Enhancing Phytoplankton Recognition Through a Hybrid Dataset and Morphological Description-Driven Prompt Learning
Abstract
1. Introduction
- Development of a Hybrid Dataset: We propose a novel dataset that combines laboratory-based data with real-world environmental samples, improving the model’s applicability in both laboratory-based and natural settings.
- Evaluation of Recognition Algorithms: We compare the performances of CNNs and Transformer models using the hybrid dataset, revealing the superior performance of CNNs in phytoplankton classification. Additionally, we demonstrate the effectiveness of incorporating enhanced prompt descriptions into a fine-tuned visual language model to improve recognition accuracy.
- Experimental Evaluation: Extensive experiments on the hybrid dataset show that the proposed approach achieves superior recognition performance, highlighting its potential for real-world applications.
2. Related Work
2.1. Phytoplankton Observation and Image Identification
2.2. Multimodal Learning and Prompt Fine-Tuning
2.3. Current Large-Scale Datasets for Phytoplankton Identification
3. Methodology
3.1. Data Preparation and Preprocessing
3.1.1. Data Collection
3.1.2. Data Annotation and Model Training
3.1.3. Model Testing and Data Expansion
3.1.4. Data Cleaning and Merging
3.2. Description Guided Prompting Learning
3.2.1. Motivation
3.2.2. Problem Definition
3.2.3. Prompt Enhancement with Morphological Descriptions
4. Results
4.1. Implementation Detail
4.2. Comparison Methods
4.3. Experiment Results
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Marzidovšek, M.; Mozetič, P.; Francé, J.; Podpečan, V. Computer Vision Techniques for Morphological Analysis and Identification of Two Pseudo-nitzschia Species. Water 2024, 16, 2160. [Google Scholar] [CrossRef]
- Yu, Y.; Li, Y.; Sun, X.; Dong, J. MPT: A large-scale multiphytoplankton tracking benchmark. Intell. Mar. Technol. Syst 2024, 2, 35. [Google Scholar] [CrossRef]
- Natchimuthu, S.; Chinnaraj, P.; Parthasarathy, S.; Senthil, K. Automatic identification of algal community from microscopic images. Bioinform. Biol. Insights 2013, 7, 327–334. [Google Scholar] [CrossRef]
- Jia, R.; Yin, G.; Zhao, N.; Chen, X.; Xu, M.; Hu, X.; Huang, P.; Liang, P.; He, Q.; Zhang, X. Phytoplankton Image Segmentation and Annotation Method Based on Microscopic Fluorescence. J. Fluoresc. 2025, 35, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Dimitrovski, I.; Kocev, D.; Loskovska, S.; Džeroski, S. Hierarchical classification of diatom images using ensembles of predictive clustering trees. Ecol. Inform. 2012, 7, 19–29. [Google Scholar] [CrossRef]
- Yann, L.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization, Ver. 9. arXiv 2017, arXiv:1412.6980. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.E.; Hinton, G. ImageNet Classification with Deep Convolutional Neural Networks. Commun 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Radford, A.; Kim, J.; Hallacy, C.; Ramesh, A.; Goh, G.; Agarwal, S.; Sastry, G.; Askell, A.; Mishkin, P.; Clark, J.; et al. Learning Transferable Visual Models From Natural Language Supervision, Ver. 1. arXiv 2021, arXiv:2103.00020. [Google Scholar]
