The Complete Mitochondrial Genome of the Chemosymbiotic Lucinid Bivalve Pillucina pisidium (Dunker, 1860) Occurring in Seagrass Zostera marina Bed in a Lagoon in Jeju Island, Korea
Abstract
1. Introduction
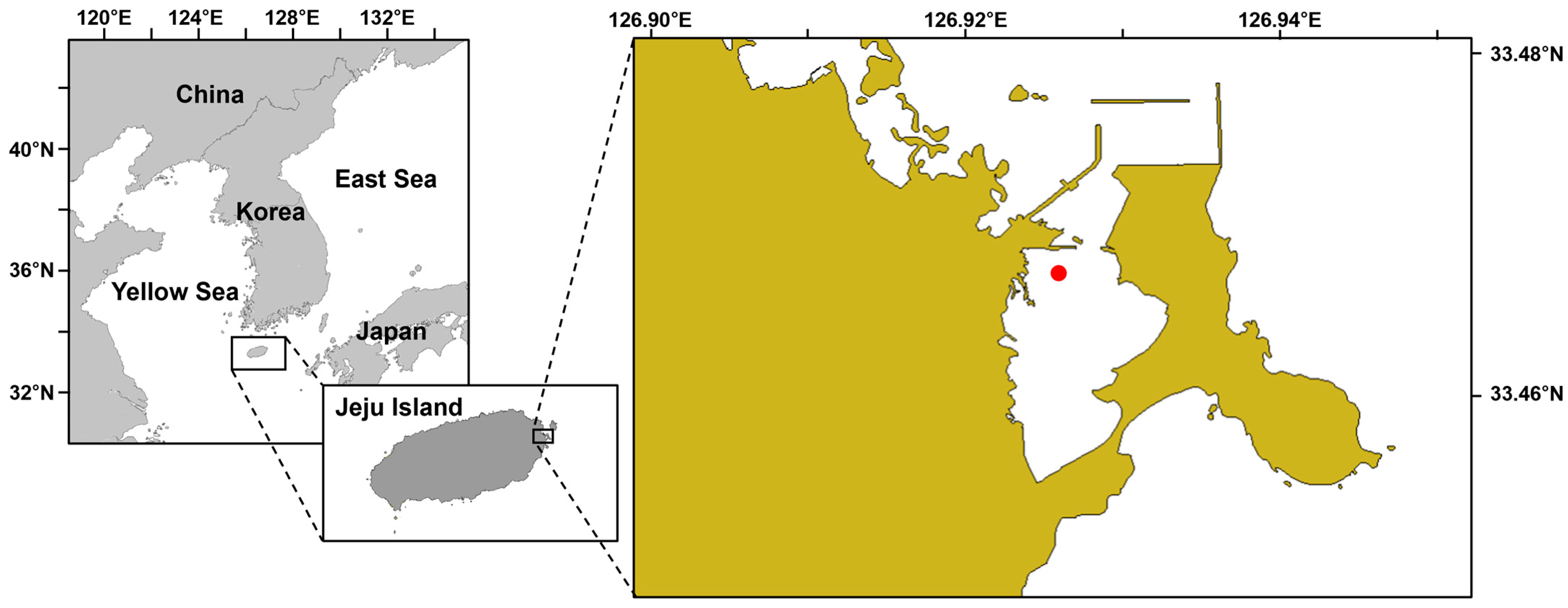
2. Materials and Methods
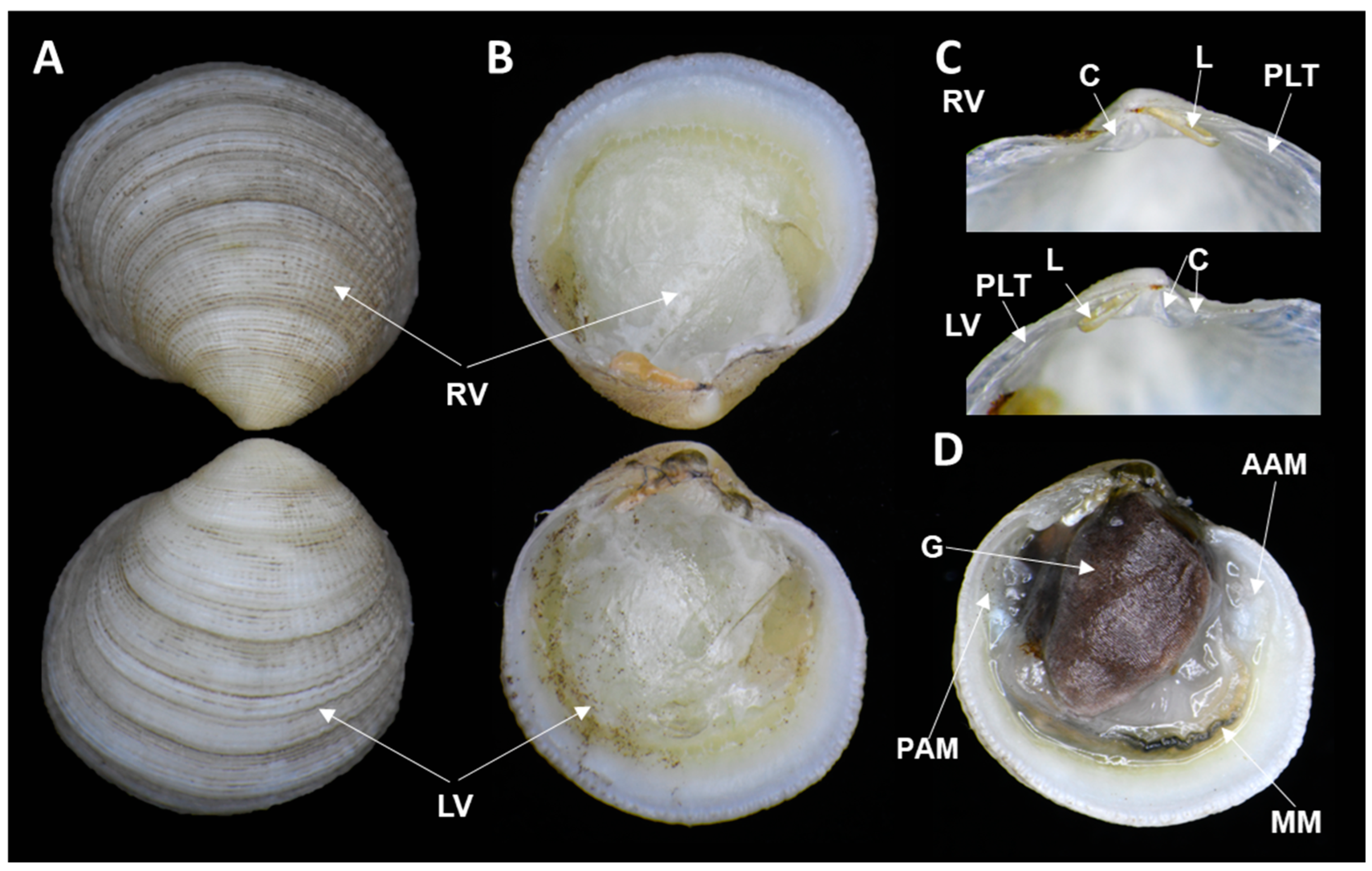
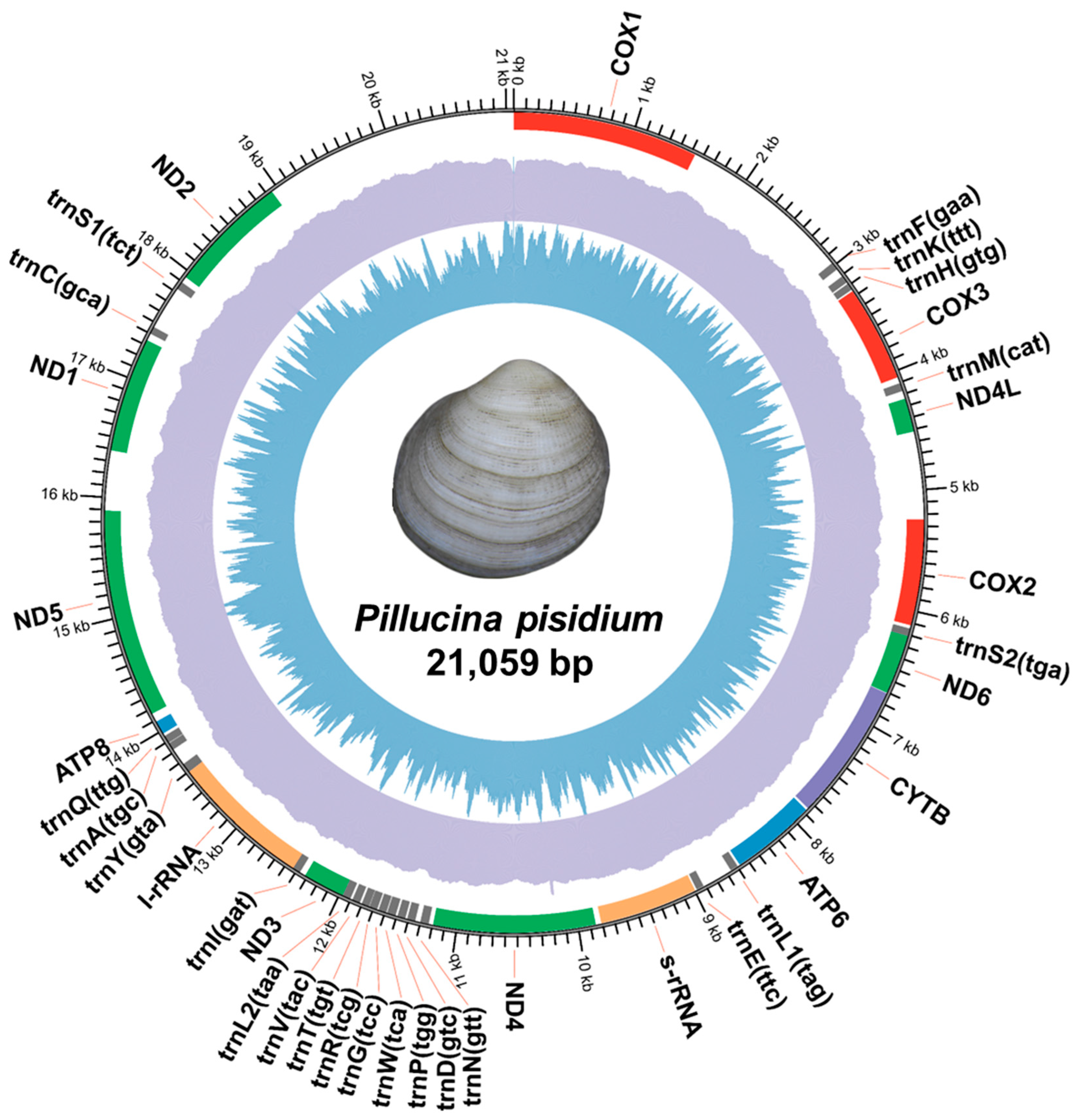
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dubilier, N.; Bergin, C.; Lott, C. Symbiotic Diversity in Marine Animals: The Art of Harnessing Chemosynthesis. Nat. Rev. Microbiol. 2008, 6, 725–740. [Google Scholar] [CrossRef]
- Cavanaugh, C.M.; Gardiner, S.L.; Jones, M.L.; Jannasch, H.W.; Waterbury, J.B. Prokaryotic Cells in the Hydrothermal Vent Tube Worm Riftia pachyptila Jones: Possible Chemoautotrophic Symbionts. Science 1981, 213, 340–342. [Google Scholar] [CrossRef] [PubMed]
- Duperron, S.; Gaudron, S.M.; Rodrigues, C.F.; Cunha, M.R.; Decker, C.; Olu, K. An Overview of Chemosynthetic Symbioses in Bivalves from the North Atlantic and Mediterranean Sea. Biogeosciences 2013, 10, 3241–3267. [Google Scholar] [CrossRef]
- DeLeo, D.M.; Morrison, C.L.; Sei, M.; Salamone, V.; Demopoulos, A.W.J.; Quattrini, A.M. Genetic Diversity and Connectivity of Chemosynthetic Cold Seep Mussels from the U.S. Atlantic Margin. BMC Ecol. Evol. 2022, 22, 76. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhang, Y.; Xu, T.; Zhang, Y.; Mu, H.; Zhang, Y.; Lan, Y.; Fields, C.J.; Hui, J.H.L.; Zhang, W.; et al. Adaptation to Deep-Sea Chemosynthetic Environments as Revealed by Mussel Genomes. Nat. Ecol. Evol. 2017, 1, 121. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.; Glover, E. Biology, Evolution and Generic Review of the Chemosymbiotic Bivalve Family Lucinidae; Ray Society: London, UK, 2021; pp. 11–12, 43–52. ISBN 978-0-903874-53-3. [Google Scholar]
- Åström, E.K.L.; Oliver, P.G.; Carroll, M.L. A New Genus and Two New Species of Thyasiridae Associated with Methane Seeps off Svalbard, Arctic Ocean. Mar. Biol. Res. 2017, 13, 402–416. [Google Scholar] [CrossRef]
- Childress, J.J.; Fisher, C.R.; Favuzzi, J.A.; Arp, A.J.; Oros, D.R. The Role of a Zinc-Based, Serum-Borne Sulphide-Binding Component in the Uptake and Transport of Dissolved Sulphide by the Chemoautotrophic Symbiont-Containing Clam Calyptogena elongata. J. Exp. Biol. 1993, 179, 131–158. [Google Scholar] [CrossRef]
