Isolation, Identification, and Biochemical Characteristics of a Cold-Tolerant Chlorella vulgaris KNUA007 Isolated from King George Island, Antarctica
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Isolation
2.2. Morphological and Molecular Identification
2.3. Temperature Testing
2.4. Biomass Characterization
2.5. Gas Chromatography/Mass Spectrometry (GC/MS) Analysis
3. Results
3.1. Identification of Strain KNUA007
3.2. Cold Tolerance of Strain KNUA007
3.3. Biomass Properties
3.4. GC/MS Analysis of Strain KNUA007
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Komárek, J.; Nedbalová, L. Green cryosestic algae. In Algae and Cyanobacteria in Extreme Envionments; Seckbach, J., Ed.; Springer: Dordrecht, The Netherlands, 2007; pp. 323–344. [Google Scholar]
- Lewis, L.A.; Lewis, P.O. Unearthing the molecular phylodiversity of desert soil green algae (Chlorophyta). Syst. Biol. 2005, 54, 936–947. [Google Scholar] [CrossRef] [PubMed]
- Vincent, W.F. Cyanobacterial dominance in the polar regions. In The Ecology of Cyanobacteria: Their Diversity in Time and Space; Whitton, B.A., Potts, M., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2000; pp. 321–340. [Google Scholar]
- Broady, P.A. Diversity, distribution and dispersal of Antarctic terrestrial algae. Biodivers. Conserv. 1996, 5, 1307–1335. [Google Scholar] [CrossRef]
- Vincent, W.F.; James, M.R. Biodiversity in extreme aquatic environments: Lakes, ponds and streams of the Ross Sea sector, Antarctica. Biodivers. Conserv. 1996, 5, 1451–1471. [Google Scholar] [CrossRef]
- Wynn-Williams, D.D. Antarctic microbial diversity: The basis of polar ecosystem processes. Biodivers. Conserv. 1996, 5, 1271–1293. [Google Scholar] [CrossRef]
- De Wever, A.; Leliaert, F.; Verleyen, E.; Vanormelingen, P.; Van Der Gucht, K.; Hodgson, D.A.; Sabbe, K.; Vyverman, W. Hidden levels of phylodiversity in Antarctic green algae: Further evidence for the existence of glacial refugia. Proc. R. Soc. B Biol. Sci. 2009, 276, 3591–3599. [Google Scholar] [CrossRef]
