A Protein Phosphatase 2A-Based Assay to Detect Okadaic Acids and Microcystins
Abstract
1. Introduction
2. Okadaic Acids as Causative Agents of DSP
3. Microcystins from Toxic Cyanobacteria
4. Structure and Function of PP2A and Inhibition by Natural Toxins
5. PP2A Inhibition Assay for Detection of OAs and MCs
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yasumoto, T. The chemistry and biological function of natural marine toxins. Chem. Rec. 2001, 1, 228–242. [Google Scholar] [CrossRef] [PubMed]
- Dell’Aversano, C.; Eaglesham, G.K.; Quilliam, M.A. Analysis of cyanobacterial toxins by hydrophilic interaction liquid chromatography-mass spectrometry. J. Chromatogr. A 2004, 1028, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Cameán, A.; Moreno, I.M.; Ruiz, M.J.; Picó, Y. Determination of microcystins in natural blooms and cyanobacterial strain cultures by matrix solid-phase dispersion and liquid chromatography–mass spectrometry. Anal. Bioanal. Chem. 2004, 380, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Quilliam, M.A. LC-MS/MS analysis of diarrhetic shellfish poisoning (DSP) toxins, okadaic acid and dinophysistoxin analogues, and other lipophilic toxins. Anal. Sci. 2011, 27, 571–584. [Google Scholar] [CrossRef]
- Louppis, A.P.; Badeka, A.V.; Katikou, P.; Paleologos, E.K.; Kontominas, M.G. Determination of okadaic acid, dinophysistoxin-1 and related esters in Greek mussels using HPLC with fluorometric detection, LC-MS/MS and mouse bioassay. Toxicon Off. J. Int. Soc. Toxinology 2010, 55, 724–733. [Google Scholar] [CrossRef]
- Ciminiello, P.; Dell’Aversano, C.; Fattorusso, E.; Forino, M.; Magno, S.; Santelia, F.; Tsoukatou, M. Investigation of the toxin profile of Greek mussels Mytilus galloprovincialis by liquid chromatography—Mass spectrometry. Toxicon Off. J. Int. Soc. Toxinology 2006, 47, 174–181. [Google Scholar] [CrossRef]
- Leonardo, S.; Toldrà, A.; Rambla-Alegre, M.; Fernández-Tejedor, M.; Andree, K.B.; Ferreres, L.; Campbell, K.; Elliott, C.T.; O’Sullivan, C.K.; Pazos, Y.; et al. Self-assembled monolayer-based immunoassays for okadaic acid detection in seawater as monitoring tools. Mar. Environ. Res. 2018, 133, 6–14. [Google Scholar] [CrossRef]
- Laycock, M.V.; Jellett, J.F.; Easy, D.J.; Donovan, M.A. First report of a new rapid assay for diarrhetic shellfish poisoning toxins. Harmful Algae 2006, 5, 74–78. [Google Scholar] [CrossRef]
- Rivasseau, C.; Racaud, P.; Deguin, A.; Hennion, M.-C. Evaluation of an elisa kit for the monitoring of microcystins (cyanobacterial toxins) in water and algae environmental samples. Environ. Sci. Technol. 1999, 33, 1520–1527. [Google Scholar] [CrossRef]
- Nagata, S.; Tsutsumi, T.; Hasegawa, A.; Yoshida, F.; Ueno, Y.; Watanabe, M.F. Enzyme immunoassay for direct determination of microcystins in environmental water. J. Aoac. Int. 2020, 80, 408–417. [Google Scholar] [CrossRef]
- Mountfort, D.O.; Holland, P.; Sprosen, J. Method for detecting classes of microcystins by combination of protein phosphatase inhibition assay and ELISA: Comparison with LC-MS. Toxicon Off. J. Int. Soc. Toxinology 2005, 45, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, A.F.; Callanan, K.R.; Donlon, J.; Palmer, R.; Forde, A.; Kane, M. A cytotoxicity assay for the detection and differentiation of two families of shellfish toxins. Toxicon Off. J. Int. Soc. Toxinology 2001, 39, 1021–1027. [Google Scholar] [CrossRef]
- Heinze, R. A biotest for hepatotoxins using primary rat hepatocytes. Phycologia 1996, 35, 89–93. [Google Scholar] [CrossRef]