- Duperron, S.; Fiala-Médioni, A.; Caprais, J.-C.; Olu, K.; Sibuet, M. Evidence for Chemoautotrophic Symbiosis in a Mediterranean Cold Seep Clam (Bivalvia: Lucinidae): Comparative Sequence Analysis of Bacterial 16S rRNA, APS Reductase and RubisCO Genes. FEMS Microbiol. Ecol. 2007, 59, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Glover, E.A.; Taylor, J.D.; Williams, S.T. Mangrove-Associated Lucinid Bivalves of the Central Indo-West Pacific: Review of the “Austriella” Group with a New Genus and Species (Mollusca: Bivalvia: Lucinidae). Raffles Bull. Zool. Suppl. 2008, 18, 25–40. [Google Scholar]
- Lim, S.J.; Davis, B.G.; Gill, D.E.; Walton, J.; Nachman, E.; Engel, A.S.; Anderson, L.C.; Campbell, B.J. Taxonomic and Functional Heterogeneity of the Gill Microbiome in a Symbiotic Coastal Mangrove Lucinid Species. ISME J. 2019, 13, 902–920. [Google Scholar] [CrossRef]
- van der Heide, T.; Govers, L.L.; de Fouw, J.; Olff, H.; van der Geest, M.; van Katwijk, M.M.; Piersma, T.; van de Koppel, J.; Silliman, B.R.; Smolders, A.J.P.; et al. A Three-Stage Symbiosis Forms the Foundation of Seagrass Ecosystems. Science 2012, 336, 1432–1434. [Google Scholar] [CrossRef] [PubMed]
- Glover, E.A.; Taylor, J.D. Lucinidae of the Philippines: Highest Known Diversity and Ubiquity of Chemosymbiotic Bivalves from Intertidal to Bathyal Depths (Mollusca: Bivalvia). Trop. Deep-Sea Benthos 2016, 29, 65–234. [Google Scholar]
- Taylor, J.D.; Glover, E.A. Lucinidae (Bivalvia)–the Most Diverse Group of Chemosymbiotic Molluscs. Zool. J. Linn. Soc. 2006, 148, 421–438. [Google Scholar] [CrossRef]
- Frenkiel, L.; Mouëza, M. Gill Ultrastructure and Symbiotic Bacteria in Codakia Orbicularis (Bivalvia, Lucinidae). Zoomorphology 1995, 115, 51–61. [Google Scholar] [CrossRef]
- Frenkiel, L.; Gros, O.; Mouëza, M. Gill Structure in Lucina Pectinata (Bivalvia: Lucinidae) with Reference to Hemoglobin in Bivalves with Symbiotic Sulphur-Oxidizing Bacteria. Mar. Biol. 1996, 125, 511–524. [Google Scholar] [CrossRef]
- Herry, A.; Diouris, M.; Le Pennec, M. Chemoautotrophic Symbionts and Translocation of Fixed Carbon from Bacteria to Host Tissues in the Littoral Bivalve Loripes lucinalis (Lucinidae). Mar. Biol. 1989, 101, 305–312. [Google Scholar] [CrossRef]
- Taylor, J.D.; Glover, E.A. Functional Anatomy, Chemosymbiosis and Evolution of the Lucinidae. In The Evolutionary Biology of the Bivalvia; Harper, E.M., Taylor, J.D., Crame, J.A., Eds.; Geological Society of London: London, UK, 2000; Volume 177, ISBN 978-1-86239-076-8. [Google Scholar]