- Goyal, S.; Kumar, A.; Garg, S.; Kolter, Z.; Raghunathan, A. Finetune like you pretrain: Improved finetuning of zero-shot vision models. In Proceedings of the CVPR, Vancouver, BC, Canada, 18–22 June 2023; pp. 19338–19347. [Google Scholar]
- Xie, Y.; Lu, H.; Yan, J.; Yang, X.; Tomizuka, M.; Zhan, W. Active Finetuning: Exploiting Annotation Budget in the Pretraining-Finetuning Paradigm. In Proceedings of the CVPR, Vancouver, BC, Canada, 18–22 June 2023; pp. 23715–23724. [Google Scholar]
- Orenstein, E.C.; Beijbom, O.; Peacock, E.E.; Sosik, H.M. WHOI-Plankton- A Large Scale Fine Grained Visual Recognition Benchmark Dataset for Plankton Classification, Ver. 1. arXiv 2015, arXiv:1510.00745. [Google Scholar]
- Li, Q.; Sun, X.; Dong, J.; Song, S.; Zhang, T.; Liu, D.; Zhang, H.; Han, S. Developing a microscopic image dataset in support of intelligent phytoplankton detection using deep learning. ICES J. Mar. Sci. 2020, 77, 1427–1439. [Google Scholar] [CrossRef]
- Bi, H.; Guo, Z.; Benfield, M.C.; Fan, C.; Ford, M.; Shahrestani, S.; Sieracki, J.M. A Semi-Automated Image Analysis Procedure for In Situ Plankton Imaging Systems. PLoS ONE 2015, 10, e0127121. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask R-CNN. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2980–2988. [Google Scholar]
- Wang, A.; Chen, H.; Liu, L.; Chen, K.; Lin, Z.; Han, J.; Ding, G. YOLOv10: Real-Time End-to-End Object Detection, Ver. 2. arXiv 2024, arXiv:2405.14458. [Google Scholar]
- Zhao, N.; Zhang, X.; Yin, G.; Yang, R.; Hu, L.; Chen, S.; Liu, J.; Liu, W. On-line analysis of algae in water by discrete three-dimensional fluorescence spectroscopy. Opt. Express 2018, 26, A251–A259. [Google Scholar] [CrossRef] [PubMed]
- Bayoudh, K.; Knani, R.; Hamdaoui, F.; Mtibaa, A. A survey on deep multimodal learning for computer vision: Advances, trends, applications, and datasets. Vis. Comput. 2022, 38, 2939–2970. [Google Scholar] [CrossRef]
- Zhou, K.; Yang, J.; Loy, C.C.; Liu, Z. Learning to Prompt for Vision-Language Models, Ver. 6. arXiv 2022, arXiv:2109.01134. [Google Scholar]
- Zheng, H.; Wang, R.; Yu, Z.; Wang, N.; Gu, Z.; Zheng, B. Automatic plankton image classification combining multiple view features via multiple kernel learning. BMC Bioinform 2017, 18, 57. [Google Scholar] [CrossRef]
- Cowen, R.K.; Guigand, C.M. In situ ichthyoplankton imaging system (ISIIS): System design and preliminary results. Limnol. Oceanogr. Methods 2008, 6, 126–132. [Google Scholar] [CrossRef]
- Brotas, V.; Tarran, G.A.; Veloso, V.; Brewin, R.; Wooodward, E.S.; Airs, R.; Beltran, C.; Ferreira, A.; Groom, S.B. Complementary Approaches to Assess Phytoplankton Groups and Size Classes on a Long Transect in the Atlantic Ocean. Sec. Mar. Ecosyst. Ecol. 2022, 8. [Google Scholar] [CrossRef]
- Bartlett, B.; Santos, M.; Dorian, T.; Moreno, M.; Trslic, P.; Dooly, G. Real-Time UAV Surveys with the Modular Detection and Targeting System: Balancing Wide-Area Coverage and High-Resolution Precision in Wildlife Monitoring. Remote Sens. 2025, 17, 879. [Google Scholar] [CrossRef]
- China Species Library. Available online: https://species.sciencereading.cn/biology/v/botanyIndex/122/ZLZK.html (accessed on 6 March 2025).
- Database of Plant Bioresources in the Coastal Zone of China (NBSDC-DB-22). Available online: http://algae.yic.ac.cn/taxa/doku.php?id=start (accessed on 8 March 2025).