- Codd, G.A.; Brooks, W.P.; Priestley, I.M.; Poon, G.K.; Bell, S.G.; Fawell, J.K. Production, detection, and quantification of cyanobacterial toxins. Toxic. Assess. 1989, 4, 499–511. [Google Scholar] [CrossRef]
- Aune, T.; Berg, K. Use of freshly prepared rat hepatocytes to study toxicity of blooms of the blue-green algae Microcystis aeruginosa and Oscillatoria agardhii. J. Toxicol. Environ. Health 1986, 19, 325–336. [Google Scholar] [CrossRef]
- Ikehara, T.; Chikanishi, K.; Oshiro, N. Specification of the okadaic acid equivalent for okadaic acid, dinophysistoxin-1, and dinophysistoxin-2 based on protein phosphatase 2A inhibition and cytotoxicity assays using neuro 2a cell line. J. Mar. Sci. Eng. 2021, 9, 1140. [Google Scholar] [CrossRef]
- Soliño, L.; Sureda, F.X.; Diogène, J. Evaluation of okadaic acid, dinophysistoxin-1 and dinophysistoxin-2 toxicity on Neuro-2a, NG108-15 and MCF-7 cell lines. Toxicol. Vitr. 2015, 29, 59–62. [Google Scholar] [CrossRef]
- Ikehara, T.; Imamura, S.; Sano, T.; Nakashima, J.; Kuniyoshi, K.; Oshiro, N.; Yoshimoto, M.; Yasumoto, T. The effect of structural variation in 21 microcystins on their inhibition of PP2A and the effect of replacing cys269 with glycine. Toxicon Off. J. Int. Soc. Toxinology 2009, 54, 539–544. [Google Scholar] [CrossRef]
- Chen, Y.-M.; Lee, T.-H.; Lee, S.-J.; Huang, H.-B.; Huang, R.; Chou, H.-N. Comparison of protein phosphatase inhibition activities and mouse toxicities of microcystins. Toxicon Off. J. Int. Soc. Toxinology 2006, 47, 742–746. [Google Scholar] [CrossRef]
- Robillot, C.; Hennion, M.-C. Issues arising when interpreting the results of the protein phosphatase 2A inhibition assay for the monitoring of microcystins. Anal. Chim. Acta 2004, 512, 339–346. [Google Scholar] [CrossRef]
- Ikehara, T.; Imamura, S.; Oshiro, N.; Ikehara, S.; Shinjo, F.; Yasumoto, T. A protein phosphatase 2A (PP2A) inhibition assay using a recombinant enzyme for rapid detection of microcystins. Toxicon Off. J. Int. Soc. Toxinology 2008, 51, 1368–1373. [Google Scholar] [CrossRef]
- Tubaro, A.; Florio, C.; Luxich, E.; Sosa, S.; Loggia, R.D.; Yasumoto, T. A protein phosphatase 2A inhibition assay for a fast and sensitive assessment of okadaic acid contamination in mussels. Toxicon Off. J. Int. Soc. Toxinology 1996, 34, 743–752. [Google Scholar] [CrossRef]
- Simon, J.F.; Vernoux, J.P. Highly sensitive assay of okadaic acid using protein phosphatase and paranitrophenyl phosphate. Nat. Toxins 1994, 2, 293–301. [Google Scholar] [CrossRef]
- Takai, A.; Mieskes, G. Inhibitory effect of okadaic acid on the p-nitrophenyl phosphate phosphatase activity of protein phosphatases. Biochem. J. 1991, 275, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Ikehara, T.; Imamura, S.; Yoshino, A.; Yasumoto, T. PP2A inhibition assay using recombinant enzyme for rapid detection of okadaic acid and its analogs in shellfish. Toxins 2010, 2, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P. The structure and regulation of protein phosphatases. Annu. Rev. Biochem. 1989, 58, 453–508. [Google Scholar] [CrossRef]
- Trinkle-Mulcahy, L.; Lamond, A.I. Mitotic phosphatases: No longer silent partners. Curr. Opin. Cell Biol. 2006, 18, 623–631. [Google Scholar] [CrossRef]
- Janssens, V.; Goris, J.; Van Hoof, C. PP2A: The expected tumor suppressor. Curr. Opin. Genet. Dev. 2005, 15, 34–41. [Google Scholar] [CrossRef]
