Machine Learning-Based Classification of Malnutrition Using Histological Biomarkers of Fish Intestine: Preliminary Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Growth Trial
2.2. Proximate Analysis of Experimental Diets
2.3. Histological Analysis

2.4. Nutritional Status Classification Using Machine Learning Algorithms
2.4.1. Data Acquisition and Preprocessing
2.4.2. Algorithms and Models
2.4.3. Training Procedure
2.4.4. Evaluation Metrics
2.4.5. Learning Performance Evaluation
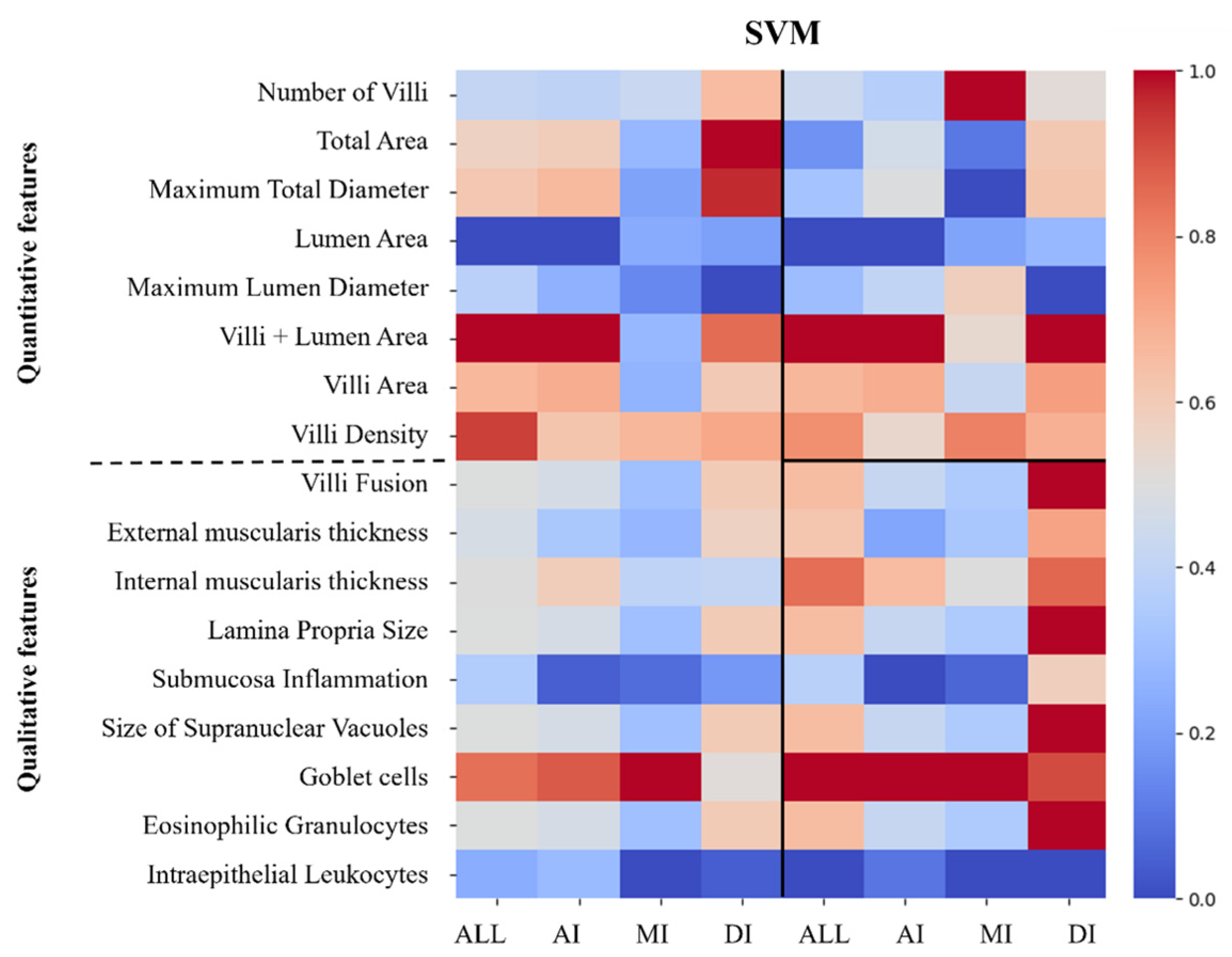
2.4.6. Feature Importance Extraction
2.5. Statistical Analysis
3. Results
3.1. Growth Performance
3.2. Gut Morphology
3.3. Nutritional Status Classification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Olmos Soto, J.; Paniagua-Michel, J.d.J.; Lopez, L.; Ochoa, L. Functional Feeds in Aquaculture. In Springer Handbook of Marine Biotechnology; Kim, S.-K., Ed.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 1303–1319. [Google Scholar]
- Gasco, L.; Gai, F.; Maricchiolo, G.; Genovese, L.; Ragonese, S.; Bottari, T.; Caruso, G. Fishmeal Alternative Protein Sources for Aquaculture Feeds. In Feeds for the Aquaculture Sector: Current Situation and Alternative Sources; Gasco, L., Gai, F., Maricchiolo, G., Genovese, L., Ragonese, S., Bottari, T., Caruso, G., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 1–28. [Google Scholar]
- Aragão, C.; Gonçalves, A.T.; Costas, B.; Azeredo, R.; Xavier, M.J.; Engrola, S. Alternative Proteins for Fish Diets: Implications beyond Growth. Animals 2022, 12, 1211. (In English) [Google Scholar] [CrossRef]
- Krogdahl, Å.; Penn, M.; Thorsen, J.; Refstie, S.; Bakke, A.M. Important antinutrients in plant feedstuffs for aquaculture: An update on recent findings regarding responses in salmonids. Aquac. Res. 2010, 41, 333–344. [Google Scholar] [CrossRef]
- Kumar, V.; Hossain, M.S.; Ragaza, J.A.; Benito, M.R. The Potential Impacts of Soy Protein on Fish Gut Health. In Soybean for Human Consumption and Animal Feed; IntechOpen: London, UK, 2020. [Google Scholar]
- Daniel, N. A review on replacing fish meal in aqua feeds using plant protein sources. Int. J. Fish. Aquat. Stud. 2018, 6, 164–179. [Google Scholar]
- Francis, G.; Makkar, H.P.S.; Becker, K. Antinutritional factors present in plant-derived alternate fish feed ingredients and their effects in fish. Aquaculture 2001, 199, 197–227. [Google Scholar] [CrossRef]
- Dawood, M.A.O. Nutritional immunity of fish intestines: Important insights for sustainable aquaculture. Rev. Aquac. 2021, 13, 642–663. [Google Scholar] [CrossRef]
- Xiong, J.B.; Nie, L.; Chen, J. Current understanding on the roles of gut microbiota in fish disease and immunity. Zool. Res. 2019, 40, 70–76. (In English) [Google Scholar] [CrossRef]
- Stosik, M.; Tokarz-Deptuła, B.; Deptuła, W. Immunity of the intestinal mucosa in teleost fish. Fish Shellfish. Immunol. 2023, 133, 108572. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ran, C.; Teame, T.; Ding, Q.; Hoseinifar, S.H.; Xie, M.; Zhang, Z.; Yang, Y.; Olsen, R.E.; Gatlin, D.M.; et al. Research progress on gut health of farmers teleost fish: A viewpoint concerning the intestinal mucosal barrier and the impact of its damage. Rev. Fish Biol. Fish. 2020, 30, 569–586. [Google Scholar] [CrossRef]
- Luan, Y.; Li, M.; Zhou, W.; Yao, Y.; Yang, Y.; Zhang, Z.; Ringø, E.; Erik Olsen, R.; Liu Clarke, J.; Xie, S.; et al. The Fish Microbiota: Research Progress and Potential Applications. Engineering 2023, 29, 137–146. [Google Scholar] [CrossRef]
- Wang, A.R.; Ran, C.; Ringø, E.; Zhou, Z.G. Progress in fish gastrointestinal microbiota research. Rev. Aquac. 2018, 10, 626–640. [Google Scholar] [CrossRef]
- Magalhães, R.; Guerreiro, I.; Santos, R.A.; Coutinho, F.; Couto, A.; Serra, C.R.; Olsen, R.E.; Peres, H.; Oliva-Teles, A. Oxidative status and intestinal health of gilthead sea bream (Sparus aurata) juveniles fed diets with different ARA/EPA/DHA ratios. Sci. Rep. 2020, 10, 13824. [Google Scholar] [CrossRef] [PubMed]