- Qian, S.; Liu, D.; Sun, J. Marine Phycology; China Ocean University Press: Qingdao, China, 2014. [Google Scholar]
- Yu, Y.; Lv, Q.; Li, Y.; Wei, Z.; Dong, J. PhyTracker: An Online Tracker for Phytoplankton. IEEE Trans. Circuits Syst. Video Technol. 2024, 35, 2932–2944. [Google Scholar] [CrossRef]
- Howard, A.; Sandler, M.; Chu, G.; Chen, L.; Chen, B.; Tan, M.; Wang, W.; Zhu, Y.; Pang, R.; Vasudevan, V.; et al. Searching for MobileNetV3. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Republic of Korea, 27 October–2 November 2019; pp. 1314–1324. [Google Scholar]
- Sieracki, C.K.; Sieracki, M.E.; Yentsch, C.S. An imaging-in-flow system for automated analysis of marine microplankton. Mar. Ecol. Prog. Ser. 1998, 168, 285–296. [Google Scholar] [CrossRef]
- Grosjean, P.; Picheral, M.; Warembourg, C.; Gorsky, G. Enumeration, measurement, and identification of net zooplankton samples using the ZOOSCAN digital imaging system. ICES J. Mar. Sci. 2004, 61, 518–525. [Google Scholar] [CrossRef]
- Olson, R.J.; Sosik, H.M. A submersible imaging-in-flow instrument to analyze nano-and microplankton: Imaging FlowCytobot. Limnol. Oceanogr. Methods 2007, 5, 195–203. [Google Scholar] [CrossRef]
- Nardelli, S.C.; Gray, P.C.; Schofield, O. A Convolutional Neural Network to Classify Phytoplankton Images Along the West Antarctic Peninsula. Mar. Technol. Soc. J. 2022, 56, 45–57. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition, Ver. 6. arXiv 2015, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition, Ver. 1. arXiv 2015, arXiv:1512.03385. [Google Scholar]