- Jungblut, A.D.; Vincent, W.F.; Lovejoy, C. Eukaryotes in Arctic and Antarctic cyanobacterial mats. FEMS Microbiol. Ecol. 2012, 82, 416–428. [Google Scholar] [CrossRef]
- Wood, S.A.; Rueckert, A.; Cowan, D.A.; Cary, S.C. Sources of edaphic cyanobacterial diversity in the Dry Valleys of Eastern Antarctica. ISME J. 2008, 2, 308–320. [Google Scholar] [CrossRef]
- Ferrara, M.; Guerriero, G.; Cardi, M.; Esposito, S. Purification and biochemical characterisation of a glucose-6-phosphate dehydrogenase from the psychrophilic green alga Koliella antarctica. Extremophiles 2013, 17, 53–62. [Google Scholar] [CrossRef]
- Pulz, O.; Gross, W. Valuable products from biotechnology of microalgae. Appl. Microbiol. Biotechnol. 2004, 65, 635–648. [Google Scholar] [CrossRef]
- Řezanka, T.; Nedbalová, L.; Sigler, K. Unusual medium-chain polyunsaturated fatty acids from the snow alga Chloromonas brevispina. Microbiol. Res. 2008, 163, 373–379. [Google Scholar] [CrossRef]
- Ru, I.T.K.; Sung, Y.Y.; Jusoh, M.; Wahid, M.E.A.; Nagappan, T. Chlorella vulgaris: A perspective on its potential for combining high biomass with high value bioproducts. Appl. Phycol. 2020, 1, 2–11. [Google Scholar] [CrossRef]
- Safi, C.; Zebib, B.; Merah, O.; Pontalier, P.Y.; Vaca-Garcia, C. Morphology, composition, production, processing and applications of Chlorella vulgaris: A review. Renew. Sustain. Energy Rev. 2014, 35, 265–278. [Google Scholar] [CrossRef]
- Cecchin, M.; Marcolungo, L.; Rossato, M.; Girolomoni, L.; Cosentino, E.; Cuine, S.; Li-Beisson, Y.; Delledonne, M.; Ballottari, M. Chlorella vulgaris genome assembly and annotation reveals the molecular basis for metabolic acclimation to high light conditions. Plant J. 2019, 100, 1289–1305. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.T.; Shariff, M.; Md. Yusoff, F.; Goh, Y.M.; Banerjee, S. Applications of microalga Chlorella vulgaris in aquaculture. Rev. Aquac. 2020, 12, 328–346. [Google Scholar] [CrossRef]
- Merchant, S.S.; Prochnik, S.E.; Vallon, O.; Harris, E.H.; Karpowicz, S.A.; Witman, G.B.; Terry, A.; Salamov, A.; Fritz-Laylin, L.K.; Maréchal-Drouard, L.; et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Natl. Inst. Heal. 2007, 318, 245–250. [Google Scholar] [CrossRef]