- Guerreiro, I.; Castro, C.; Serra, C.R.; Coutinho, F.; Couto, A.; Peres, H.; Pousão-Ferreira, P.; Corraze, G.; Oliva-Teles, A.; Enes, P. Feeding Yellow Worms to Meagre: Effects on Whole-Body Fatty Acid Profile and Hepatic and Intestine Oxidative Status. Antioxidants 2023, 12, 1031. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.A.M.; Dehler, C.E.; Król, E. Transcriptomic responses in the fish intestine. Dev. Comp. Immunol. 2016, 64, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Lilleeng, E.; Frøystad, M.K.; Vekterud, K.; Valen, E.C.; Krogdahl, Å. Comparison of intestinal gene expression in Atlantic cod (Gadus morhua) fed standard fish meal or soybean meal by means of suppression subtractive hybridization and real-time PCR. Aquaculture 2007, 267, 269–283. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Yu, Y.; Li, X.; Poolsawat, L.; Yang, P.; Bian, Y.; Guo, Z.; Leng, X. An evaluation of replacing fish meal with fermented soybean meal in the diets of largemouth bass (Micropterus salmoides): Growth, nutrition utilization and intestinal histology. Aquac. Res. 2020, 51, 4302–4314. [Google Scholar] [CrossRef]
- Lin, H.; Tan, B.; Ray, G.W.; Zeng, M.; Li, M.; Chi, S.; Yang, Q. A Challenge to Conventional Fish Meal: Effects of Soy Protein Peptides on Growth, Histomorphology, Lipid Metabolism and Intestinal Health for Juvenile Pompano Trachinotus ovatus. Front. Mar. Sci. 2022, 8, 815323. (In English) [Google Scholar] [CrossRef]
- Hoseinifar, S.H.; Shakouri, M.; Yousefi, S.; Van Doan, H.; Shafiei, S.; Yousefi, M.; Mazandarani, M.; Torfi Mozanzadeh, M.; Tulino, M.G.; Faggio, C. Humoral and skin mucosal immune parameters, intestinal immune related genes expression and antioxidant defense in rainbow trout (Oncorhynchus mykiss) fed olive (Olea europea L.) waste. Fish Shellfish Immunol. 2020, 100, 171–178. [Google Scholar] [CrossRef]
- Ruiz, A.; Andree, K.B.; Furones, D.; Holhorea, P.G.; Calduch-Giner, J.À.; Viñas, M.; Pérez-Sánchez, J.; Gisbert, E. Modulation of gut microbiota and intestinal immune response in gilthead seabream (Sparus aurata) by dietary bile salt supplementation. Front. Microbiol. 2023, 14, 1123716. (In English) [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Li, X.-F.; Tian, H.-Y.; Jiang, G.-Z.; Liu, W.-B. Feeding rates affect growth, intestinal digestive and absorptive capabilities and endocrine functions of juvenile blunt snout bream Megalobrama amblycephala. Fish Physiol. Biochem. 2016, 42, 689–700. [Google Scholar] [CrossRef]
- Santigosa, E.; García-Meilán, I.; Valentin, J.M.; Pérez-Sánchez, J.; Médale, F.; Kaushik, S.; Gallardo, M.A. Modifications of intestinal nutrient absorption in response to dietary fish meal replacement by plant protein sources in sea bream (Sparus aurata) and rainbow trout (Onchorynchus mykiss). Aquaculture 2011, 317, 146–154. [Google Scholar] [CrossRef]
- Raskovic, B.; Stankovic, M.; Markovic, Z.; Poleksic, V. Histological methods in the assessment of different feed effects on liver and intestine of fish. J. Agric. Sci. 2011, 56, 87–100. [Google Scholar] [CrossRef]
- Couto, A.; Barroso, C.; Guerreiro, I.; Pousão-Ferreira, P.; Matos, E.; Peres, H.; Oliva-Teles, A.; Enes, P. Carob seed germ meal in diets for meagre (Argyrosomus regius) juveniles: Growth, digestive enzymes, intermediary metabolism, liver and gut histology. Aquaculture 2016, 451, 396–404. [Google Scholar] [CrossRef]
- Couto, A.; Serra, C.R.; Guerreiro, I.; Coutinho, F.; Castro, C.; Rangel, F.; Lavrador, A.S.; Monteiro, M.; Santos, R.; Peres, H.; et al. Black soldier fly meal effects on meagre health condition: Gut morphology, gut microbiota and humoral immune response. J. Insects Food Feed. 2022, 8, 1281–1295. [Google Scholar] [CrossRef]
- Saavedra, M.; Barata, M.; Matias, A.C.; Couto, A.; Salem, A.; Ribeiro, L.; Pereira, T.G.; Gamboa, M.; Lourenço-Marques, C.; Soares, F.; et al. Effect of Dietary Incorporation of Yellow Mealworm as a Partial Fishmeal Replacer on Growth, Metabolism, and Intestinal Histomorphology in Juvenile Meagre (Argyrosomus regius). Aquac. Nutr. 2023, 2023, 6572421. [Google Scholar] [CrossRef]