- Janssens, V.; Goris, J. Protein phosphatase 2A: A highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 2001, 353, 417–439. [Google Scholar] [CrossRef]
- Eichhorn, P.J.; Creyghton, M.P.; Bernards, R. Protein phosphatase 2A regulatory subunits and cancer. Biochim. Et. Biophys. Acta 2009, 1795, 1–15. [Google Scholar] [CrossRef]
- Sablina, A.A.; Hahn, W.C. The role of PP2A A subunits in tumor suppression. Cell Adhes. Migr. 2007, 1, 140–141. [Google Scholar] [CrossRef]
- Arroyo, J.D.; Hahn, W.C. Involvement of PP2A in viral and cellular transformation. Oncogene 2005, 24, 7746–7755. [Google Scholar] [CrossRef]
- Sontag, E.; Luangpirom, A.; Hladik, C.; Mudrak, I.; Ogris, E.; Speciale, S.; White, C.L., III. Altered expression levels of the protein phosphatase 2A ABαC enzyme are associated with alzheimer disease pathology. J. Neuropathol. Exp. Neurol. 2004, 63, 287–301. [Google Scholar] [CrossRef]
- Gong, C.X.; Shaikh, S.; Wang, J.Z.; Zaidi, T.; Grundke-Iqbal, I.; Iqbal, K. Phosphatase activity toward abnormally phosphorylated tau: Decrease in Alzheimer disease brain. J. Neurochem. 1995, 65, 732–738. [Google Scholar] [CrossRef]
- Bialojan, C.; Takai, A. Inhibitory effect of a marine-sponge toxin, okadaic acid, on protein phosphatases. Specificity and kinetics. Biochem. J. 1988, 256, 283–290. [Google Scholar] [CrossRef]
- Yasumoto, T.; Murata, M. Marine toxins. Chem. Rev. 1993, 93, 1897–1909. [Google Scholar] [CrossRef]
- Honkanen, R.E.; Zwiller, J.; Moore, R.E.; Daily, S.L.; Khatra, B.S.; Dukelow, M.; Boynton, A.L. Characterization of microcystin-LR, a potent inhibitor of type 1 and type 2A protein phosphatases. J. Biol. Chem. 1990, 265, 19401–19404. [Google Scholar] [CrossRef] [PubMed]
- Smienk, H.; Domínguez, E.; Rodríguez-Velasco, M.L.; Clarke, D.; Kapp, K.; Katikou, P.; Cabado, A.G.; Otero, A.; Vieites, J.M.; Razquin, P.; et al. Quantitative determination of the okadaic acid toxins group by a colorimetric phosphatase inhibition assay: Interlaboratory study. J. Aoac. Int. 2013, 96, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Garibo, D.; Dàmaso, E.; Eixarch, H.; de la Iglesia, P.; Fernández-Tejedor, M.; Diogène, J.; Pazos, Y.; Campàs, M. Protein phosphatase inhibition assays for okadaic acid detection in shellfish: Matrix effects, applicability and comparison with LC–MS/MS analysis. Harmful Algae 2012, 19, 68–75. [Google Scholar] [CrossRef]
- Li, Y.M.; Casida, J.E. Cantharidin-binding protein: Identification as protein phosphatase 2A. Proc. Natl. Acad. Sci. USA 1992, 89, 11867–11870. [Google Scholar] [CrossRef]
- Ishihara, H.; Martin, B.L.; Brautigan, D.L.; Karaki, H.; Ozaki, H.; Kato, Y.; Fusetani, N.; Watabe, S.; Hashimoto, K.; Uemura, D.; et al. Calyculin A and okadaic acid: Inhibitors of protein phosphatase activity. Biochem. Biophys. Res. Commun. 1989, 159, 871–877. [Google Scholar] [CrossRef]
- Walsh, A.H.; Cheng, A.; Honkanen, R.E. Fostriecin, an antitumor antibiotic with inhibitory activity against serine/threonine protein phosphatases types 1 (PP1) and 2A (PP2A), is highly selective for PP2A. FEBS Lett. 1997, 416, 230–234. [Google Scholar] [CrossRef]
- MacKintosh, C.; Klumpp, S. Tautomycin from the bacterium Streptomyces verticillatus. Another potent and specific inhibitor of protein phosphatases 1 and 2A. FEBS Lett. 1990, 277, 137–140. [Google Scholar] [CrossRef]
- Honkanen, R.E.; Dukelow, M.; Zwiller, J.; Moore, R.E.; Khatra, B.S.; Boynton, A.L. Cyanobacterial nodularin is a potent inhibitor of type 1 and type 2A protein phosphatases. Mol. Pharmacol. 1991, 40, 577. [Google Scholar]