- Blanc, G.; Duncan, G.; Agarkova, I.; Borodovsky, M.; Gurnon, J.; Kuo, A.; Lindquist, E.; Lucas, S.; Pangilinan, J.; Polle, J.; et al. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell 2010, 22, 2943–2955. [Google Scholar] [CrossRef]
- Vieler, A.; Wu, G.; Tsai, C.H.; Bullard, B.; Cornish, A.J.; Harvey, C.; Reca, I.B.; Thornburg, C.; Achawanantakun, R.; Buehl, C.J.; et al. Correction: Genome, functional gene annotation, and nuclear transformation of the Heterokont oleaginous alga Nannochloropsis oceanica CCMP1779. PLoS Genet. 2017, 13, e1006802. [Google Scholar] [CrossRef]
- Arriola, M.B.; Velmurugan, N.; Zhang, Y.; Plunkett, M.H.; Hondzo, H.; Barney, B.M. Genome sequences of Chlorella sorokiniana UTEX 1602 and Micractinium conductrix SAG 241.80: Implications to maltose excretion by a green alga. Plant J. 2018, 93, 566–586. [Google Scholar] [CrossRef]
- Luo, W.; Pflugmacher, S.; Pröschold, T.; Walz, N.; Krienitz, L. Genotype versus phenotype variability in Chlorella and Micractinium (Chlorophyta, Trebouxiophyceae). Protist 2006, 157, 315–333. [Google Scholar] [CrossRef]
- Luo, W.; Krienitz, L.; Pflugmacher, S.; Walz, N. Genus and species concept in Chlorella and Micractinium (Chlorophyta, Chlorellaceae): Genotype versus phenotypical variability under ecosystem conditions. SIL Proc. 2005, 29, 170–173. [Google Scholar] [CrossRef]
- Rippka, R.; Deruelles, J.; Waterbury, J.B.; Herdman, M.; Stanier, R.Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1979, 111, 1–61. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR protocols: A Guide to Method and Applications; Academic Press: Cambridge, MA, USA, 1990; pp. 315–322. [Google Scholar]
- Marshall, M.N.; Cocolin, L.; Mills, D.A.; VanderGheynst, J.S. Evaluation of PCR primers for denaturing gradient gel electrophoresis analysis of fungal communities in compost. J. Appl. Microbiol. 2003, 95, 934–948. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Germond, A.; Hata, H.; Fujikawa, Y.; Nakajima, T. The phylogenetic position and phenotypic changes of a Chlorella-like alga during 5-year microcosm culture. Eur. J. Phycol. 2013, 48, 485–496. [Google Scholar] [CrossRef]