- Meyerholz, D.K.; Beck, A.P. Fundamental Concepts for Semiquantitative Tissue Scoring in Translational Research. ILAR J. 2018, 59, 13–17. (In English) [Google Scholar] [CrossRef]
- Ribeiro, L.; Moura, J.; Santos, M.; Colen, R.; Rodrigues, V.; Bandarra, N.; Soares, F.; Ramalho, P.; Barata, M.; Moura, P.; et al. Effect of vegetable based diets on growth, intestinal morphology, activity of intestinal enzymes and haematological stress indicators in meagre (Argyrosomus regius). Aquaculture 2015, 447, 116–128. [Google Scholar] [CrossRef]
- Silva, P.F.; McGurk, C.; Knudsen, D.L.; Adams, A.; Thompson, K.D.; Bron, J.E. Histological evaluation of soya bean-induced enteritis in Atlantic salmon (Salmo salar L.): Quantitative image analysis vs. semi-quantitative visual scoring. Aquaculture 2015, 445, 42–56. [Google Scholar] [CrossRef]
- Vatsos, I.N. Planning and Reporting of the Histomorphometry Used to Assess the Intestinal Health in Fish Nutrition Research—Suggestions to Increase Comparability of the Studies. Front. Vet. Sci. 2021, 8, 666044. (In English) [Google Scholar] [CrossRef]
- Mokhtar, D.M. The Digestive System. In Fish Histology: From Cells to Organs; Mokhtar, D.M., Ed.; Apple Academic Press Inc.: Burlington, ON, Canada, 2017. [Google Scholar]
- Verdile, N.; Pasquariello, R.; Scolari, M.; Scirè, G.; Brevini, T.; Gandolfi, F. A Detailed Study of Rainbow Trout (Onchorhynchus mykiss) Intestine Revealed That Digestive and Absorptive Functions Are Not Linearly Distributed along Its Length. Animals 2020, 10, 745. [Google Scholar] [CrossRef]
- Ferreira, M.; Sousa, V.; Oliveira, B.; Canadas-Sousa, A.; Abreu, H.; Dias, J.; Kiron, V.; Valente, L.M.P. An in-depth characterisation of European seabass intestinal segments for assessing the impact of an algae-based functional diet on intestinal health. Sci. Rep. 2023, 13, 11686. [Google Scholar] [CrossRef]
- Baeza-Ariño, R.; Martínez-Llorens, S.; Nogales-Mérida, S.; Jover-Cerda, M.; Tomás-Vidal, A. Study of liver and gut alterations in sea bream, Sparus aurata L., fed a mixture of vegetable protein concentrates. Aquac. Res. 2016, 47, 460–471. [Google Scholar] [CrossRef]
- Flores, A.; Wiff, R.; Donovan, C.R.; Gálvez, P. Applying machine learning to predict reproductive condition in fish. Ecol. Inform. 2024, 80, 102481. [Google Scholar] [CrossRef]
- Spanou, D.S.; Petroudi, P.; Dimou, E.; Kokkinos, K.; Klaoudatos, D. Walleye (Sander vitreus, Mitchill 1818) age and sex classification using innovative supervised and unsupervised machine learning and soft computing methodologies. Fish. Res. 2024, 275, 107031. [Google Scholar] [CrossRef]
- Graham, C.A.; Shamkhalichenar, H.; Browning, V.E.; Byrd, V.J.; Liu, Y.; Gutierrez-Wing, M.T.; Novelo, N.; Choi, J.-W.; Tiersch, T.R. A practical evaluation of machine learning for classification of ultrasound images of ovarian development in channel catfish (Ictalurus punctatus). Aquaculture 2022, 552, 738039. [Google Scholar] [CrossRef] [PubMed]
- Palaiokostas, C. Predicting for disease resistance in aquaculture species using machine learning models. Aquac. Rep. 2021, 20, 100660. [Google Scholar] [CrossRef]
- Rahman, L.F.; Marufuzzaman, M.; Alam, L.; Bari, M.A.; Sumaila, U.R.; Sidek, L.M. Developing an Ensembled Machine Learning Prediction Model for Marine Fish and Aquaculture Production. Sustainability 2021, 13, 9124. [Google Scholar] [CrossRef]
- Mahesh, B. Machine Learning Algorithms—A Review. Int. J. Sci. Res. (IJSR) 2019, 9, 381–386. [Google Scholar] [CrossRef]
- FAO. Fishery and Aquaculture Statistics—Yearbook 2021. In FAO Yearbook of Fishery and Aquaculture Statistics; FAO: Rome, Italy, 2024. [Google Scholar]
- Monfort, M.C. Present Market Situation and Prospects of Meagre (Argyrosomus regius), as an Emerging Species in Mediterranean Aquaculture; Food and Agriculture Organization of the United Nations (FAO): Rome, Italy, 2010. [Google Scholar]
- Piccolo, G.; Bovera, F.; De Riu, N.; Marono, S.; Salati, F.; Cappuccinelli, R.; Moniello, G. Effect of two different protein/fat ratios of the diet on meagre (Argyrosomus regius) traits. Ital. J. Anim. Sci. 2010, 7, 363–371. [Google Scholar] [CrossRef]