- König, S.; Gros, O.; Heiden, S.E.; Hinzke, T.; Thuermer, A.; Poehlein, A.; Meyer, S.; Vatin, M.; Tocny, J.; Ponnudurai, R. Nitrogen Fixation in a Chemoautotrophic Lucinid Symbiosis. Nat. Microbiol. 2016, 2, 16193. [Google Scholar] [CrossRef]
- Petersen, J.M.; Kemper, A.; Gruber-Vodicka, H.; Cardini, U.; Van Der Geest, M.; Kleiner, M.; Bulgheresi, S.; Mußmann, M.; Herbold, C.; Seah, B.K. Chemosynthetic Symbionts of Marine Invertebrate Animals Are Capable of Nitrogen Fixation. Nat. Microbiol. 2016, 2, 16195. [Google Scholar] [CrossRef] [PubMed]
- Glover, E.; Taylor, J. Systematic Revision of Australian and Indo-Pacific Lucinidae (Mollusca: Bivalvia): Pillucina, Wallucina and Descriptions of Two New Genera and Four New Species. Rec. Aust. Mus. 2001, 53, 263–292. [Google Scholar] [CrossRef][Green Version]
- Rodionov, I.A.; Yushin, V.V. Procaryotic Symbionts in Gill Cells of the Bivalve Mollusc Pillucina Pisidium. Biol. Morya 1991, 1, 39–46. [Google Scholar]
- Taylor, J.D.; Glover, E.A.; Williams, S.T. Diversification of Chemosymbiotic Bivalves: Origins and Relationships of Deeper Water Lucinidae. Biol. J. Linn. Soc. 2014, 111, 401–420. [Google Scholar] [CrossRef]
- Uede, T.; Yamauchi, M.; Takahashi, Y. Distribution and Habitat Environment of Pillucina pisidium (Bivalvia, Licinidae) in Zostera Japonica Beds in the Intertidal Zone at Uchinoura, Tanabe Bay, Wakayama, Japan. Jpn. J. Benthol. 2013, 68, 28–36. [Google Scholar] [CrossRef]
- Min, D.K.; Lee, J.S.; Koh, D.B.; Je, J.G. Mollusks in Korea; Min Molluscan Research Institute: Seoul, Korea, 2004; p. 566. [Google Scholar]
- Zhukova, N.V.; Kharlamenko, V.I.; Svetashev, V.I.; Rodionov, I.A. Fatty Acids as Markers of Bacterial Symbionts of Marine Bivalve Molluscs. J. Exp. Mar. Biol. Ecol. 1992, 162, 253–263. [Google Scholar] [CrossRef]
- Kharlamenko, V.I.; Kiyashko, S.I.; Imbs, A.B.; Vyshkvartzev, D.I. Identification of Food Sources of Invertebrates from the Seagrass Zostera Marina Community Using Carbon and Sulfur Stable Isotope Ratio and Fatty Acid Analyses. Mar. Ecol. Prog. Ser. 2001, 220, 103–117. [Google Scholar] [CrossRef]
- Lee, H.-J.; Noseworthy, R.G.; Park, S.; Hong, H.-K.; Lee, B.-G.; Choi, K.-S. Report on the Molluscan Fauna in Tongbatarl Lagoon on the East Coast of Jeju, Korea. Korean J. Malacol. 2014, 30, 95–99. [Google Scholar] [CrossRef][Green Version]
- Lutaenko, K.A.; Je, J.-G.; Shin, S.-H. Bivalve Mollusks in Yeongil Bay, Korea. 2. Faunal Analysis. Korean J. Malacol. 2006, 22, 63–86. [Google Scholar]
