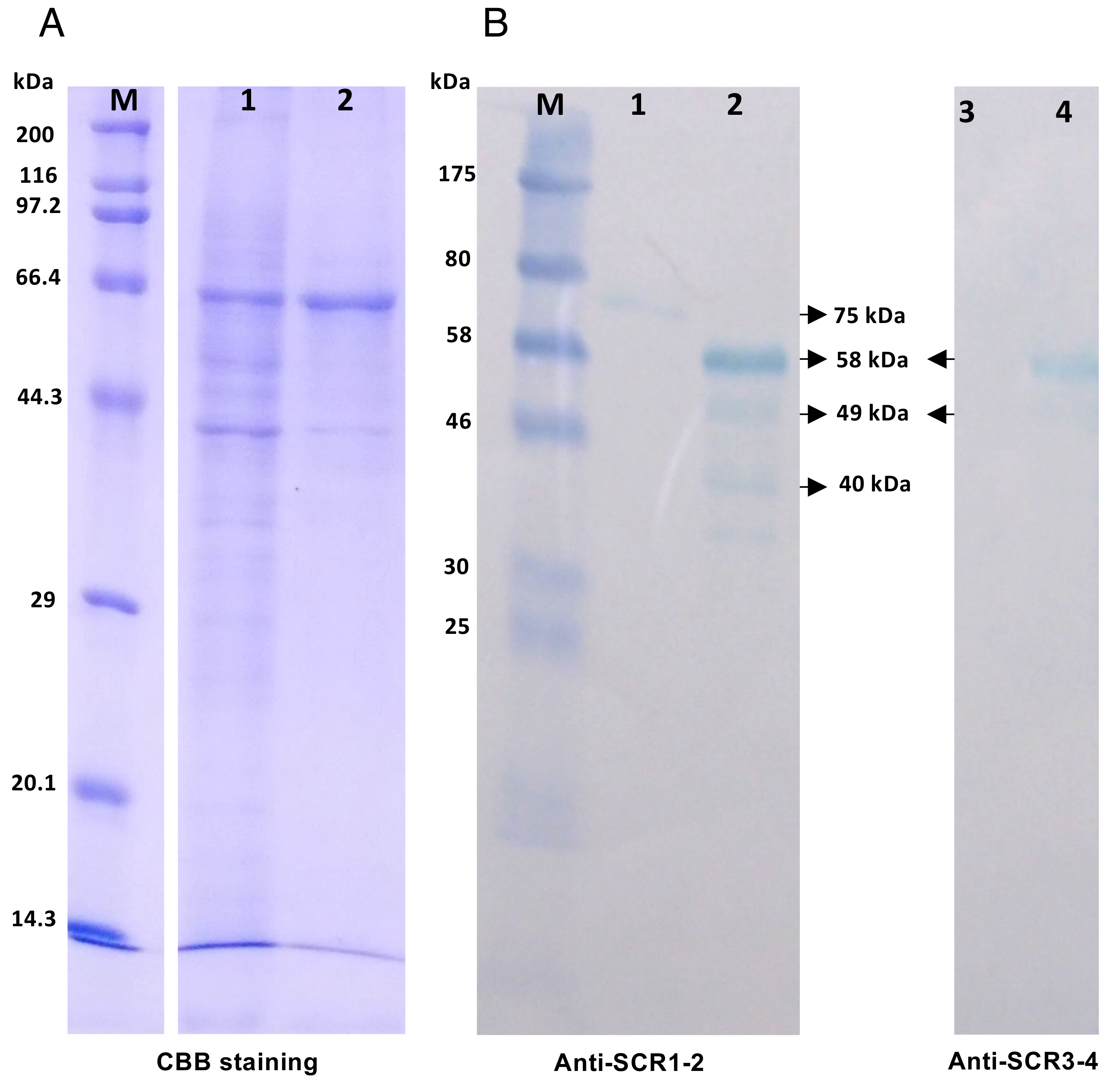
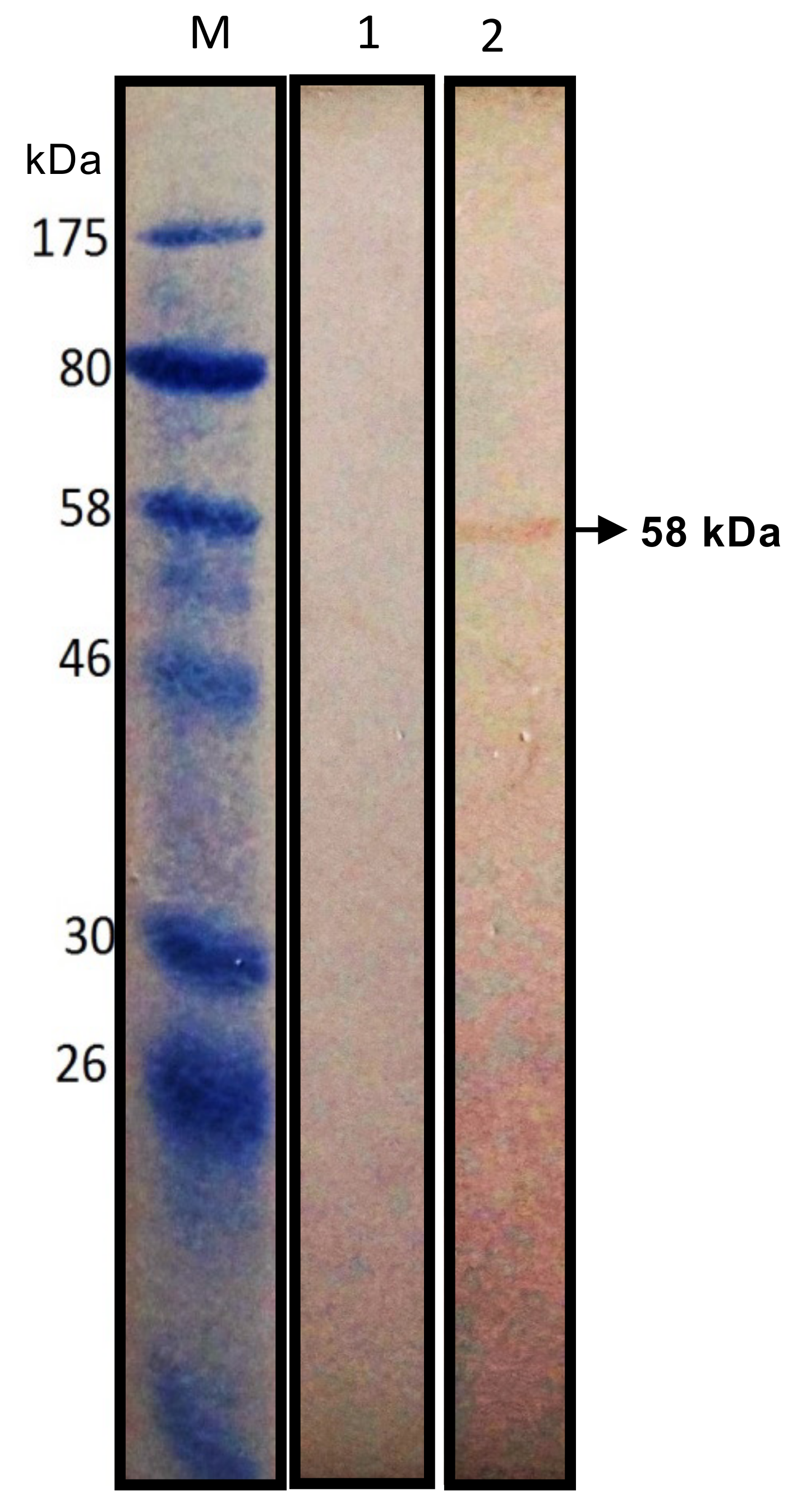
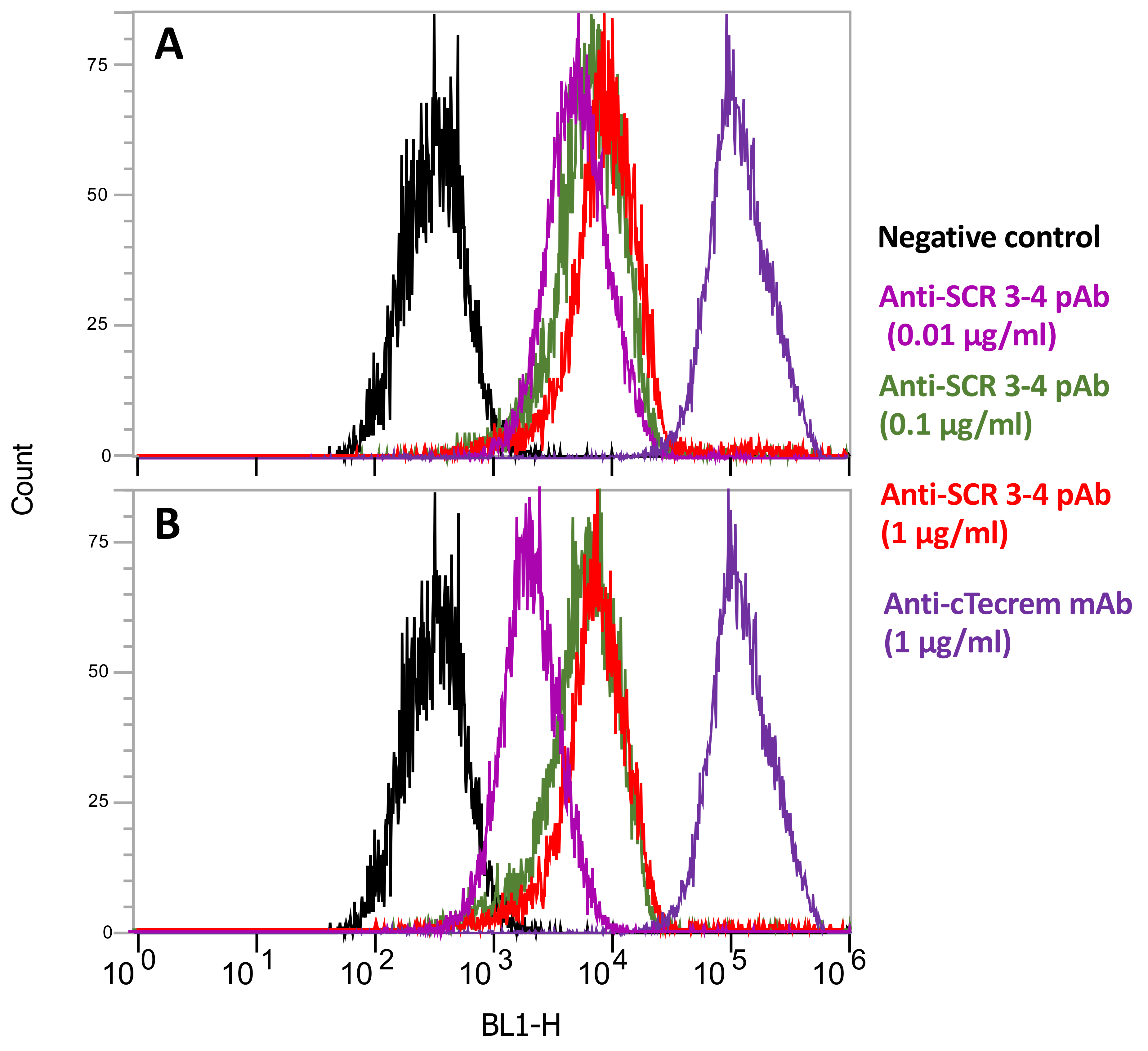



| Name | Nucleotide Sequence 1 | Amino Acids Encoded | Strand |
|---|---|---|---|
| SCR1-2F | gcctcgagGCGGAATGTACACAGCCTTC | AECTQP | Sense |
| SCR1-2R | cgctgcagttaCGATTCACAGAGAGGATCTC | DPLCES | Antisense |
| SCR3-4F | gcctcgagGTAAAATGTTCAGCACCTCCAGC | VKCSAPP | Sense |
| SCR3-4R | cgctgcagttaGACAATGCATTGTGGTGGTTCAG | EPPQCIV | Antisense |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prakash, H.; Motobe, S.; Nagasawa, T.; Somamoto, T.; Nakao, M. Homeostatic