- Suquet, M.; Divanach, P.; Hussenot, J.; Coves, D.; Christian, F. Marine fish culture of “new species” farmed in Europe. Cah. Agric. 2009, 18, 148–156. [Google Scholar] [CrossRef]
- Saavedra, M.; Pereira, T.G.; Barata, M.; Aragão, C.; Requeijo, B.; Conceição, L.E.C.; Pousão-Ferreira, P. Plant-based diets fed to juvenile meagre Argyrosomus regius with low methionine and taurine supplementation led to an overall reduction in fish performance and to an increase in muscle fibre recruitment. J. Fish Biol. 2022, 101, 1182–1188. [Google Scholar] [CrossRef]
- Carvalho, M.; Izquierdo, M.; Valdés, M.; Montero, D.; Farías, A. Oils Combination with Microalgal Products as a Strategy for Increasing the N-3 Long-Chain Polyunsaturated Fatty Acid Content in Fish Oil-Free Diets for Meagre (Argyrosomus regius). Aquac. Nutr. 2022, 2022, 5275570. [Google Scholar] [CrossRef]
- Katsika, L.; Tasbozan, O.; Mastoraki, M.; Karapanagiotis, S.; Zalamitsou, C.; Feidantsis, K.; Antonopoulou, E.; Chatzifotis, S. Effects of fish oil substitution by hazelnut oil on growth performance, whole-body fatty acid composition and enzymes of intermediary metabolism of juvenile meagre (Argyrosomus regius Asso, 1801). Aquac. Res. 2021, 52, 5760–5776. [Google Scholar] [CrossRef]
- de Moura, L.B.; Diógenes, A.F.; Campelo, D.A.V.; de Almeida, F.L.A.; Pousão-Ferreira, P.M.; Furuya, W.M.; Peres, H.; Oliva-Teles, A. Nutrient digestibility, digestive enzymes activity, bile drainage alterations and plasma metabolites of meagre (Argyrosomus regius) feed high plant protein diets supplemented with taurine and methionine. Aquaculture 2019, 511, 734231. [Google Scholar] [CrossRef]
- Asencio-Alcudia, G.; Andree, K.B.; Giraldez, I.; Tovar-Ramirez, D.; Alvarez-González, A.; Herrera, M.; Gisbert, E. Stressors Due to Handling Impair Gut Immunity in Meagre (Argyrosomus regius): The Compensatory Role of Dietary L-Tryptophan. Front. Physiol. 2019, 10, 547. (In English) [Google Scholar] [CrossRef] [PubMed]
- Urán, P.A.; Schrama, J.W.; Rombout, J.H.W.M.; Obach, A.; Jensen, L.; Koppe, W.; Verreth, J.A.J. Soybean meal-induced enteritis in Atlantic salmon (Salmo salar L.) at different temperatures. Aquac. Nutr. 2008, 14, 324–330. [Google Scholar] [CrossRef]
- Oliva, M.; Unceta, C.; Canales, M. Histomorphology and histochemistry of the digestive tract in meagre (Argyrosomus regius). Biochem. Indian J. 2011, 5, 10–17. [Google Scholar]
- Zhao, S.; Zhang, S.; Liu, J.; Wang, H.; Zhu, J.; Li, D.; Zhao, R. Application of machine learning in intelligent fish aquaculture: A review. Aquaculture 2021, 540, 736724. [Google Scholar] [CrossRef]
- Ray, S. A Quick Review of Machine Learning Algorithms. In Proceedings of the 2019 International Conference on Machine Learning, Big Data, Cloud and Parallel Computing (COMITCon), Faridabad, India, 14–16 February 2019; pp. 35–39. [Google Scholar]
- Habbat, N.; Anoun, H.; Hassouni, L.; Hicham, N. Analyzing Booking’s Comments Using Stacking Ensemble Deep Learning Model and Neural Topic Model. SSRN Electron. J. 2022. [Google Scholar] [CrossRef]
- Araújo, F.; Morado, C.; Parente, T.; Jose Roma Paumgartten, F.; Gomes, I. Biomarkers and bioindicators of the environmental condition using a fish species (Pimelodus maculatus Lacepède, 1803) in a tropical reservoir in Southeastern Brazil. Braz. J. Biol. 2017, 78, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Goede, R.W. Organismic indices and an autopsy-based assessment as indicator of health and condition of fish. Am. Fish. Soc. Symp. 1990, 8, 93–108. [Google Scholar]
- Krogdahl, A.; Bakke, A.M.; Baeverfjord, G. Effects of graded levels of standard soybean meal on intestinal structure, mucosal enzyme activities, and pancreatic response in Atlantic salmon (Salmo salar L.). Aquac. Nutr. 2003, 9, 361–371. [Google Scholar] [CrossRef]
- Penn, M.H.; Bendiksen, E.Å.; Campbell, P.; Krogdahl, Å. High level of dietary pea protein concentrate induces enteropathy in Atlantic salmon (Salmo salar L.). Aquaculture 2011, 310, 267–273. [Google Scholar] [CrossRef]