- Huang, G.; Liu, Z.; Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks, Ver. 5. arXiv 2018, arXiv:1608.06993. [Google Scholar]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks, Ver. 5. arXiv 2020, arXiv:1905.11946. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale, Ver. 2. arXiv 2021, arXiv:2010.11929. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer using Shifted Windows, Ver. 2. arXiv 2021, arXiv:2103.14030. [Google Scholar]
- Touvron, H.; Cord, M.; Douze, M.; Massa, F.; Sablayrolles, A.; Jégou, H. Training data-efficient image transformers & distillation through attention, Ver. 2. arXiv 2021, arXiv:2012.12877. [Google Scholar]
- Liu, Z.; Mao, H.; Wu, C.; Feichtenhofer, C.; Darrell, T.; Xie, S. A ConvNet for the 2020s, Ver. 2. arXiv 2022, arXiv:2201.03545. [Google Scholar]
- Li, Z.; Wu, X.; Du, H.; Liu, F.; Nghiem, H.; Shi, G. A Survey of State of the Art Large Vision Language Models: Alignment, Benchmark, Evaluations and Challenges, Ver. 6. arXiv 2025, arXiv:2501.02189. [Google Scholar]
- Li, J.; Li, D.; Xiong, C.; Hoi, S. BLIP: Bootstrapping Language-Image Pre-training for Unified Vision-Language Understanding and Generation, Ver. 2. arXiv 2022, arXiv:2201.12086. [Google Scholar]
- O’Shea, K.; Nash, R. An Introduction to Convolutional Neural Networks, Ver. 2. arXiv 2015, arXiv:1511.08458. [Google Scholar]
- Zhou, K.; Yang, J.; Loy, C.C.; Liu, Z. Conditional Prompt Learning for Vision-Language Models, Ver. 2. arXiv 2022, arXiv:2203.05557. [Google Scholar]
- Zhang, Y.; Wei, D.; Qin, C.; Wang, H.; Pfister, H.; Fu, Y. Context Reasoning Attention Network for Image Super-Resolution. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, BC, Canada, 11–17 October 2021; pp. 4258–4267. [Google Scholar]