- Heo, J.; Cho, D.-H.; Ramanan, R.; Oh, H.-M.; Kim, H.-S. PhotoBiobox: A tablet sized, low-cost, high throughput photobioreactor for microalgal screening and culture optimization for growth, lipid content and CO2 sequestration. Biochem. Eng. J. 2015, 103, 193–197. [Google Scholar] [CrossRef]
- Friedl, A.; Padouvas, E.; Rotter, H.; Varmuza, K. Prediction of heating values of biomass fuel from elemental composition. Anal. Chim. Acta 2005, 544, 191–198. [Google Scholar] [CrossRef]
- Aussant, J.; Guihéneuf, F.; Stengel, D.B. Impact of temperature on fatty acid composition and nutritional value in eight species of microalgae. Appl. Microbiol. Biotechnol. 2018, 102, 5279–5297. [Google Scholar] [CrossRef]
- Chae, H.; Lim, S.; Kim, H.S.; Choi, H.-G.; Kim, J.H. Morphology and phylogenetic relationships of Micractinium (Chlorellaceae, trebouxiophyceae) taxa, including three new species from antarctica. Algae 2019, 34, 267–275. [Google Scholar] [CrossRef]
- Illman, A.M.; Scragg, A.H.; Shales, S.W. Increase in Chlorella strains calorific values when grown in low nitrogen medium. Enzyme Microb. Technol. 2000, 27, 631–635. [Google Scholar] [CrossRef]
- Do, J.-M.; Jo, S.-W.; Kim, I.-S.; Na, H.; Lee, J.H.; Kim, H.S.; Yoon, H.-S. A feasibility study of wastewater treatment using domestic microalgae and analysis of biomass for potential applications. Water 2019, 11, 2294. [Google Scholar] [CrossRef]
- Lunch, C.K.; Lafountain, A.M.; Thomas, S.; Frank, H.A.; Lewis, L.A.; Cardon, Z.G. The xanthophyll cycle and NPQ in diverse desert and aquatic green algae. Photosynth. Res. 2013, 115, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Müller, P.; Li, X.-P.; Niyogi, K.K. Non-photochemical quenching. A response to excess light energy. Plant Physiol. 2001, 125, 1558–1566. [Google Scholar] [CrossRef] [PubMed]
- Chong, G.-L.; Chu, W.-L.; Othman, R.Y.; Phang, S.-M. Differential gene expression of an Antarctic Chlorella in response to temperature stress. Polar Biol. 2011, 34, 637–645. [Google Scholar] [CrossRef]