- Secombes, C.J. Cytokines and Immunity. In Principles of Fish Immunology: From Cells and Molecules to Host Protection; Buchmann, K., Secombes, C.J., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 301–353. [Google Scholar]
- Farrell, A.P. Encyclopedia of Fish Physiology; Academic Press: Cambridge, MA, USA, 2011. [Google Scholar]
- Rašković, B.; Stanković, M.; Markelić, M.; Poleksić, V.; Božić, G.; Janković, S.; Marković, Z. Growth, feed utilization, and quantitative histological assessment of the distal intestine and liver of common carp (Cyprinus carpio L.) fed formulated diets containing grains of different soybean cultivars. Aquac. Int. 2024, 32, 6903–6921. [Google Scholar] [CrossRef]
- Aidos, L.; Mirra, G.; Pallaoro, M.; Herrera Millar, V.R.; Radaelli, G.; Bazzocchi, C.; Modina, S.C.; Di Giancamillo, A. How Do Alternative Protein Resources Affect the Intestine Morphology and Microbiota of Atlantic Salmon? Animals 2023, 13, 1922. [Google Scholar] [CrossRef]
- Panettieri, V.; Chatzifotis, S.; Messina, C.M.; Olivotto, I.; Manuguerra, S.; Randazzo, B.; Ariano, A.; Bovera, F.; Santulli, A.; Severino, L.; et al. Honey Bee Pollen in Meagre (Argyrosomus regius) Juvenile Diets: Effects on Growth, Diet Digestibility, Intestinal Traits, and Biochemical Markers Related to Health and Stress. Animals 2020, 10, 231. (In English) [Google Scholar] [CrossRef]
- Turchini, G.M.; Torstensen, B.E.; Ng, W.-K. Fish oil replacement in finfish nutrition. Rev. Aquac. 2009, 1, 10–57. [Google Scholar] [CrossRef]
- Olsen, R.E.; Myklebust, R.; Ringø, E.; Mayhew, T.M. The influences of dietary linseed oil and saturated fatty acids on caecal enterocytes in Arctic char (Salvelinus alpinus L.): A quantitative ultrastructural study. Fish Physiol. Biochem. 2000, 22, 207–216. [Google Scholar] [CrossRef]
- Berg, J.M.; Tymoczko, J.L.; Stryer, L. The Biosynthesis of Membrane Lipids and Steroids. In Biochemistry, 8th ed.; W. H. Freeman and Company: New York, NY, USA, 2015. [Google Scholar]
- Li, Y.; Kortner, T.M.; Chikwati, E.M.; Munang’andu, H.M.; Lock, E.-J.; Krogdahl, Å. Gut health and vaccination response in pre-smolt Atlantic salmon (Salmo salar) fed black soldier fly (Hermetia illucens) larvae meal. Fish Shellfish. Immunol. 2019, 86, 1106–1113. [Google Scholar] [CrossRef]
- Glover, C.N.; Petri, D.; Tollefsen, K.-E.; Jørum, N.; Handy, R.D.; Berntssen, M.H.G. Assessing the sensitivity of Atlantic salmon (Salmo salar) to dietary endosulfan exposure using tissue biochemistry and histology. Aquat. Toxicol. 2007, 84, 346–355. [Google Scholar] [CrossRef]
- Caimi, C.; Gasco, L.; Biasato, I.; Malfatto, V.; Varello, K.; Prearo, M.; Pastorino, P.; Bona, M.C.; Francese, D.R.; Schiavone, A.; et al. Could Dietary Black Soldier Fly Meal Inclusion Affect the Liver and Intestinal Histological Traits and the Oxidative Stress Biomarkers of Siberian Sturgeon (Acipenser baerii) Juveniles? Animals 2020, 10, 155. [Google Scholar] [CrossRef]
- Faccioli, C.K.; Chedid, R.A.; Amaral, A.C.d.; Franceschini Vicentini, I.B.; Vicentini, C.A. Morphology and histochemistry of the digestive tract in carnivorous freshwater Hemisorubim platyrhynchos (Siluriformes: Pimelodidae). Micron 2014, 64, 10–19. [Google Scholar] [CrossRef] [PubMed]
- L⊘kka, G.; Austb⊘, L.; Falk, K.; Bjerkås, I.; Koppang, E.O. Intestinal morphology of the wild atlantic salmon (Salmo salar). J. Morphol. 2013, 274, 859–876. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-r.; Wang, L.; Zhang, C.-x.; Song, K. Effects of substituting fishmeal with soybean meal on growth performance and intestinal morphology in orange-spotted grouper (Epinephelus coioides). Aquac. Rep. 2017, 5, 52–57. [Google Scholar] [CrossRef]
- Adeoye, A.A.; Jaramillo-Torres, A.; Fox, S.W.; Merrifield, D.L.; Davies, S.J. Supplementation of formulated diets for tilapia (Oreochromis niloticus) with selected exogenous enzymes: Overall performance and effects on intestinal histology and microbiota. Anim. Feed. Sci. Technol. 2016, 215, 133–143. [Google Scholar] [CrossRef]




| Experimental Diets | |||