Functions of Tecrem, a CD46-Like Regulatory Protein of Complement Activation, on Epithelial Cells in Carp Fish. J. Mar. Sci. Eng. 2021, 9, 687. https://doi.org/10.3390/jmse9070687
Prakash H, Motobe S, Nagasawa T, Somamoto T, Nakao M. Homeostatic Functions of Tecrem, a CD46-Like Regulatory Protein of Complement Activation, on Epithelial Cells in Carp Fish. Journal of Marine Science and Engineering. 2021; 9(7):687. https://doi.org/10.3390/jmse9070687
Chicago/Turabian StylePrakash, Harsha, Shiori Motobe, Takahiro Nagasawa, Tomonori Somamoto, and Miki Nakao. 2021. "Homeostatic Functions of Tecrem, a CD46-Like Regulatory Protein of Complement Activation, on Epithelial Cells in Carp Fish" Journal of Marine Science and Engineering 9, no. 7: 687. https://doi.org/10.3390/jmse9070687
APA StylePrakash, H., Motobe, S., Nagasawa, T., Somamoto, T., & Nakao, M. (2021). Homeostatic Functions of Tecrem, a CD46-Like Regulatory Protein of Complement Activation, on Epithelial Cells in Carp Fish. Journal of Marine Science and Engineering, 9(7), 687. https://doi.org/10.3390/jmse9070687