- Noseworthy, R.G.; Lim, N.-R.; Choi, K.-S. A Catalogue of the Mollusks of Jeju Island, South Korea. Korean J. Malacol. 2007, 23, 65–104. [Google Scholar]
- Taylor, J.D.; Glover, E.A.; Smith, L.; Dyal, P.; Williams, S.T. Molecular Phylogeny and Classification of the Chemosymbiotic Bivalve Family Lucinidae (Mollusca: Bivalvia). Zool. J. Linn. Soc. 2011, 163, 15–49. [Google Scholar] [CrossRef]
- Friedman, J.R.; Nunnari, J. Mitochondrial Form and Function. Nature 2014, 505, 335–343. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect Mitochondrial Genomics: Implications for Evolution and Phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef]
- Kim, M.; Choi, H.; Kim, H.; Kang, J.; Jeong, H.G.; Eyun, S.; Kang, J.-H. Characterization of the Mitochondrial Genome, Ecological Distribution, and Morphological Features of the Marine Gastropod Mollusc Lophocochlias parvissimus (Gastropoda, Tornidae). J. Mar. Sci. Eng. 2023, 11, 2307. [Google Scholar] [CrossRef]
- Ma, C.; Yang, P.; Jiang, F.; Chapuis, M.; Shali, Y.; Sword, G.A.; Kang, L. Mitochondrial Genomes Reveal the Global Phylogeography and Dispersal Routes of the Migratory Locust. Mol. Ecol. 2012, 21, 4344–4358. [Google Scholar] [CrossRef]
- Osvatic, J.T.; Wilkins, L.G.E.; Leibrecht, L.; Leray, M.; Zauner, S.; Polzin, J.; Camacho, Y.; Gros, O.; van Gils, J.A.; Eisen, J.A.; et al. Global Biogeography of Chemosynthetic Symbionts Reveals Both Localized and Globally Distributed Symbiont Groups. Proc. Natl. Acad. Sci. USA 2021, 118, e2104378118. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Meng, G.; Li, Y.; Yang, C.; Liu, S. MitoZ: A Toolkit for Animal Mitochondrial Genome Assembly, Annotation and Visualization. Nucleic Acids Res. 2019, 47, e63. [Google Scholar] [CrossRef] [PubMed]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. metaSPAdes: A New Versatile Metagenomic Assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Jeon, D.; Park, Y.; Woo, H.; Eyun, S. Dietary Exposure of the Water Flea Daphnia Galeata to Microcystin-LR. Anim. Cells Syst. 2024, 28, 25–36. [Google Scholar] [CrossRef]