|---|---|---|---|
| Control (CTRL) | Challenge (CD) | Extreme Challenge (ED) | |
| Ingredients (% dry weight basis) | |||
| Fish meal a | 55.1 | 15.0 | 5.0 |
| CPSP b | 5.0 | 5.0 | 5.0 |
| Wheat gluten c | - | 10.0 | 12.0 |
| Corn gluten d | - | 11.6 | 14.3 |
| Soybean meal e | - | 20.0 | 25.0 |
| Rapeseed meal f | - | 7.5 | 11.0 |
| Sunflower meal g | - | 5.0 | 7.5 |
| Wheat meal h | 24.7 | 7.1 | - |
| Fish oil | 11.3 | 7.0 | 5.0 |
| Soy oil | - | 3.8 | 5.2 |
| Rapeseed oil | - | 3.8 | 5.2 |
| Phosphate | - | 0.4 | 1.1 |
| Vitamin i | 1.0 | 1.0 | 1.0 |
| Mineral j | 1.0 | 1.0 | 1.0 |
| Choline | 0.5 | 0.5 | 0.5 |
| Binder | 1.0 | 1.0 | 1.0 |
| Taurine | 0.3 | 0.3 | 0.3 |
| Proximate analysis (% dry matter basis) | |||
| Dry matter | 92.3 | 95.0 | 93.0 |
| Crude protein | 49.0 | 46.4 | 47.3 |
| Crude fat | 18.1 | 18.1 | 17.9 |
| Ash | 10.0 | 7.1 | 7.0 |
| Gross energy (kJ g−1 DM) | 23.0 | 24.1 | 23.9 |
| CTRL | CD | ED | p-Value | |
|---|---|---|---|---|
| Final weight (g) | 48.7 ± 1.0 c | 30.0 ± 0.6 b | 19.3 ± 1.0 a | <0.001 |
| Weight gain (g) | 44.2 ± 1.0 c | 25.4 ± 0.6 b | 14.9 ± 1.1 a | <0.001 |
| Feed intake (g kg ABW−1 day−1) | 39.1 ± 1.1 a | 38.7 ± 1.2 a | 46.98 ± 1.7 b | <0.001 |
| Feed efficiency | 0.85 ± 0.03 c | 0.76 ± 0.03 b | 0.53 ± 0.04 a | <0.001 |
| Daily growth index | 3.99 ± 0.05 c | 2.90 ± 0.04 b | 2.07 ± 0.11 a | <0.001 |
| CTRL | CD | ED | p-Value | |
|---|---|---|---|---|
| Hepatosomatic index (HSI) | 1.5 ± 0.3 a | 2.0 ± 0.2 b | 2.6 ± 0.4 c | <0.001 |
| Enterosomatic index (ESI) | 1.2 ± 0.2 a | 1.5 ± 0.2 ab | 1.6 ± 0.2 b | 0.005 |
| Viscerosomatic index (VSI) | 4.3 ± 1.0 a | 5.1 ± 0.9 ab | 6.0 ± 0.9 b | 0.004 |
| Intestine Section | Semi-Quantitative Feature | Control (CTRL) | Challenge (CD) | Extreme Challenge (ED) | Kruskal–Wallis |
|---|---|---|---|---|---|
| Villi fusion | 1.45 ± 0.54 | 1.50 ± 0.67 | 1.52 ± 0.66 | 0.940 | |
| Lamina propria size | 1.90 ± 0.50 | 1.89 ± 0.45 | 1.97 ± 0.39 | 0.647 | |
| Submucosa inflammation | 1.93 ± 0.53 | 2.17 ± 0.50 | 2.07 ± 0.85 | 0.131 | |
| Anterior | Supranuclear vacuoles size | 1.27 ± 0.37 a | 1.54 ± 062 ab | 1.86 ± 0.71 b | 0.000 |
| Goblet cells | 1.45 ± 0.28 b | 1.14 ± 0.28 a | 1.08 ± 0.21 a | 0.000 | |
| Eosinophilic granulocytes | 2.13 ± 0.42 a | 2.45 ± 0.60 b | 2.39 ± 0.87 b | 0.046 | |
| Intraepithelial leukocytes | 2.15 ± 0.40 a | 2.41 ± 0.48 b | 2.47 ± 0.62 b | 0.006 | |
| External muscularis thickness | 1.17 ± 0.26 | 1.26 ± 0.35 | 1.21 ± 0.31 | 0.537 | |
| Internal muscularis thickness | 1.82 ± 0.32 | 1.81 ± 0.41 | 1.89 ± 0.33 | 0.489 | |
| Villi fusion | 1.59 ± 0.65 a | 1.95 ± 0.65 b | 1.62 ± 0.68 a | 0.015 | |
| Lamina propria size | 2.08 ± 0.49 a | 2.74 ± 0.60 b | 2.57 ± 0.67 b | 0.000 | |
| Submucosa inflammation | 1.73 ± 0.57 a | 2.19 ± 0.77 b | 2.61 ± 0.77 b | 0.000 | |
| Intermediate | Supranuclear vacuoles size | 1.20 ± 0.43 a | 2.00 ± 0.93 b | 2.32 ± 0.99 b | 0.000 |
| Goblet cells | 2.21 ± 0.48 b | 1.62 ± 0.29 a | 1.5 ± 0.29 a | 0.000 | |
| Eosinophilic granulocytes | 1.40 ± 0.52 a | 1.66 ± 0.58 b | 1.37 ± 0.59 a | 0.006 | |
| Intraepithelial leukocytes | 2.01 ± 0.52 a | 2.50 ± 0.51 b | 2.45 ± 0.42 b | 0.000 | |
| External muscularis thickness | 1.07 ± 0.17 a | 1.11 ± 0.23 a | 1.23 ± 0.28 b | 0.003 | |
| Internal muscularis thickness | 1.75 ± 0.42 | 1.71 ± 0.35 | 1.85 ± 0.26 | 0.165 | |
| Villi fusion | 1.38 ± 0.50 a | 2.22 ± 0.69 b | 2.46 ± 0.90 b | 0.000 | |
| Lamina propria size | 1.83 ± 0.58 a | 2.98 ± 0.47 c | 2.35 ± 0.47 b | 0.000 | |
| Distal | Submucosa inflammation | 1.56 ± 0.34 a | 2.28 ± 0.65 c | 1.68 ± 0.56 b | 0.000 |
| Supranuclear vacuoles size | 1.84 ± 0.67 a | 2.47 ± 0.66 b | 2.40 ± 0.96 b | 0.000 | |
| Goblet cells | 1.45 ± 0.43 a | 2.03 ± 0.52 b | 1.97 ± 0.40 b | 0.000 | |
| Eosinophilic granulocytes | 1.24 ± 0.41 a | 2.17 ± 0.67 c | 1.83 ± 0.75 b | 0.000 | |
| Intraepithelial leukocytes | 1.44 ± 0.43 a | 2.89 ± 0.57 b | 2.60 ± 0.49 b | 0.000 | |
| External muscularis thickness | 1.41 ± 0.49 | 1.56 ± 0.40 | 1.44 ±0.32 | 0.079 | |
| Internal muscularis thickness | 1.76 ± 0.34 | 1.84 ± 0.36 | 1.71 ± 0.31 | 0.061 |