- Li, H.; Liu, X.; Wang, Y.; Hu, H.; Xu, X. Enhanced expression of antifreeze protein genes drives the development of freeze tolerance in an Antarctica isolate of Chlorella vulgaris. Prog. Nat. Sci. 2009, 19, 1059–1062. [Google Scholar] [CrossRef]
- Machida, T.; Murase, H.; Kato, E.; Honjoh, K.; Matsumoto, K.; Miyamoto, T.; Iio, M. Isolation of cDNAs for hardening-induced genes from Chlorella vulgaris by suppression subtractive hybridization. Plant Sci. 2008, 175, 238–246. [Google Scholar] [CrossRef]
- Spijkerman, E.; Wacker, A.; Weithoff, G.; Leya, T. Elemental and fatty acid composition of snow algae in Arctic habitats. Front. Microbiol. 2012, 3, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Lyon, B.R.; Mock, T. Polar microalgae: New approaches towards understanding adaptations to an extreme and changing environment. Biology 2014, 3, 56–80. [Google Scholar] [CrossRef]
- Harris, P.; James, A.T. The effect of low temperatures on fatty acid biosynthesis in plants. Biochem. J. 1969, 112, 325–330. [Google Scholar] [CrossRef]
- Suga, K.; Honjoh, K.I.; Furuya, N.; Shimizu, H.; Nishi, K.; Shinohara, F.; Hirabaru, Y.; Maruyama, I.; Miyamoto, T.; Hatano, S.; et al. Two low-temperature-inducible Chlorella genes for Δ12 and ω-3 fatty acid desaturase (FAD): Isolation of Δ12 and ω-3 fad cDNA clones, expression of Δ12 fad in Saccharomyces cerevisiae, and expression of ω-3 fad in Nicotiana tabacum. Biosci. Biotechnol. Biochem. 2002, 66, 1314–1327. [Google Scholar] [CrossRef] [PubMed]
- El-sheekh, M.; Abomohra, A.E.; El-azim, M.A.; Abou-shanab, R. Effect of temperature on growth and fatty acids profile of the biodiesel producing microalga Scenedesmus acutus. Biotechno. Agron. Soc. Environ. 2017, 21, 233–239. [Google Scholar]
- Mehta, L.R.; Dworkin, R.H.; Schwid, S.R. Polyunsaturated fatty acids and their potential therapeutic role in multiple sclerosis. Nat. Clin. Pract. Neurol. 2009, 5, 82–92. [Google Scholar] [CrossRef]