- Jeon, M.-S.; Jeong, D.M.; Doh, H.; Kang, H.A.; Jung, H.; Eyun, S. A Practical Comparison of the Next-Generation Sequencing Platform and Assemblers Using Yeast Genome. Life Sci. Alliance 2023, 6, e202201744. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.; Ventura, T.; Chung, J.S.; Kim, W.-J.; Nam, B.-H.; Kong, H.J.; Kim, Y.-O.; Jeon, M.-S.; Eyun, S. Twelve Quick Steps for Genome Assembly and Annotation in the Classroom. PLoS Comput. Biol. 2020, 16, e1008325. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de Novo Metazoan Mitochondrial Genome Annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, İ.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An Information Aesthetic for Comparative Genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML-VI-HPC: Maximum Likelihood-Based Phylogenetic Analyses with Thousands of Taxa and Mixed Models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A Fast, Scalable and User-Friendly Tool for Maximum Likelihood Phylogenetic Inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian Phylogenetic Inference under Mixed Models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Dunker, W.B.R.H. Mollusca Japonica Descripta et Tabulis Tribus Iconum; Schweizerbart: Stuttgart, Germany, 1861; p. 28. [Google Scholar]
- Allen, J.A.; Yonge, M. On the Basic Form and Adaptations to Habitat in the Lucinacea (Eulamellibranchia). Philos. Trans. R. Soc. Lond. B Biol. Sci. 1997, 241, 421–484. [Google Scholar] [CrossRef]
- Distel, D.L.; Felbeck, H. Endosymbiosis in the Lucinid Clams Lucinoma aequizonata, Lucinoma annulata and Lucina floridana: A Reexamination of the Functional Morphology of the Gills as Bacteria-Bearing Organs. Mar. Biol. 1987, 96, 79–86. [Google Scholar] [CrossRef]
- Johnson, M.A.; Fernandez, C.; Pergent, G. The Ecological Importance of an Invertebrate Chemoautotrophic Symbiosis to Phanerogam Seagrass Beds. Bull. Mar. Sci. 2002, 71, 1343–1351. [Google Scholar]
- Han, J.; Kim, J.G.; Kwon, O.-N.; Park, J.J.C.; Lee, K.-W.; Choi, Y.-U. On the Species Identification of Korean Geoduck Clam (Panopea Sp. 1) Based on the Morphological and Molecular Evidence. J. Mar. Sci. Eng. 2023, 11, 2115. [Google Scholar] [CrossRef]
- Smedley, G.D.; Audino, J.A.; Grula, C.; Porath-Krause, A.; Pairett, A.N.; Alejandrino, A.; Lacey, L.; Masters, F.; Duncan, P.F.; Strong, E.E.; et al. Molecular Phylogeny of the Pectinoidea (Bivalvia) Indicates Propeamussiidae to Be a Non-Monophyletic Family with One Clade Sister to the Scallops (Pectinidae). Mol. Phylogenet. Evol. 2019, 137, 293–299. [Google Scholar] [CrossRef] [PubMed]

| Subclass | Infraclass | Order | Family | Species Name | Accession Number |
|---|---|---|---|---|---|
| Autobranchia | Heteroconchia | Adapedonta | Pharidae | Sinohyriopsis schlegelii | AP018551.1 |