- Feng, Z.; Yang, J.; Chen, L.; Chen, Z.; Li, L. An Intelligent Waste-Sorting and Recycling Device Based on Improved EfficientNet. Int. J. Environ. Res. Public Health 2022, 19, 15987. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, M.; Ishibashi, R.; Lin, M. Norm-Regularized Token Compression in Vision Transformer Networks. In Proceedings of the 6th International Symposium on Advanced Technologies and Applications in the Internet of Things, Shiga, Japan, 19–22 August 2024. [Google Scholar]
- Wei, C.; Duke, B.; Jiang, R.; Aarabi, P.; Taylor, G.W.; Shkurti, F. Sparsifiner: Learning Sparse Instance-Dependent Attention for Efficient Vision Transformers, Ver. 1. arXiv 2023, arXiv:2303.13755. [Google Scholar]
- Xie, S.; Girshick, R.; Dollár, P.; Tu, Z.; He, K. Aggregated Residual Transformations for Deep Neural Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 5987–5995. [Google Scholar]

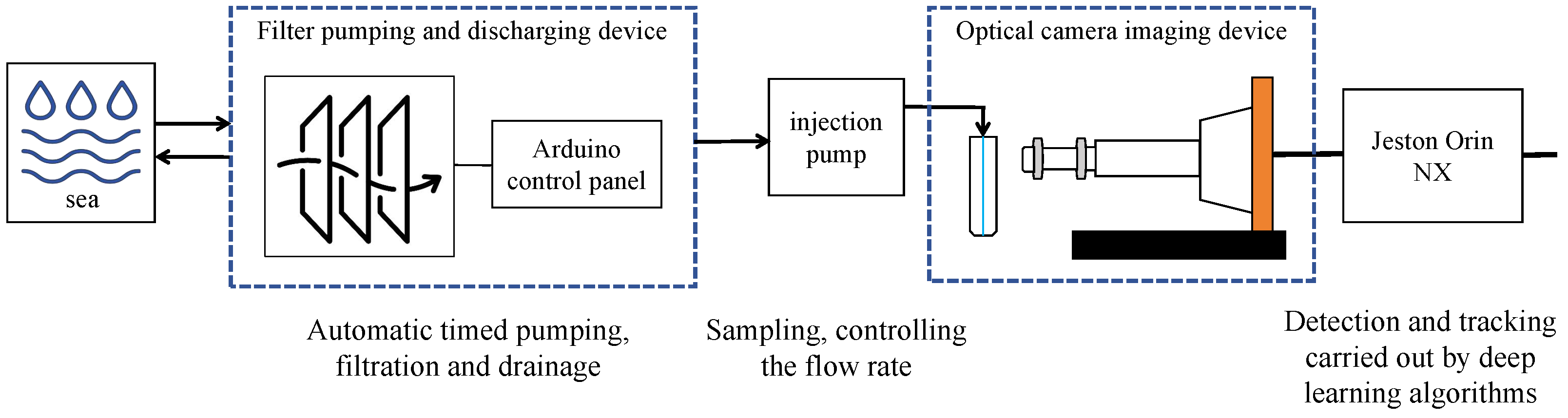

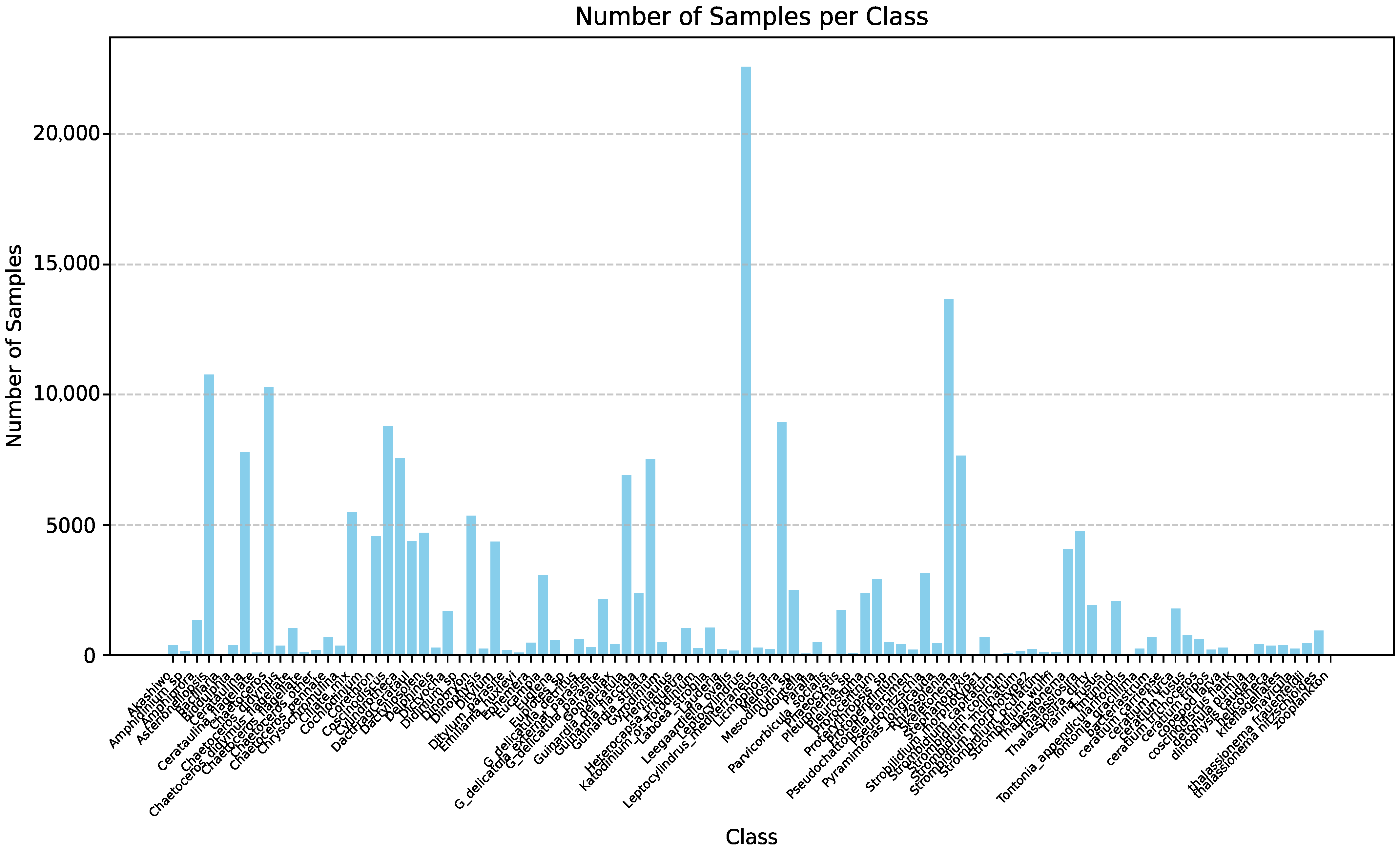
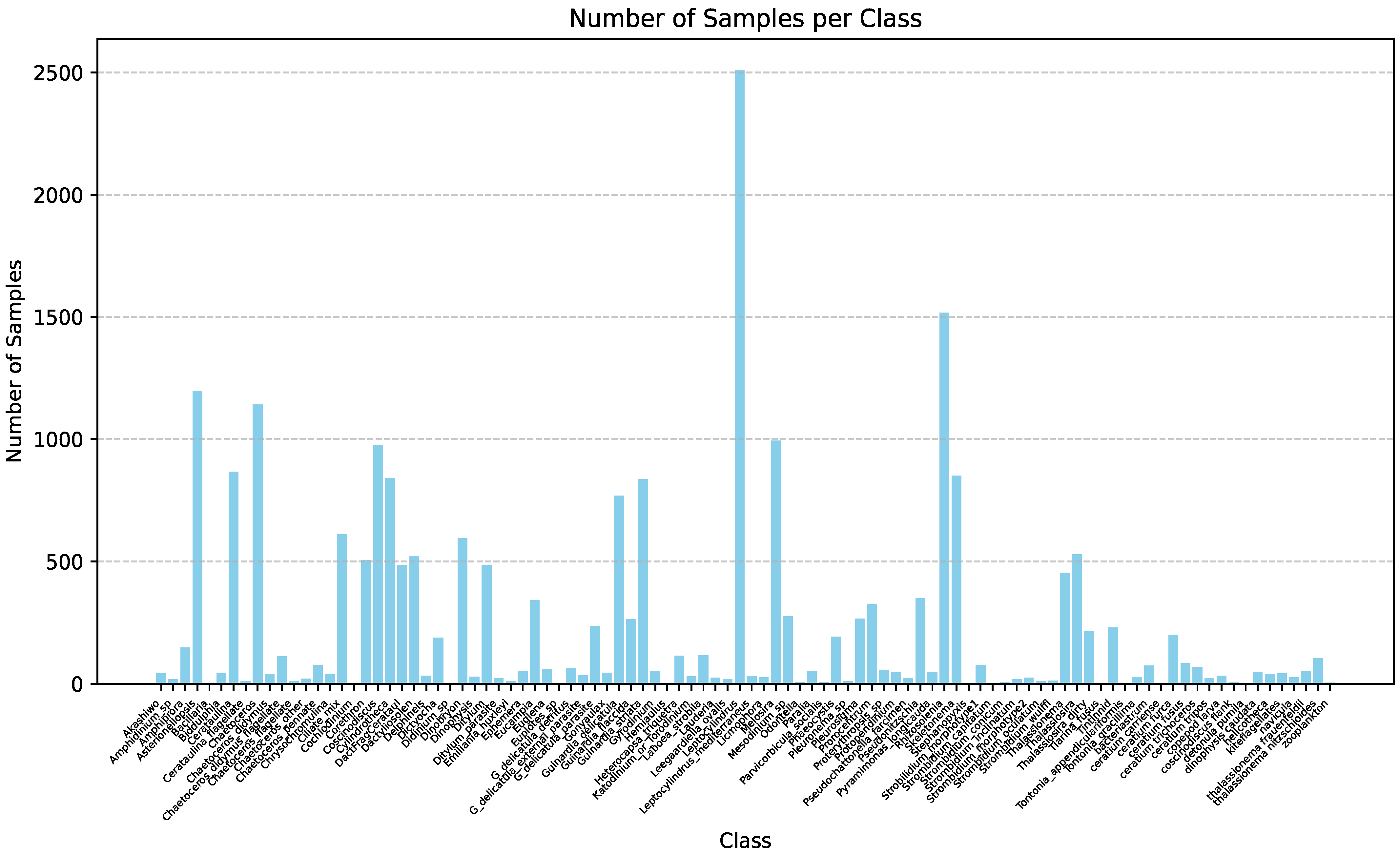

| Method | mAP | Parameter | Infer Time (640) | FPS | Infer Time (640 Orin NX) | FPS |
|---|---|---|---|---|---|---|
| Yolov10s | 32.80 | 7.2 M | 31 ms | 32.3 | 54 ms | 18.5 |
| Yolov10m | 35.90 | 15.4 M | 36 ms | 27.8 | 55 ms | 18.2 |
| Yolov10b | 43.22 | 19.1 M | 38 ms | 26.3 | 57 ms | 17.5 |
| Yolov10l | 45.77 | 24.4 M | 42 ms | 23.8 | 62 ms | 16.1 |
| Yolov10x | 48.60 | 29.5 M | 45 ms | 22.2 | 64 ms | 15.6 |
| Model | mAcc. | oAcc. | Cls8 | Cls15 | Cls48 | Cls92 |
|---|---|---|---|---|---|---|