- Hu, T.; Doyle, J.; Jackson, D.; Marr, J.; Nixon, E.; Pleasance, S.; Quilliam, M.A.; Walter, J.A.; Wright, J.L.C. Isolation of a new diarrhetic shellfish poison from Irish mussels. J. Chem. Soc. Chem. Commun. 1992, 1, 39–41. [Google Scholar] [CrossRef]
- Lee, J.-S.; Igarashi, T.; Fraga, S.; Dahl, E.; Hovgaard, P.; Yasumoto, T. Determination of diarrhetic shellfish toxins in various dinoflagellate species. J. Appl. Phycol. 1989, 1, 147–152. [Google Scholar] [CrossRef]
- Murakami, Y.; Oshima, Y.; Yasumoto, T. Identification of okadaic acid as a toxic component of a marine dinoflagellate Prorocentrum lima. Bull. Jpn. Soc. Sci. Fish. 1982, 48, 69–72. [Google Scholar] [CrossRef]
- Yasumoto, T.; Oshima, Y.; Sugawara, W.; Fukuyo, Y.; Oguri, H.; Igarashi, T.; Fujita, N. Identification of Dinophysis fortii as the causative organism of diarrhetic shellfish poisoning. Bull. Jpn. Soc. Sci. Fish. 1980, 46, 1405–1411. [Google Scholar] [CrossRef]
- Suzuki, T.; Kamiyama, T.; Okumura, Y.; Ishihara, K.; Matsushima, R.; Kaneniwa, M. Liquid-chromatographic hybrid triple–quadrupole linear-ion-trap MS/MS analysis of fatty-acid esters of dinophysistoxin-1 in bivalves and toxic dinoflagellates in Japan. Fish. Sci. 2009, 75, 1039–1048. [Google Scholar] [CrossRef]
- Yasumoto, T.; Murata, M.; Oshima, Y.; Sano, M.; Matsumoto, G.K.; Clardy, J. Diarrhetic shellfish toxins. Tetrahedron 1985, 41, 1019–1025. [Google Scholar] [CrossRef]
- Suzuki, T.; Ota, H.; Yamasaki, M. Direct evidence of transformation of dinophysistoxin-1 to 7-O-acyl-dinophysistoxin-1 (dinophysistoxin-3) in the scallop Patinopecten yessoensis. Toxicon Off. J. Int. Soc. Toxinology 1999, 37, 187–198. [Google Scholar] [CrossRef]
- Garcia, C.; Truan, D.; Lagos, M.; Santelices, J.P.; Carlos, D.J.; Lagos, N. Metabolic transformation of dinophysistoxin-3 into dinophysistoxin-1 causes human intoxication by consumption of O-acyl-derivatives dinophysistoxins contaminated shellfish. J. Toxicol. Sci. 2005, 30, 287–296. [Google Scholar] [CrossRef]
- Doucet, E.; Ross, N.N.; Quilliam, M.A. Enzymatic hydrolysis of esterified diarrhetic shellfish poisoning toxins and pectenotoxins. Anal. Bioanal. Chem. 2007, 389, 335–342. [Google Scholar] [CrossRef]
- Braga, A.C.; Alves, R.N.; Maulvault, A.L.; Barbosa, V.; Marques, A.; Costa, P.R. In vitro bioaccessibility of the marine biotoxin okadaic acid in shellfish. Food Chem. Toxicol. 2016, 89, 54–59. [Google Scholar] [CrossRef]
- European Food Safety Authority. Marine biotoxins in shellfish-okadaic acid and analogues-scientific opinion of the panel on contaminants in the food chain. EFSA J. 2008, 6, 589. [Google Scholar] [CrossRef]
- Hamano, Y.; Kinoshita, Y.; Yasumoto, T. Toxic Dinoflagellates; Anderson, D.M., White, A.W., Baden, D.G., Eds.; Elsevier: New York, NY, USA, 1985; pp. 383–388. [Google Scholar]
- Fujiki, H.; Suganuma, M.; Suguri, H.; Yoshizawa, S.; Ojika, M.; Wakamatsu, K.; Yamada, K.; Sugimura, T. Induction of ornithine decarboxylase activity in mouse skin by a possible tumor promoter, okadaic acid. Proc. Jpn. Acad. Ser. B 1987, 63, 51–53. [Google Scholar] [CrossRef]
- European Commission. Commission Regulation no. 15/2011 of January 10, 2011. Off. J. Eur. Communities 2011, 3–6. [Google Scholar]
- European Commission. Regulation (EC) No 853/2004. Off. J. Eur. Union L139 2004, 47, 55–105. [Google Scholar]
- Bartram, J.; Chorus, I. Toxic Cyanobacteria in Water—A Guide to Their Public Health Consequences, Monitoring and Management; E&FN SPON: London, UK, 1999. [Google Scholar]