| Sinonovacula constricta | EU880278.1 | ||||
| Solenidae | Solen grandis | HQ703012.1 | |||
| Lucinida | Lucinidae | Loripes lacteus | EF043341.1 | ||
| Lucinella divaricata | EF043342.1 | ||||
| Pillucina pacifica | BK067723 | ||||
| Pillucina pisidium | NC_071184.1 | ||||
| Thyasiridae | Conchocele bisecta | LC126312.1 | |||
| Myida | Dreissenidae | Dreissena polymorpha | MT483676.1 | ||
| Myidae | Mya arenaria | MW727516.1 | |||
| Venerida | Veneridae | Ruditapes decussatus | KP089983.1 | ||
| Vesicomyidae | Calyptogena magnifica | KR862368.1 | |||
| Calyptogena pacifica | MT947386.1 | ||||
| Calyptogena rectimargo | MT947387.1 | ||||
| Pteriomorphia | Arcida | Arcidae | Tegillarca granosa | KJ607173.1 | |
| Mytilida | Mytilidae | Bathymodiolus brooksi | MT916743.1 | ||
| Gigantidas platifrons | AP014561.1 | ||||
| Mytilus galloprovincialis | DQ399833.1 | ||||
| Ostreida | Ostreidae | Crassostrea gigas | AF177226.1 | ||
| Pectinida | Pectinidae | Argopecten irradians | EU023915.1 | ||
| Mizuhopecten yessoensis | AB271769.1 | ||||
| Pecten maximus | KP900975.1 | ||||
| Protobranchia | - | Solemyida | Solemyidae | Solemya elarraichensis | KY244079.1 |
| Solemya pervernicosa | KY244080.1 | ||||
| Solemya velesiana | NC_034906.1 | ||||
| Solemya velum | JQ728447.1 |
| Gene | Position | Length (bp) | Initiation Codon | Stop Codon | Intergenic Nucleotide (bp) |
|---|---|---|---|---|---|
| Cytochrome c oxidase subunit I (COX1) | 1–1536 | 1536 | ATG | TAA | 2112 |
| tRNA-Phe (trnF) | 2973–3039 | 67 | 1436 | ||
| tRNA-Lys (trnK) | 3109–3173 | 65 | 69 | ||
| tRNA-His (trnH) | 3185–3244 | 60 | 14 | ||
| Cytochrome c oxidase subunit III (COX3) | 3257–4054 | 798 | ATG | TAG | 12 |
| tRNA-Met (trnM) | 4105–4173 | 69 | 50 | ||
| NADH dehydrogenase subunit 4L (ND4L) | 4239–4517 | 279 | ATG | TAA | 65 |
| Cytochrome c oxidase subunit II (COX2) | 5243–6118 | 876 | ATA | TAA | 725 |
| tRNA-Ser (trnS2) | 6145–6209 | 65 | 26 | ||
| NADH dehydrogenase subunit 6 (NAD6) | 6210–6704 | 495 | ATT | TAG | 0 |
| Cytochrome b (COB) | 6709–7851 | 1143 | ATG | TAA | 4 |
| ATP synthase F0 subunit 6 (ATP6) | 7870–8574 | 705 | ATT | TAA | 18 |
| tRNA-Leu (trnL1) | 8594–8660 | 67 | 19 | ||
| tRNA-Glu (trnE) | 8916–8980 | 65 | 255 | ||
| 12S ribosomal RNA (rrnS) | 8994–9808 | 815 | 13 | ||
| NADH dehydrogenase subunit 4 (NAD4) | 9854–11,194 | 1341 | ATT | TAA | 45 |
| tRNA-Asn (trnN) | 11,230–11,301 | 72 | 35 | ||