| ResNet50 [34] | 83.53 | 94.48 | 91.67 | 70.00 | 93.36 | 85.92 |
| ResNet101 [34] | 84.11 | 94.48 | 75.00 | 85.00 | 93.36 | 91.55 |
| ResNeXt50 [49] | 84.36 | 94.41 | 58.33 | 85.00 | 94.01 | 86.85 |
| ResNeXt101 [49] | 77.62 | 92.14 | 41.67 | 65.00 | 94.66 | 89.20 |
| MobileNet [28] | 83.33 | 93.92 | 66.67 | 82.50 | 94.01 | 89.20 |
| Efficient-b0 [36] | 83.74 | 94.40 | 83.33 | 92.50 | 94.14 | 84.98 |
| Efficient-b4 [36] | 84.54 | 94.54 | 83.33 | 80.00 | 93.75 | 88.73 |
| Ours-ResNet50 | 84.14 | 93.20 | 91.67 | 72.50 | 88.54 | 90.12 |
| Ours-ResNet101 | 85.27 | 93.76 | 91.67 | 75.00 | 88.54 | 89.67 |
| Model | mAcc. | oAcc. | Cls8 | Cls15 | Cls48 | Cls92 |
|---|---|---|---|---|---|---|
| ViT-Small [37] | 80.44 | 94.03 | 66.67 | 67.50 | 94.01 | 84.51 |
| ViT-Base [37] | 80.66 | 93.60 | 58.33 | 52.50 | 93.10 | 85.92 |
| Swin-Small [38] | 83.39 | 94.19 | 83.33 | 67.50 | 94.53 | 91.08 |
| Swin-Base [38] | 84.12 | 94.61 | 84.33 | 68.03 | 94.98 | 92.02 |
| Convnext-Small [40] | 84.71 | 94.68 | 66.67 | 82.50 | 93.36 | 90.14 |
| Convnext-Base [40] | 85.72 | 95.05 | 66.67 | 82.50 | 93.88 | 89.20 |
| Ours-ResNet50 | 84.14 | 93.20 | 91.67 | 72.50 | 88.54 | 90.12 |
| Ours-ResNet101 | 85.27 | 93.76 | 91.67 | 75.00 | 88.54 | 89.67 |
| Ours-ViTBase | 72.62 | 90.10 | 66.67 | 50.00 | 88.80 | 92.02 |
| Backbone | mAcc. | oAcc. | Cls8 | Cls15 | Cls48 | Cls92 |
|---|---|---|---|---|---|---|
| CLIP-Res50 [9] | 83.74 | 93.12 | 83.33 | 75.00 | 87.22 | 90.12 |
| CLIP-Res101 [9] | 84.92 | 93.55 | 100.0 | 80.00 | 88.28 | 88.73 |
| CLIP-ViT-Base [9] | 72.14 | 89.08 | 66.67 | 50.00 | 87.92 | 86.85 |