- Bouaicha, N.; Miles, C.O.; Beach, D.G.; Labidi, Z.; Djabri, A.; Benayache, N.Y.; Nguyen-Quang, T. Structural diversity, characterization and toxicology of microcystins. Toxins 2019, 11, 714. [Google Scholar] [CrossRef] [PubMed]
- Harke, M.J.; Steffen, M.M.; Gobler, C.J.; Otten, T.G.; Wilhelm, S.W.; Wood, S.A.; Paerl, H.W. A review of the global ecology, genomics, and biogeography of the toxic cyanobacterium, Microcystis spp. Harmful Algae 2016, 54, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Paerl, H.W.; Otten, T.G. Blooms bite the hand that feeds them. Science 2013, 342, 433–434. [Google Scholar] [CrossRef] [PubMed]
- Carmichael, W.W.; Boyer, G.L. Health impacts from cyanobacteria harmful algae blooms: Implications for the North American Great Lakes. Harmful Algae 2016, 54, 194–212. [Google Scholar] [CrossRef] [PubMed]
- Meriluoto, J.; Spoof, L.; Codd, G. Handbook of Cyanobacterial Monitoring and Cyanotoxin Analysis; John Wiley & Sons: San Francisco, CA, USA, 2017; Volume 8. [Google Scholar]
- Jochimsen, E.M.; Carmichael, W.W.; An, J.S.; Cardo, D.M.; Cookson, S.T.; Holmes, C.E.; Antunes, M.B.; de Melo Filho, D.A.; Lyra, T.M.; Barreto, V.S.; et al. Liver failure and death after exposure to microcystins at a hemodialysis center in Brazil. N. Engl. J. Med. 1998, 338, 873–878. [Google Scholar] [CrossRef] [PubMed]
- Beasley, V.R.; Cook, W.O.; Dahlem, A.M.; Hooser, S.B.; Lovell, R.A.; Valentine, W.M. Algae intoxication in livestock and waterfowl. Vet. Clin. North Am. Food Anim. Pract. 1989, 5, 345–361. [Google Scholar] [CrossRef]
- Miller, M.A.; Kudela, R.M.; Mekebri, A.; Crane, D.; Oates, S.C.; Tinker, M.T.; Staedler, M.; Miller, W.A.; Toy-Choutka, S.; Dominik, C. Evidence for a novel marine harmful algal bloom: Cyanotoxin (microcystin) transfer from land to sea otters. PLoS ONE 2010, 5, e12576. [Google Scholar] [CrossRef]
- Pouria, S.; de Andrade, A.; Barbosa, J.; Cavalcanti, R.L.; Barreto, V.T.; Ward, C.J.; Preiser, W.; Poon, G.K.; Neild, G.H.; Codd, G.A. Fatal microcystin intoxication in haemodialysis unit in Caruaru, Brazil. Lancet 1998, 352, 21–26. [Google Scholar] [CrossRef]
- Guidelines for Drinking-Water Quality: Fourth Edition Incorporating the First Addendum; World Health Organization: Geneva, Switzerland, 2017; Available online: https://www.who.int/publications/i/item/9789241549950 (accessed on 26 January 2024).
- Massey, I.Y.; Wu, P.; Wei, J.; Luo, J.; Ding, P.; Wei, H.; Yang, F. A mini-review on detection methods of microcystins. Toxins 2020, 12, 641. [Google Scholar] [CrossRef]
- Teta, R.; Romano, V.; Sala, G.D.; Picchio, S.; Sterlich, C.D.; Mangoni, A.; Tullio, G.D.; Costantino, V.; Lega, M. Cyanobacteria as indicators of water quality in Campania coasts, Italy: A monitoring strategy combining remote/proximal sensing and in situ data. Environ. Res. Lett. 2017, 12, 024001. [Google Scholar] [CrossRef]
- Esposito, G.; Teta, R.; Marrone, R.; De Sterlich, C.; Casazza, M.; Anastasio, A.; Lega, M.; Costantino, V. A fast detection strategy for cyanobacterial blooms and associated cyanotoxins (FDSCC) reveals the occurrence of lyngbyatoxin A in campania (South Italy). Chemosphere 2019, 225, 342–351. [Google Scholar] [CrossRef]
- Ikehara, T.; Kuniyoshi, K.; Yamaguchi, H.; Tanabe, Y.; Sano, T.; Yoshimoto, M.; Oshiro, N.; Nakashima, S.; Yasumoto-Hirose, M. First report of microcystis strains producing MC-FR and -WR toxins in Japan. Toxins 2019, 11, 521. [Google Scholar] [CrossRef]