- Nadeau, T.-L.; Castenholz, R.W. Characterization of psychrophilic Oscillatorians (cyanobacteria) from antarctic meltwater ponds. J. Phycol. 2000, 36, 914–923. [Google Scholar] [CrossRef]
- Tang, E.P.Y.; Vincent, W.F. Strategies of thermal adaptation by high-latitude cyanobacteria. New Phytol. 1999, 142, 315–323. [Google Scholar] [CrossRef]
- Chevalier, P.; Proulx, D.; Lessard, P.; Vincent, W.F.; De La Noüe, J. Nitrogen and phosphorus removal by high latitude mat-forming cyanobacteria for potential use in tertiary wastewater treatment. J. Appl. Phycol. 2000, 12, 105–112. [Google Scholar] [CrossRef]
- Tang, E.P.Y.; Tremblay, R.; Vincent, W.F. Cyanobacterial dominance of polar freshwater ecosystems: Are high-latitude mat-formers adapted to low temperature? J. Phycol. 1997, 33, 171–181. [Google Scholar] [CrossRef]
- Hu, H.; Li, H.; Xu, X. Alternative cold response modes in Chlorella (Chlorophyta, Trebouxiophyceae) from Antarctica. Phycologia 2008, 47, 28–34. [Google Scholar] [CrossRef]
- Hong, J.W.; Kim, O.H.; Jo, S.-W.; Kim, H.; Jeong, M.R.; Park, K.M.; Lee, K.I.; Yoon, H.-S. Biochemical composition of a Korean domestic microalga Chlorella vulgaris KNUA027. Microbiol. Biotechnol. Lett. 2016, 44, 400–407. [Google Scholar] [CrossRef]

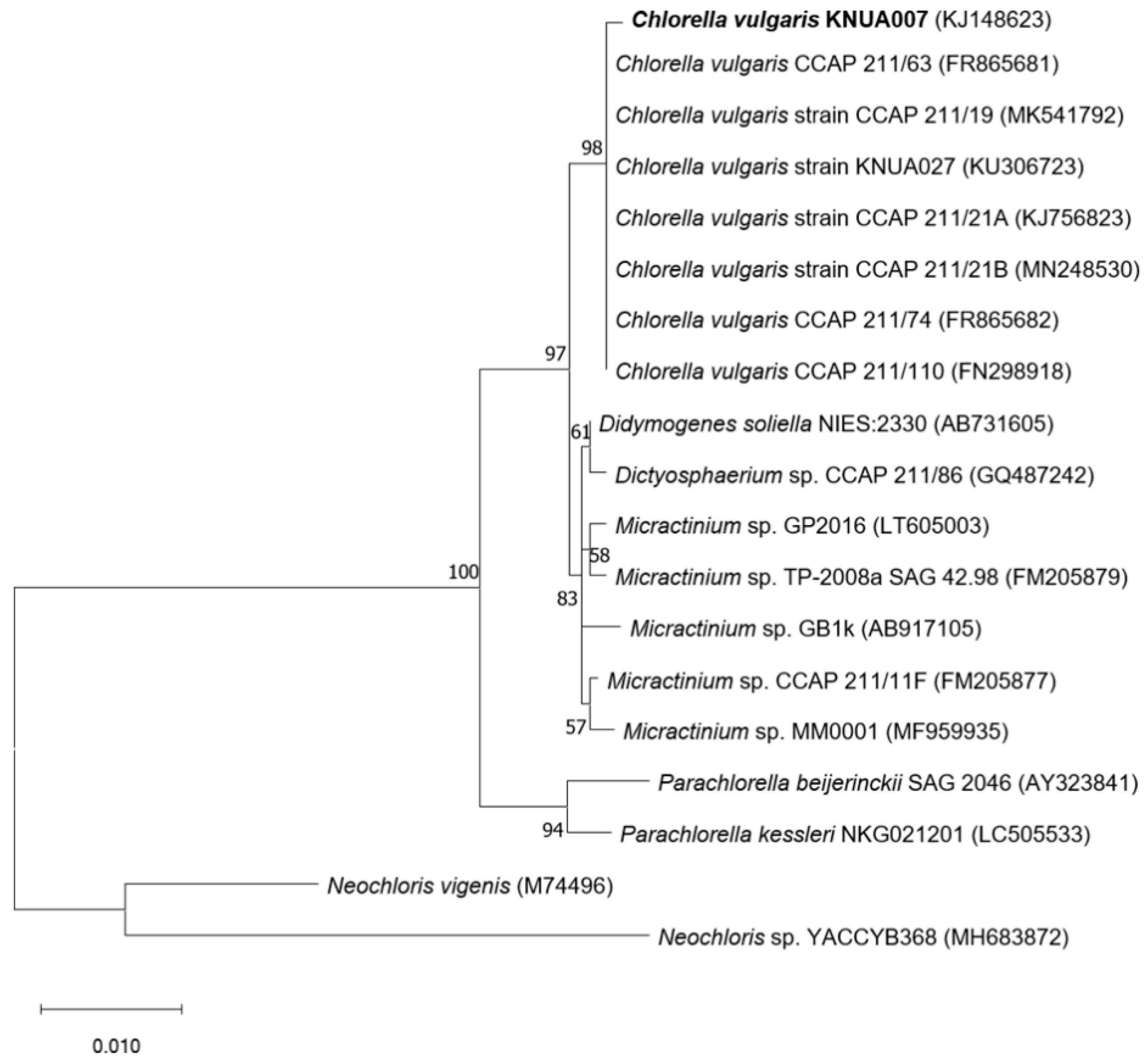
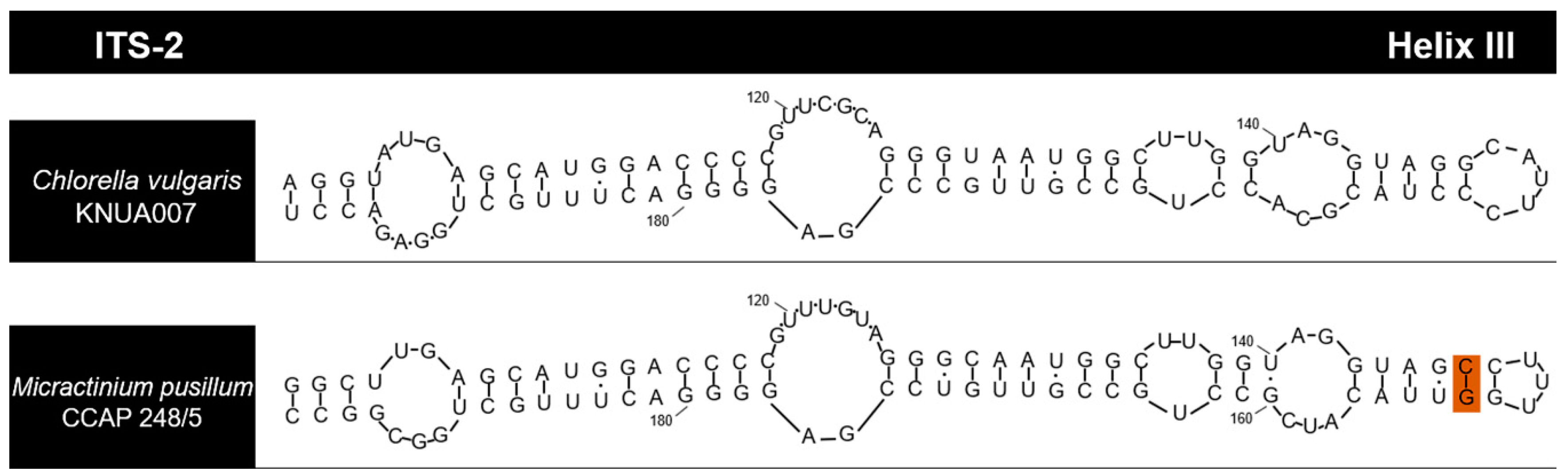
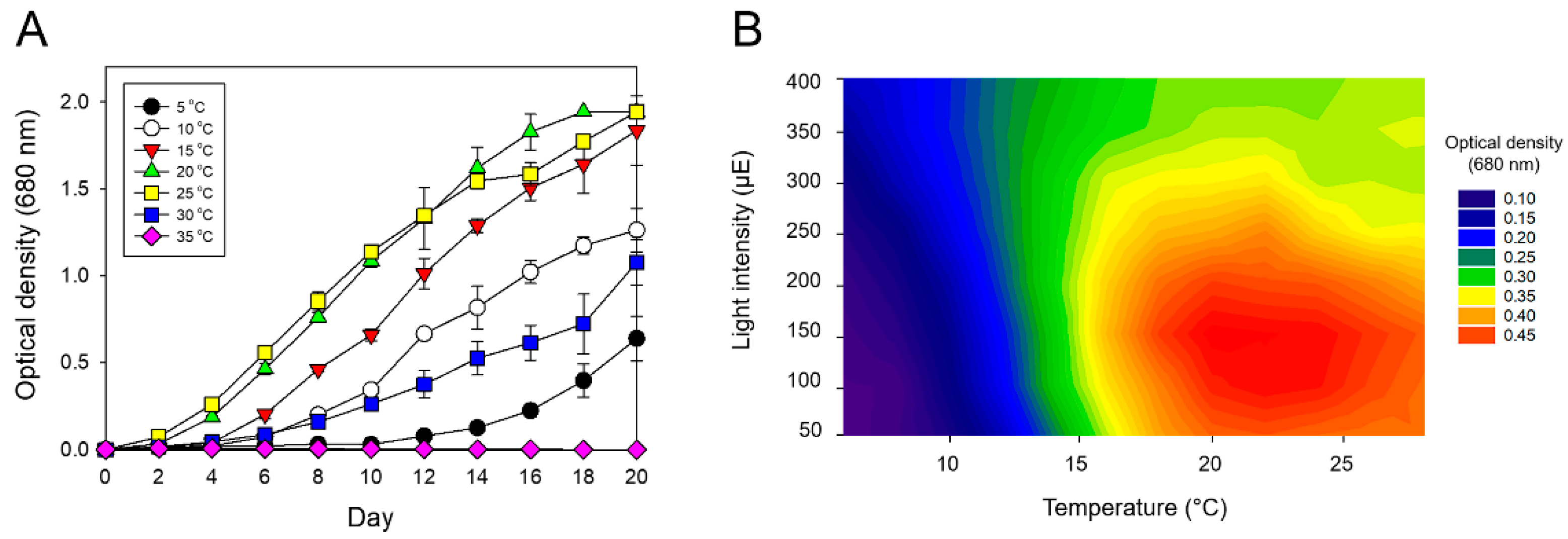
| Marker Gene | Accession no. | Product Size (bp) | Closet Match (GenBank Accession no.) | Query Cover (%) | Identification (%) |
|---|---|---|---|---|---|
| 18S rRNA | KJ148623 | 1771 | C. vulgaris CCAP 211/19 (MK541792) | 100 | 99.89 |
| ITS | KJ148624 | 783 | C. vulgaris ATFG2 (MT137382) | 100 | 99.87 |
| LSU | KJ148625 | 612 | C. vulgaris NIES:227 (AB237642) | 100 | 99.84 |
| Contents (wt%) | |||
|---|---|---|---|
| 10 °C | 20 °C | 30 °C | |
| C | 45.36 ± 0.06 | 42.94 ± 0.15 | 42.72 ± 0.26 |
| H | 6.64 ± 0.03 | 6.42 ± 0.05 | 6.54 ± 0.07 |
| N | 6.17 ± 0.00 | 5.84 ± 0.03 | 5.25 ± 0.02 |
| S | 0.76 ± 0.07 | 1.10 ± 0.03 | 1.14 ± 0.02 |
| Gross calorific value (MJ kg−1) | 18.7 ± 0.02 | 17.7 ± 0.06 | 17.5 ± 0.11 |
| Contents (wt%) | |||
|---|---|---|---|