| tRNA-Asp (trnD) | 11,343–11,411 | 69 | 41 | ||
| tRNA-Pro (trnP) | 11,426–11,493 | 68 | 14 | ||
| tRNA-Trp (trnW) | 11,509–11,582 | 74 | 15 | ||
| tRNA-Gly (trnG) | 11,589–11,656 | 68 | 6 | ||
| tRNA-Arg (trnR) | 11,667–11,735 | 69 | 10 | ||
| tRNA-Thr (trnT) | 11,738–11,802 | 65 | 202 | ||
| tRNA-Val (trnV) | 11,819–11,885 | 67 | 16 | ||
| tRNA-Leu (trnL2) | 11,903–11,971 | 69 | 17 | ||
| NADH dehydrogenase subunit 3 (NAD3) | 11,972–12,319 | 348 | ATT | TAG | 0 |
| tRNA-Ile (trnI) | 12,369–12,435 | 67 | 49 | ||
| 16S ribosomal RNA | 12,435–13,596 | 1162 | −1 | ||
| tRNA-Tyr (trnY) | 13,597–13,676 | 80 | 0 | ||
| RNA-Ala (trnA) | 13,820–13,883 | 64 | 143 | ||
| tRNA-Gln (trnQ) | 13,887–13,952 | 66 | 3 | ||
| ATP synthase F0 subunit 8 (ATP8) | 13,975–14,088 | 114 | ATT | TAA | 22 |
| NADH dehydrogenase subunit 5 (NAD5) | 14,161–15,891 | 1731 | ATA | TAA | 72 |
| NADH dehydrogenase subunit 1 (NAD1) | 16,389–17,342 | 954 | ATG | TAA | 497 |
| tRNA-Cys (trnC) | 17,402–17,464 | 63 | 59 | ||
| tRNA-Ser (trnS1) | 17,830–17,897 | 68 | 366 | ||
| NADH dehydrogenase subunit 2 (NAD 2) | 17,949–18,947 | 999 | ATA | TAA | 51 |
| Gene | Position | Length (bp) | Initiation Codon | Stop Codon | Intergenic Nucleotide (bp) |
|---|---|---|---|---|---|
| Cytochrome c oxidase subunit I (COX1) | 1–1557 | 1557 | TTG | TAA | 4 |
| tRNA-Thr (trnT) | 1610–1679 | 70 | 52 | ||
| tRNA-Pro (trnP) | 1681–1747 | 67 | 1 | ||
| tRNA-Phe (trnF) | 1770–1836 | 67 | 22 | ||
| tRNA-Lys (trnK) | 1851–1914 | 64 | 14 | ||
| tRNA-His (trnH) | 1921–1985 | 65 | 6 | ||
| Cytochrome c oxidase subunit III (COX3) | 1996–2793 | 798 | ATG | TAA | 10 |
| tRNA-Met (trnM) | 2814–2883 | 70 | 20 | ||
| NADH dehydrogenase subunit 4L (ND4L) | 2947–3234 | 288 | ATG | TAA | 63 |
| tRNA-Ser (trnS1) | 3248–3316 | 69 | 13 | ||
| NADH dehydrogenase subunit 2 (NAD2) | 3369–4370 | 1002 | ATT | TAG | 52 |
| Cytochrome c oxidase subunit II (COX2) | 4377–5312 | 936 | TTG | TAG | 6 |
| tRNA-Val (trnV) | 5331–5395 | 65 | 18 | ||
| tRNA-Ser (trnS2) | 5410–5472 | 63 | 14 | ||
| NADH dehydrogenase subunit 6 (NAD6) | 5480–5972 | 493 | ATT | TAA | 7 |
| Cytochrome b (COB) | 5973–7106 | 1134 | ATG | TAG | 0 |
| ATP synthase F0 subunit 6 (ATP6) | 7136–7781 | 646 | ATA | TAA | 29 |
| tRNA-Glu (trnE) | 8780–8845 | 66 | 998 | ||
| tRNA-Trp (trnW) | 8889–8954 | 66 | 43 | ||
| NADH dehydrogenase subunit 4 (NAD4) | 8973–10,331 | 1359 | ATT | TAA | 18 |
| tRNA-Tyr (trnY) | 10,521–10,584 | 64 | 189 | ||