- Carmichael, W.W. The toxins of cyanobacteria. Sci. Am. 1994, 270, 78–86. [Google Scholar] [CrossRef]
- Pinchart, P.E.; Leruste, A.; Pasqualini, V.; Mastroleo, F. Microcystins and cyanobacterial contaminants in the french small-scale productions of spirulina (Limnospira sp.). Toxins 2023, 15, 354. [Google Scholar] [CrossRef]
- Lawrence, J.F.; Niedzwiadek, B.; Menard, C.; Lau, B.P.; Lewis, D.; Kuper-Goodman, T.; Carbone, S.; Holmes, C. Comparison of liquid chromatography/mass spectrometry, ELISA, and phosphatase assay for the determination of microcystins in blue-green algae products. J. Aoac. Int. 2001, 84, 1035–1044. [Google Scholar] [CrossRef] [PubMed]
- Gilroy, D.J.; Kauffman, K.W.; Hall, R.A.; Huang, X.; Chu, F.S. Assessing potential health risks from microcystin toxins in blue-green algae dietary supplements. Environ. Health Perspect. 2000, 108, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Wera, S.; Hemmings, B.A. Serine/threonine protein phosphatases. Biochem. J. 1995, 311, 17–29. [Google Scholar] [CrossRef]
- Mayer, R.E.; Hendrix, P.; Cron, P.; Matthies, R.; Stone, S.R.; Goris, J.; Merlevede, W.; Hofsteenge, J.; Hemmings, B.A. Structure of the 55-kDa regulatory subunit of protein phosphatase 2A: Evidence for a neuronal-specific isoform. Biochemistry 1991, 30, 3589–3597. [Google Scholar] [CrossRef]
- McCright, B.; Virshup, D.M. identification of a new family of protein phosphatase 2A regulatory subunits. J. Biol. Chem. 1995, 270, 26123–26128. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, P.; Mayer-Jackel, R.E.; Cron, P.; Goris, J.; Hofsteenge, J.; Merlevede, W.; Hemmings, B.A. Structure and expression of a 72-kDa regulatory subunit of protein phosphatase 2A. Evidence for different size forms produced by alternative splicing. J. Biol. Chem. 1993, 268, 15267–15276. [Google Scholar] [CrossRef]
- Moreno, C.S.; Park, S.; Nelson, K.; Ashby, D.; Hubalek, F.; Lane, W.S.; Pallas, D.C. Wd40 repeat proteins striatin and s/g2 nuclear autoantigen are members of a novel family of calmodulin-binding proteins that associate with protein phosphatase 2A. J. Biol. Chem. 2000, 275, 5257–5263. [Google Scholar] [CrossRef]
- Li, X.; Virshup, D.M. Two conserved domains in regulatory B subunits mediate binding to the A subunit of protein phosphatase 2A. Eur. J. Biochem. 2002, 269, 546–552. [Google Scholar] [CrossRef]
- Leonard, D.; Huang, W.; Izadmehr, S.; O’Connor, C.M.; Wiredja, D.D.; Wang, Z.; Zaware, N.; Chen, Y.; Schlatzer, D.M.; Kiselar, J.; et al. Selective PP2A enhancement through biased heterotrimer stabilization. Cell 2020, 181, 688–701.e616. [Google Scholar] [CrossRef]
- Morita, K.; He, S.; Nowak, R.P.; Wang, J.; Zimmerman, M.W.; Fu, C.; Durbin, A.D.; Martel, M.W.; Prutsch, N.; Gray, N.S.; et al. RETRACTED: Allosteric activators of protein phosphatase 2a display broad antitumor activity mediated by dephosphorylation of MYBL2. Cell 2020, 181, 702–715.e720. [Google Scholar] [CrossRef] [PubMed]
- Vervoort, S.J.; Welsh, S.A.; Devlin, J.R.; Barbieri, E.; Knight, D.A.; Offley, S.; Bjelosevic, S.; Costacurta, M.; Todorovski, I.; Kearney, C.J.; et al. The PP2A-Integrator-CDK9 axis fine-tunes transcription and can be targeted therapeutically in cancer. Cell 2021, 184, 3143–3162.e3132. [Google Scholar] [CrossRef] [PubMed]
- Haanen, T.J.; O’Connor, C.M.; Narla, G. Biased holoenzyme assembly of protein phosphatase 2A (PP2A): From cancer to small molecules. J. Biol. Chem. 2022, 298, 102656. [Google Scholar] [CrossRef]