| 10 °C | 20 °C | 30 °C | |
| C15:0 (Pentadecanoic acid) | 0.12 ± 0.01 | 0.15 ± 0.03 | N.D |
| C16:0 (Palmitic acid) | 18.55 ± 0.02 | 22.35 ± 0.48 | 25.02 ± 0.03 |
| C16:1 (Palmitoleic acid) | 0.51 ± 0.02 | 0.53 ± 0.08 | 0.55 ± 0.04 |
| C16:2 (Hexadecadienoic acid) | 2.29 ± 0.07 | 4.25 ± 0.33 | 5.16 ± 0.04 |
| C16:3 (Hexadecatrienoic acid) | 17.31 ± 0.19 | 8.08 ± 0.31 | 2.93 ± 0.11 |
| C18:0 (Stearic acid) | 1.30 ± 0.10 | 1.52 ± 0.05 | 2.36 ± 0.28 |
| C18:1 (Oleic acid) | 8.05 ± 0.16 | 9.00 ± 0.12 | 13.78 ± 0.46 |
| C18:2 (Linoleic acid) | 8.52 ± 0.08 | 17.97 ± 0.84 | 29.54 ± 0.17 |
| C18:3 (α-Linolenic acid) | 43.35 ± 0.05 | 36.14 ± 2.01 | 20.67 ± 0.42 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jo, S.-W.; Do, J.-M.; Kang, N.S.; Park, J.M.; Lee, J.H.; Kim, H.S.; Hong, J.W.; Yoon, H.-S. Isolation, Identification, and Biochemical Characteristics of a Cold-Tolerant Chlorella vulgaris KNUA007 Isolated from King George Island, Antarctica. J. Mar. Sci. Eng. 2020, 8, 935. https://doi.org/10.3390/jmse8110935
Jo S-W, Do J-M, Kang NS, Park JM, Lee JH, Kim HS, Hong JW, Yoon H-S. Isolation, Identification, and Biochemical Characteristics of a Cold-Tolerant Chlorella vulgaris KNUA007 Isolated from King George Island, Antarctica. Journal of Marine Science and Engineering. 2020; 8(11):935. https://doi.org/10.3390/jmse8110935
Chicago/Turabian StyleJo, Seung-Woo, Jeong-Mi Do, Nam Seon Kang, Jong Myong Park, Jae Hak Lee, Han Soon Kim, Ji Won Hong, and Ho-Sung Yoon. 2020. "Isolation, Identification, and Biochemical Characteristics of a Cold-Tolerant Chlorella vulgaris KNUA007 Isolated from King George Island, Antarctica" Journal of Marine Science and Engineering 8, no. 11: 935. https://doi.org/10.3390/jmse8110935
APA StyleJo, S.-W., Do, J.-M., Kang, N. S., Park, J. M., Lee, J. H., Kim, H. S., Hong, J. W., & Yoon, H.-S. (2020). Isolation, Identification, and Biochemical Characteristics of a Cold-Tolerant Chlorella vulgaris KNUA007 Isolated from King George Island, Antarctica. Journal of Marine Science and Engineering, 8(11), 935. https://doi.org/10.3390/jmse8110935