| Intestine Section | Quantitative Feature | Control (CTRL) | Challenge (CD) | Extreme Challenge (ED) | ANOVA p-Value |
|---|---|---|---|---|---|
| Total Area (mm2) | 1.75 ± 0.59 c | 1.49 ± 0.54 b | 1.02 ± 0.38 a | 0.000 | |
| Total Maximum Diameter (µm) | 1597 ± 313 c | 1455 ± 270 b | 1221 ± 260 a | 0.000 | |
| Lumen Area (mm2) | 0.29 ± 0.27 ab | 0.33 ± 0.28 b | 0.20 ± 0.18 a | 0.039 | |
| Anterior | Lumen Maximum Diameter (µm) | 709 ± 340 | 704 ± 266 | 581 ± 259 | 0.059 |
| Villi Area (mm2) | 1.18 ± 0.31 c | 0.91 ± 0.26 b | 0.65 ± 0.20 a | 0.000 | |
| Villi + Lumen Area (mm2) | 1.52 ± 0.54 c | 1.26 ± 0.50 b | 0.86 ± 0.34 a | 0.000 | |
| Number of Villi | 37 ± 5 b | 36 ± 4 b | 33 ± 5 a | 0.001 | |
| Villi Density | 33 ± 7 a | 42 ± 9 b | 54 ± 13 c | 0.000 | |
| Total Area (mm2) | 0.90 ± 0.64 b | 1.66 ± 0.56 c | 0.50 ± 0.45 a | 0.000 | |
| Total Maximum Diameter (µm) | 1080 ± 430 b | 1560 ± 293 c | 782 ± 374 a | 0.000 | |
| Lumen Area (mm2) | 0.29 ± 0.30 a | 0.69 ± 0.44 b | 0.14 ± 0.19 a | 0.000 | |
| Intermediate | Lumen Maximum Diameter (µm) | 635 ± 306 b | 1000 ± 326 c | 425 ± 263 a | 0.000 |
| Villi Area (mm2) | 0.51 ± 0.35 b | 0.82 ± 0.22 c | 0.28 ± 0.23 a | 0.000 | |
| Villi + Lumen Area (mm2) | 0.80 ± 0.58 b | 1.51 ± 0.57 c | 0.42 ± 0.39 a | 0.000 | |
| Number of Villi | 39 ± 5 b | 37 ± 5 b | 31 ± 5 a | 0.000 | |
| Villi Density | 152 ± 136 b | 49 ± 13 a | 221 ± 137 c | 0.000 | |
| Total Area (mm2) | 2.19 ± 0.80 ab | 2.55 ± 0.73 b | 1.94 ± 0.84 a | 0.001 | |
| Total Maximum Diameter (µm) | 1758 ± 336 ab | 1941 ± 322 b | 1664 ± 403 a | 0.001 | |
| Distal | Lumen Area (mm2) | 0.40 ± 0.36 a | 0.78 ± 0.41 b | 0.58 ± 0.45 ab | 0.000 |
| Lumen Maximum Diameter (µm) | 772 ± 322 a | 1138 ± 327 c | 936 ± 362 b | 0.000 | |
| Villi Area (mm2) | 1.34 ± 0.43 b | 1.30 ± 0.32 b | 1.00 ± 0.35 a | 0.000 | |
| Villi + Lumen Area (mm2) | 1.74 ± 0.73 ab | 2.10 ± 0.65 b | 1.56 ± 0.74 a | 0.002 | |
| Number of Villi | 48 ± 10 ab | 50 ± 6 b | 46 ± 9 a | 0.028 | |
| Villi Density | 38 ± 10 a | 41 ± 10 a | 51 ± 17 b | 0.000 |
| Sections | ALL | AI | MI | DI | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Features/ Algorithms | ALL | QT | SQT | ALL | QT | SQT | ALL | QT | SQT | ALL | QT | SQT | |
| Accuracy | AB | 72.0 ± 4.6 | 53.9 ± 11.1 | 65.0 ±6.8 | 61.1 ± 10.8 | 53.8 ± 13.2 | 63.7 ± 11.2 | 84.1 ± 9.4 | 64.9 ± 12.3 | 67.4 ± 17.6 | 74.0 ± 9.9 | 57.4 ± 9.9 | 71.0 ± 9.7 |
| DT | 66.5 ± 6.4 | 52.9 ± 8 | 61.3 ± 8.3 | 56.1 ± 15.4 | 52.3 ± 13.6 | 58.6 ± 10 | 74.7 ± 10.2 | 66.7 ± 10.7 | 69.2 ± 6.2 | 75.4 ± 16.8 | 48.6 ± 13.0 | 74.0 ± 11.8 | |
| ES | 78.2 ± 5.1 | 61.7 ± 6.4 | 69.1 ± 5.5 | 65.4 ± 18.3 | 63.5 ± 13.1 | 60.5 ± 10.3 | 89.6 ± 7.1 | 71.6 ± 12.9 | 73.5 ± 5.9 | 83.9 ± 8.5 | 56.2 ± 5.6 | 81.4 ± 9.7 | |
| KNN | 72.0 ± 5.7 | 57.4 ± 5.6 | 67.0 ± 7.8 | 64.1 ± 10.7 | 64.7 ± 12.6 | 59.3 ± 8.8 | 85.2 ± 9.7 | 71.0 ± 13.0 | 74.7 ± 8.4 | 84.5 ± 7.9 | 52.5 ± 10.8 | 79.5 ± 10.4 | |
| LR | 76.3 ± 4.2 | 58.9 ± 6.1 | 67 ± 7.4 | 72.8 ± 13.2 | 64 ± 15.8 | 62.9 ± 8.3 | 90.1 ± 8.8 | 74.1 ± 14.0 | 74.7 ± 8.9 | 84.5 ± 10.4 | 61.2 ± 11.5 | 83.2 ± 9.9 | |
| NB | 66.1 ± 5.4 | 50.2 ± 10 | 67.1 ± 7.4 | 62.4 ± 16 | 55 ± 17.4 | 59.3 ± 10.7 | 85.8 ± 11.0 | 68.6 ± 10.3 | 72.8 ± 8.4 | 78.4 ± 9.4 | 46.4 ± 10.9 | 85.6 ± 12.2 | |
| RF | 74.1 ± 4.3 | 60.9 ± 8.5 | 68.3 ± 7.8 | 67.2 ± 15.1 | 59.7 ± 18.3 | 61.7 ± 10.9 | 80.9 ± 11.1 | 70.3 ± 12.6 | 74.7 ± 8.8 | 81.4 ± 12.2 | 55.6 ± 8.9 | 80.8 ± 12.4 | |
| SVM | 78.4 ± 5.9 | 60.7 ± 6.4 | 69.5 ± 6.4 | 64.8 ± 14.7 | 61.7 ± 18 | 62.4 ± 13.8 | 87.1 ± 9.9 | 74.7 ± 14.5 | 75.3 ± 7.2 | 80.8 ± 9.1 | 63.1 ± 8.7 | 82.0 ± 8.2 | |
| Precision CTRL | AB | 0.84 ± 0.08 | 0.52 ± 0.17 | 0.73 ± 0.14 | 0.74 ± 0.33 | 0.48 ± 0.37 | 0.83 ± 0.15 | 0.88 ± 0.15 | 0.58 ± 0.23 | 0.90 ± 0.11 | 0.91 ± 0.10 | 0.78 ± 0.13 | 0.88 ± 0.09 |
| DT | 0.72 ± 0.13 | 0.54 ± 0.20 | 0.69 ± 0.18 | 0.68 ± 0.30 | 0.64 ± 0.25 | 0.76 ± 0.22 | 0.75 ± 0.22 | 0.65 ± 0.23 | 0.85 ± 0.15 | 0.83 ± 0.15 | 0.62 ± 0.14 | 0.86 ± 0.15 | |
| ES | 0.85 ± 0.12 | 0.65 ± 0.16 | 0.73 ± 0.15 | 0.80 ± 0.16 | 0.71 ± 0.21 | 0.76 ± 0.14 | 0.94 ± 0.12 | 0.68 ± 0.22 | 0.90 ± 0.09 | 0.94 ± 0.08 | 0.69 ± 0.09 | 0.91 ± 0.10 | |