- Takemoto, A.; Maeshima, K.; Ikehara, T.; Yamaguchi, K.; Murayama, A.; Imamura, S.; Imamoto, N.; Yokoyama, S.; Hirano, T.; Watanabe, Y.; et al. The chromosomal association of condensin II is regulated by a noncatalytic function of PP2A. Nat. Struct. Mol. Biol. 2009, 16, 1302–1308. [Google Scholar] [CrossRef]
- Xing, Y.; Li, Z.; Chen, Y.; Stock, J.B.; Jeffrey, P.D.; Shi, Y. Structural mechanism of demethylation and inactivation of protein phosphatase 2A. Cell 2008, 133, 154–163. [Google Scholar] [CrossRef]
- Xu, Y.; Xing, Y.; Chen, Y.; Chao, Y.; Lin, Z.; Fan, E.; Yu, J.W.; Strack, S.; Jeffrey, P.D.; Shi, Y. Structure of the protein phosphatase 2A holoenzyme. Cell 2006, 127, 1239–1251. [Google Scholar] [CrossRef]
- Xing, Y.; Xu, Y.; Chen, Y.; Jeffrey, P.D.; Chao, Y.; Lin, Z.; Li, Z.; Strack, S.; Stock, J.B.; Shi, Y. Structure of protein phosphatase 2A core enzyme bound to tumor-inducing toxins. Cell 2006, 127, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Cho, U.S.; Xu, W. Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature 2007, 445, 53–57. [Google Scholar] [CrossRef]
- Murata, M.; Shimatani, M.; Sugitani, H.; Oshima, Y.; Yasumoto, T. Isolation and structural elucidation of the causative toxin of the diarrhetic shellfish poisoning. Bull. Jpn. Soc. Sci. Fish. 1982, 48, 549–552. [Google Scholar] [CrossRef]
- Huhn, J.; Jeffrey, P.D.; Larsen, K.; Rundberget, T.; Rise, F.; Cox, N.R.; Arcus, V.; Shi, Y.; Miles, C.O. A structural basis for the reduced toxicity of dinophysistoxin-2. Chem. Res. Toxicol. 2009, 22, 1782–1786. [Google Scholar] [CrossRef]
- Swiatek, W.; Sugajska, E.; Lankiewicz, L.; Hemmings, B.A.; Zolnierowicz, S. Biochemical characterization of recombinant subunits of type 2A protein phosphatase overexpressed in Pichia pastoris. Eur. J. Biochem. 2000, 267, 5209–5216. [Google Scholar] [CrossRef]
- Evans, D.R.H.; Myles, T.; Hofsteenge, J.; Hemmings, B.A. Functional expression of human PP2Ac in yeast permits the identification of novel c-terminal and dominant-negative mutant forms. J. Biol. Chem. 1999, 274, 24038–24046. [Google Scholar] [CrossRef] [PubMed]
- Wadzinski, B.E.; Eisfelder, B.J.; Peruski, L.F.; Mumby, M.C.; Johnson, G.L. NH2-terminal modification of the phosphatase 2A catalytic subunit allows functional expression in mammalian cells. J. Biol. Chem. 1992, 267, 16883–16888. [Google Scholar] [CrossRef] [PubMed]
- Kamibayashi, C.; Estes, R.; Lickteig, R.L.; Yang, S.I.; Craft, C.; Mumby, M.C. Comparison of heterotrimeric protein phosphatase 2A containing different B subunits. J. Biol. Chem. 1994, 269, 20139–20148. [Google Scholar] [CrossRef]
- Ikehara, T.; Shinjo, F.; Ikehara, S.; Imamura, S.; Yasumoto, T. Baculovirus expression, purification, and characterization of human protein phosphatase 2A catalytic subunits α and β. Protein Expr. Purif. 2006, 45, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Ikehara, T.; Nakashima, S.; Nakashima, J.; Kinoshita, T.; Yasumoto, T. Efficient production of recombinant PP2A at a low temperature using a baculovirus expression system. Biotechnol. Rep. 2016, 11, 86–89. [Google Scholar] [CrossRef]
- Ikehara, T.; Kinoshita, T.; Kurokawa, A.; Nakashima, S.; Maekawa, K.; Ohshiro, N.; Yasumoto, T. Evaluation of protein phosphatase 2A (PP2A) inhibition assay for rapid detection of DSP toxins in scallop. Nippon Suisan Gakkaishi 2017, 83, 367–372. [Google Scholar] [CrossRef][Green Version]