| CoOp-Res50 [19] | 40.82 | 68.06 | 0.00 | 7.50 | 74.34 | 65.73 |
| CoOp-Res101 [19] | 40.88 | 69.23 | 0.00 | 7.50 | 72.34 | 63.85 |
| CoOp-ViT/Base [19] | 10.72 | 40.85 | 0.00 | 0.00 | 62.50 | 12.68 |
| Ours-ResNet50 | 84.14 | 93.20 | 91.67 | 72.50 | 88.54 | 90.12 |
| Ours-ResNet101 | 85.27 | 93.76 | 91.67 | 75.00 | 88.54 | 89.67 |
| Ours-ViTBase | 72.62 | 90.10 | 66.67 | 50.00 | 88.80 | 92.02 |
| Model | mAcc. | oAcc. | Cls8 | Cls15 | Cls48 | Cls92 |
|---|---|---|---|---|---|---|
| 83.74 | 93.12 | 83.33 | 75.00 | 87.22 | 90.12 | |
| 84.11 | 94.48 | 91.67 | 72.50 | 88.54 | 90.12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huo, Y.; Lv, Q.; Dong, J. Enhancing Phytoplankton Recognition Through a Hybrid Dataset and Morphological Description-Driven Prompt Learning. J. Mar. Sci. Eng. 2025, 13, 1680. https://doi.org/10.3390/jmse13091680
Huo Y, Lv Q, Dong J. Enhancing Phytoplankton Recognition Through a Hybrid Dataset and Morphological Description-Driven Prompt Learning. Journal of Marine Science and Engineering. 2025; 13(9):1680. https://doi.org/10.3390/jmse13091680
Chicago/Turabian StyleHuo, Yubo, Qingxuan Lv, and Junyu Dong. 2025. "Enhancing Phytoplankton Recognition Through a Hybrid Dataset and Morphological Description-Driven Prompt Learning" Journal of Marine Science and Engineering 13, no. 9: 1680. https://doi.org/10.3390/jmse13091680
APA StyleHuo, Y., Lv, Q., & Dong, J. (2025). Enhancing Phytoplankton Recognition Through a Hybrid Dataset and Morphological Description-Driven Prompt Learning. Journal of Marine Science and Engineering, 13(9), 1680. https://doi.org/10.3390/jmse13091680