| tRNA-Leu (trnL1) | 10,595–10,661 | 67 | 10 | ||
| 12S ribosomal RNA (rrnS) | 10,682–11,511 | 830 | 20 | ||
| tRNA-Arg (trnR) | 11,578–11,648 | 71 | 66 | ||
| tRNA-Leu (trnL2) | 11,766–11,833 | 68 | 117 | ||
| NADH dehydrogenase subunit 3 (NAD3) | 11,833–12,180 | 348 | ATA | TAG | -1 |
| tRNA-Ile (trnI) | 12,191–12,258 | 68 | 10 | ||
| 16S ribosomal RNA (rrnL) | 12,289–13,455 | 1167 | 30 | ||
| NADH dehydrogenase subunit 1 (NAD1) | 13,483–14,442 | 960 | ATA | TAA | 27 |
| tRNA-Asn (trnN) | 14,449–14,513 | 65 | 6 | ||
| tRNA-Gly (trnG) | 14,517–14,583 | 67 | 3 | ||
| tRNA-Gln (trnQ) | 14,616–14,694 | 79 | 32 | ||
| ATP synthase F0 subunit 8 (ATP8) | 14,702–14,815 | 114 | ATT | TAG | 7 |
| tRNA-Ala (trnA) | 14,862–14,928 | 67 | 46 | ||
| tRNA-Cys (trnC) | 14,961–15,023 | 63 | 32 | ||
| NADH dehydrogenase subunit 5 (NAD5) | 15,025–16,740 | 1716 | TTG | TAG | 1 |
| tRNA-Asp (trnD) | 16,748–16,814 | 67 | 7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, J.-S.; Song, C.-u.; Choi, H.; Yang, S.H.; Kwon, K.K.; Eyun, S.-i.; Choi, K.-S. The Complete Mitochondrial Genome of the Chemosymbiotic Lucinid Bivalve Pillucina pisidium (Dunker, 1860) Occurring in Seagrass Zostera marina Bed in a Lagoon in Jeju Island, Korea. J. Mar. Sci. Eng. 2024, 12, 847. https://doi.org/10.3390/jmse12050847
Shin J-S, Song C-u, Choi H, Yang SH, Kwon KK, Eyun S-i, Choi K-S. The Complete Mitochondrial Genome of the Chemosymbiotic Lucinid Bivalve Pillucina pisidium (Dunker, 1860) Occurring in Seagrass Zostera marina Bed in a Lagoon in Jeju Island, Korea. Journal of Marine Science and Engineering. 2024; 12(5):847. https://doi.org/10.3390/jmse12050847
Chicago/Turabian StyleShin, Jong-Seop, Chi-une Song, Hyeongwoo Choi, Sung Hyun Yang, Kae Kyoung Kwon, Seong-il Eyun, and Kwang-Sik Choi. 2024. "The Complete Mitochondrial Genome of the Chemosymbiotic Lucinid Bivalve Pillucina pisidium (Dunker, 1860) Occurring in Seagrass Zostera marina Bed in a Lagoon in Jeju Island, Korea" Journal of Marine Science and Engineering 12, no. 5: 847. https://doi.org/10.3390/jmse12050847
APA StyleShin, J.-S., Song, C.-u., Choi, H., Yang, S. H., Kwon, K. K., Eyun, S.-i., & Choi, K.-S. (2024). The Complete Mitochondrial Genome of the Chemosymbiotic Lucinid Bivalve Pillucina pisidium (Dunker, 1860) Occurring in Seagrass Zostera marina Bed in a Lagoon in Jeju Island, Korea. Journal of Marine Science and Engineering, 12(5), 847. https://doi.org/10.3390/jmse12050847