- Ikehara, T.; Yasumoto, T. High sensitive detection of microcystins (MCs) and okadaic acids (OAs) by recombinant proptein phosphatase 2A (PP2A) inhibition. Int. Soc. Toxinology Newsl. 2009. Available online: https://www.toxinology.org/shelrecm/Media/Newsletters/IST-Newsletter-Dec09.pdf (accessed on 26 January 2009).
- Takai, A.; Murata, M.; Torigoe, K.; Isobe, M.; Mieskes, G.; Yasumoto, T. Inhibitory effect of okadaic acid derivatives on protein phosphatases. A study on structure-affinity relationship. Biochem. J. 1992, 284, 539–544. [Google Scholar] [CrossRef]
- Munday, R. Is protein phosphatase inhibition responsible for the toxic effects of okadaic acid in animals? Toxins 2013, 5, 267–285. [Google Scholar] [CrossRef]
- Deeds, J.R.; Stutts, W.L.; Celiz, M.D.; MacLeod, J.; Hamilton, A.E.; Lewis, B.J.; Miller, D.W.; Kanwit, K.; Smith, J.L.; Kulis, D.M.; et al. Dihydrodinophysistoxin-1 produced by dinophysis norvegica in the gulf of maine, USA and its accumulation in shellfish. Toxins 2020, 12, 533. [Google Scholar] [CrossRef] [PubMed]
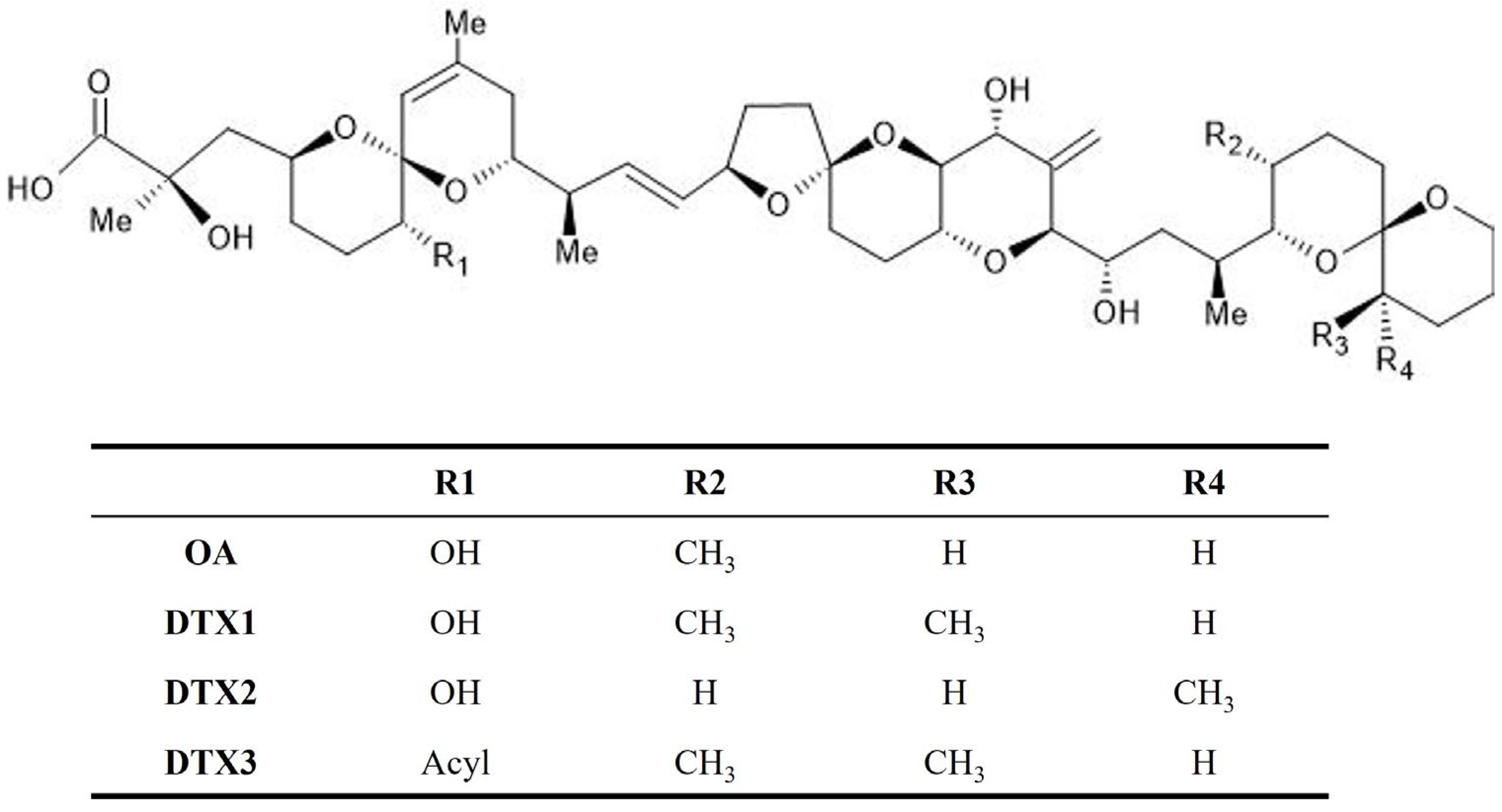



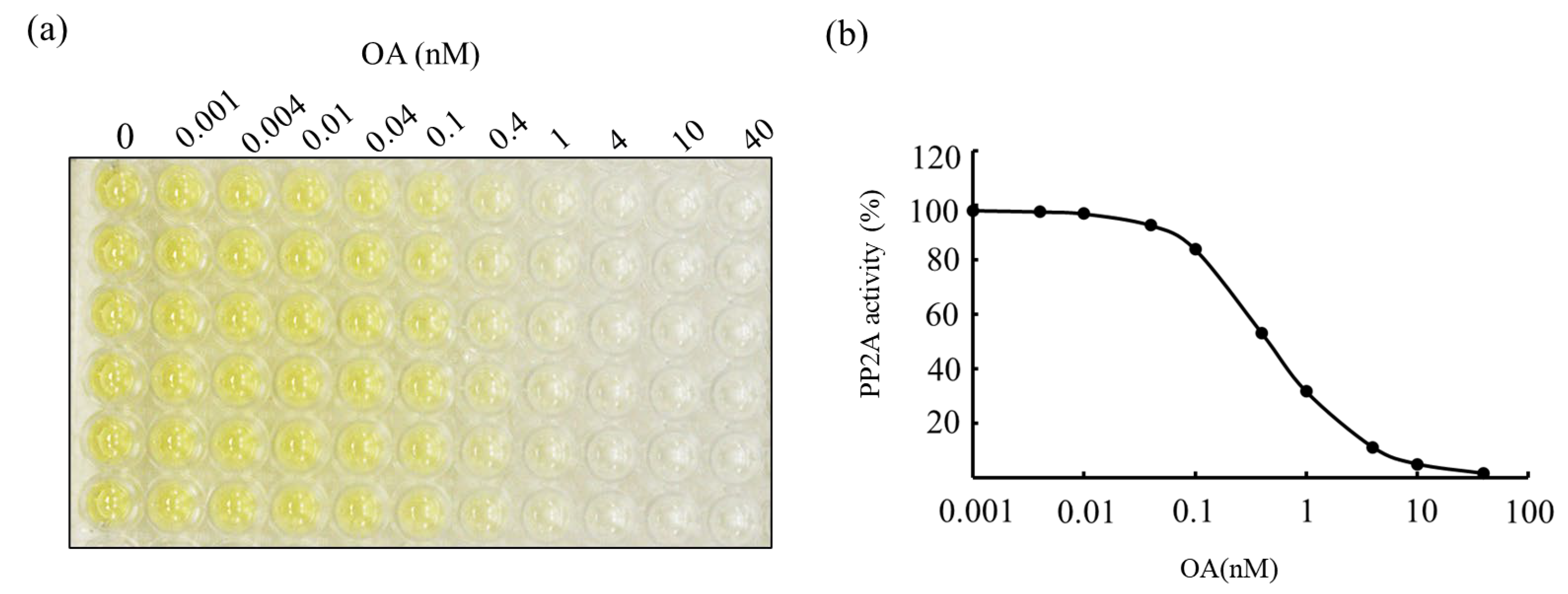
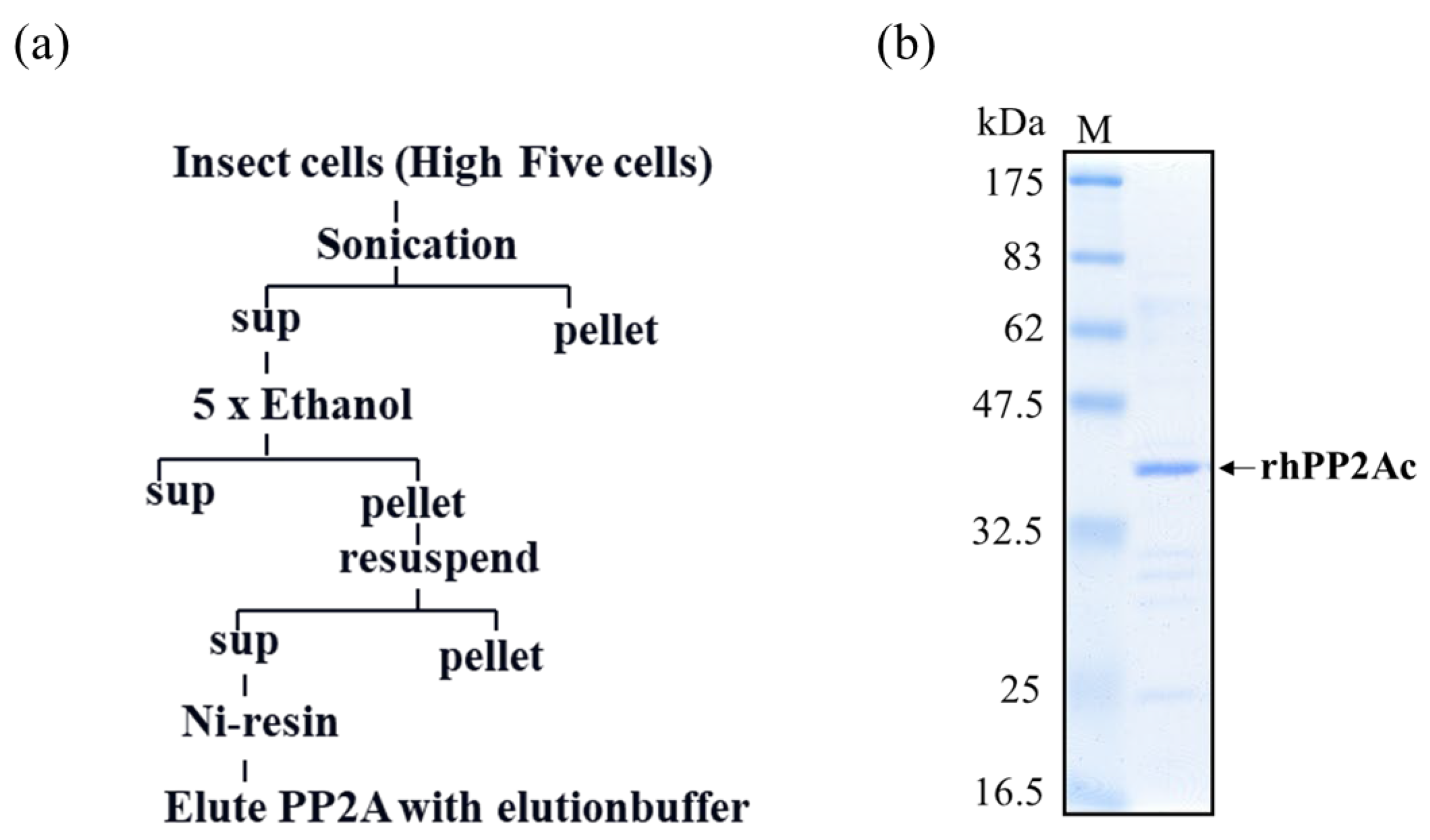

| Matrix | LOD (µg/g) | LOQ (µg/g) | Reference |
|---|---|---|---|
| Mussels | |||
| Unhydrolyzed | 0.0387 | 0.0765 | [16] |
| 0.0424 | 0.0725 | [25] | |
| Hydrolyzed | 0.0646 | 0.0989 | [16] |
| 0.0476 | 0.0932 | [25] | |
| Scallops | |||
| Unhydrolyzed | 0.0217 | 0.0372 | [25] |
| 0.0262 | 0.0470 | [102] | |
| Hydrolyzed | 0.0274 | 0.0415 | [25] |
| 0.0432 | 0.0780 | [102] |
| Toxins | PP2A Inhibition | Reference | |
|---|---|---|---|
| OAs | IC50 (nM) | OApp2a a | |
| OA | 0.14 | 1 | [16] |
| DTX1 | 0.09 | 1.6 | [16] |
| DTX2 | 0.45 | 0.3 | [16] |
| MCs | IC50 (nM) | Conversion factor b | |
| MC-LR | 0.032 | 1.000 | [18] |
| MC-RR | 0.056 | 0.571 | [18] |
| MC-FR | 0.069 | 0.464 | [18] |
| MC-LF | 0.096 | 0.333 | [18] |
| [D-Asp3]MC-HtyR | 0.098 | 0.327 | [18] |
| [D-Asp3, (Z)-Dhb7]MC-HtyR | 0.110 | 0.291 | [18] |
| MC-LW | 0.114 | 0.281 | [18] |
| [D-Asp3, (E)-Dhb7]MC-HtyR | 0.122 | 0.262 | [18] |
| MC-YR | 0.125 | 0.256 | [18] |
| MC-LA | 0.161 | 0.199 | [18] |
| [D-Asp3, (Z)-Dhb7]MC-LR | 0.164 | 0.195 | [18] |
| [Dha7]MC-LR | 0.167 | 0.192 | [18] |
| MC-WR | 0.179 | 0.179 | [18] |
| [D-Asp3, (E)-Dhb7]MC-LR | 0.201 | 0.159 | [18] |
| [D-Asp3, Dha7]MC-RR | 0.220 | 0.145 | [18] |
| [D-Asp3, Dha7]MC-LR | 0.254 | 0.126 | [18] |
| [Dha7]MC-RR | 0.293 | 0.109 | [18] |
| [D-Asp3]MC-RR | 0.300 | 0.107 | [18] |
| [Dha7]MC-YR | 0.379 | 0.084 | [18] |
| Nodularin | 0.540 | 0.059 | [18] |
| [6-(Z)-Adda5]MC-RR | 10.126 | 0.003 | [18] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ikehara, T.; Oshiro, N. A Protein Phosphatase 2A-Based Assay to Detect Okadaic Acids and Microcystins. J. Mar. Sci. Eng. 2024, 12, 244. https://doi.org/10.3390/jmse12020244
Ikehara T, Oshiro N. A Protein Phosphatase 2A-Based Assay to Detect Okadaic Acids and Microcystins. Journal of Marine Science and Engineering. 2024; 12(2):244. https://doi.org/10.3390/jmse12020244
Chicago/Turabian StyleIkehara, Tsuyoshi, and Naomasa Oshiro. 2024. "A Protein Phosphatase 2A-Based Assay to Detect Okadaic Acids and Microcystins" Journal of Marine Science and Engineering 12, no. 2: 244. https://doi.org/10.3390/jmse12020244
APA StyleIkehara, T., & Oshiro, N. (2024). A Protein Phosphatase 2A-Based Assay to Detect Okadaic Acids and Microcystins. Journal of Marine Science and Engineering, 12(2), 244. https://doi.org/10.3390/jmse12020244