| KNN | 0.74 ± 0.15 | 0.54 ± 0.11 | 0.68 ± 0.13 | 0.74 ± 0.15 | 0.69 ± 0.22 | 0.71 ± 0.16 | 0.83 ± 0.18 | 0.62 ± 0.19 | 0.85 ± 0.11 | 0.95 ± 0.08 | 0.55 ± 0.10 | 0.89 ± 0.12 | |
| LR | 0.83 ± 0.13 | 0.60 ± 0.15 | 0.76 ± 0.13 | 0.88 ± 0.12 | 0.68 ± 0.29 | 0.76 ± 0.09 | 0.93 ± 0.12 | 0.74 ± 0.23 | 0.89 ± 0.10 | 0.92 ± 0.10 | 0.72 ± 0.14 | 0.90 ± 0.09 | |
| NB | 0.78 ± 0.15 | 0.52 ± 0.28 | 0.71 ± 0.13 | 0.72 ± 0.21 | 0.61 ± 0.33 | 0.74 ± 0.14 | 0.93 ± 0.14 | 0.63 ± 0.33 | 0.78 ± 0.15 | 0.95 ± 0.08 | 0.44 ± 0.11 | 0.92 ± 0.10 | |
| RF | 0.78 ± 0.15 | 0.61 ± 0.16 | 0.74 ± 0.15 | 0.81 ± 0.19 | 0.63 ± 0.31 | 0.72 ± 0.12 | 0.85 ± 0.20 | 0.68 ± 0.21 | 0.82 ± 0.14 | 0.89 ± 0.12 | 0.68 ± 0.14 | 0.90 ± 0.11 | |
| SVM | 0.82 ± 0.16 | 0.60 ± 0.14 | 0.74 ± 0.15 | 0.79 ± 0.20 | 0.67 ± 0.22 | 0.80 ± 0.17 | 0.90 ± 0.15 | 0.70 ± 0.20 | 0.83 ± 0.11 | 0.93 ± 0.10 | 0.68 ± 0.13 | 0.91 ± 0.10 | |
| Precision CD | AB | 0.61 ± 0.08 | 0.57 ± 0.17 | 0.57 ± 0.06 | 0.46 ± 0.12 | 0.46 ± 0.16 | 0.51 ± 0.18 | 0.80 ± 0.16 | 0.67 ± 0.16 | 0.55 ± 0.36 | 0.65 ± 0.12 | 0.52 ± 0.19 | 0.72 ± 0.17 |
| DT | 0.61 ± 0.11 | 0.49 ± 0.13 | 0.58 ± 0.10 | 0.48 ± 0.23 | 0.37 ± 0.19 | 0.48 ± 0.21 | 0.75 ± 0.20 | 0.68 ± 0.19 | 0.60 ± 0.13 | 0.76 ± 0.21 | 0.44 ± 0.27 | 0.72 ± 0.17 | |
| ES | 0.73 ± 0.10 | 0.58 ± 0.11 | 0.67 ± 0.09 | 0.55 ± 0.29 | 0.55 ± 0.24 | 0.43 ± 0.19 | 0.87 ± 0.12 | 0.69 ± 0.18 | 0.65 ± 0.20 | 0.80 ± 0.13 | 0.49 ± 0.17 | 0.74 ± 0.13 | |
| KNN | 0.65 ± 0.04 | 0.54 ± 0.09 | 0.62 ± 0.11 | 0.48 ± 0.35 | 0.54 ± 0.23 | 0.52 ± 0.21 | 0.88 ± 0.15 | 0.68 ± 0.16 | 0.69 ± 0.21 | 0.79 ± 0.15 | 0.51 ± 0.21 | 0.73 ± 0.13 | |
| LR | 0.69 ± 0.10 | 0.58 ± 0.12 | 0.60 ± 0.09 | 0.62 ± 0.20 | 0.46 ± 0.24 | 0.58 ± 0.22 | 0.92 ± 0.14 | 0.69 ± 0.15 | 0.71 ± 0.22 | 0.79 ± 0.13 | 0.51 ± 0.21 | 0.83 ± 0.16 | |
| NB | 0.56 ± 0.10 | 0.49 ± 0.13 | 0.62 ± 0.08 | 0.52 ± 0.35 | 0.44 ± 0.32 | 0.47 ± 0.12 | 0.79 ± 0.17 | 0.69 ± 0.17 | 0.77 ± 0.26 | 0.67 ± 0.12 | 0.51 ± 0.19 | 0.83 ± 0.14 | |
| RF | 0.67 ± 0.06 | 0.58 ± 0.13 | 0.63 ± 0.11 | 0.63 ± 0.26 | 0.51 ± 0.23 | 0.46 ± 0.17 | 0.76 ± 0.14 | 0.69 ± 0.15 | 0.76 ± 0.18 | 0.76 ± 0.18 | 0.45 ± 0.21 | 0.82 ± 0.15 | |
| SVM | 0.74 ± 0.10 | 0.60 ± 0.13 | 0.64 ± 0.07 | 0.57 ± 0.30 | 0.52 ± 0.26 | 0.48 ± 0.21 | 0.83 ± 0.13 | 0.72 ± 0.19 | 0.72 ± 0.22 | 0.74 ± 0.13 | 0.55 ± 0.16 | 0.78 ± 0.14 | |
| Precision ED | AB | 0.75 ± 0.08 | 0.55 ± 0.17 | 0.65 ± 0.17 | 0.77 ± 0.18 | 0.65 ± 0.18 | 0.67 ± 0.18 | 0.92 ± 0.11 | 0.80 ± 0.22 | 0.65 ± 0.26 | 0.69 ± 0.24 | 0.50 ± 0.16 | 0.64 ± 0.22 |
| DT | 0.66 ± 0.12 | 0.55 ± 0.13 | 0.55 ± 0.11 | 0.57 ± 0.17 | 0.60 ± 0.22 | 0.57 ± 0.18 | 0.79 ± 0.19 | 0.78 ± 0.16 | 0.65 ± 0.18 | 0.73 ± 0.27 | 0.40 ± 0.19 | 0.66 ± 0.15 | |
| ES | 0.76 ± 0.08 | 0.65 ± 0.14 | 0.65 ± 0.12 | 0.70 ± 0.22 | 0.72 ± 0.20 | 0.63 ± 0.28 | 0.92 ± 0.11 | 0.90 ± 0.13 | 0.76 ± 0.17 | 0.80 ± 0.17 | 0.51 ± 0.17 | 0.85 ± 0.21 | |
| KNN | 0.77 ± 0.14 | 0.66 ± 0.10 | 0.70 ± 0.13 | 0.72 ± 0.24 | 0.75 ± 0.18 | 0.68 ± 0.24 | 0.90 ± 0.11 | 0.94 ± 0.11 | 0.74 ± 0.18 | 0.82 ± 0.15 | 0.49 ± 0.35 | 0.75 ± 0.21 | |
| LR | 0.78 ± 0.09 | 0.58 ± 0.15 | 0.64 ± 0.09 | 0.76 ± 0.18 | 0.71 ± 0.16 | 0.63 ± 0.23 | 0.93 ± 0.13 | 0.90 ± 0.13 | 0.73 ± 0.17 | 0.82 ± 0.17 | 0.57 ± 0.30 | 0.82 ± 0.20 | |
| NB | 0.71 ± 0.12 | 0.56 ± 0.16 | 0.65 ± 0.13 | 0.64 ± 0.18 | 0.57 ± 0.20 | 0.70 ± 0.24 | 0.89 ± 0.11 | 0.72 ± 0.17 | 0.74 ± 0.13 | 0.80 ± 0.20 | 0.50 ± 0.34 | 0.85 ± 0.22 | |
| RF | 0.77 ± 0.13 | 0.64 ± 0.14 | 0.66 ± 0.09 | 0.72 ± 0.23 | 0.70 ± 0.20 | 0.67 ± 0.22 | 0.90 ± 0.13 | 0.82 ± 0.16 | 0.73 ± 0.25 | 0.78 ± 0.16 | 0.55 ± 0.15 | 0.76 ± 0.24 | |
| SVM | 0.78 ± 0.13 | 0.65 ± 0.16 | 0.69 ± 0.13 | 0.69 ± 0.18 | 0.72 ± 0.20 | 0.67 ± 0.23 | 0.95 ± 0.12 | 0.92 ± 0.11 | 0.77 ± 0.15 | 0.77 ± 0.20 | 0.68 ± 0.17 | 0.80 ± 0.18 | |
| Three-Way ANOVA | Section | Features | Algorithm | Section × Feature | Section × Algorithm | Feature × Algorithm | Section × Feature × Algorithm |
|---|---|---|---|---|---|---|---|
| Accuracy | <0.001 | <0.001 | <0.001 | <0.001 | ns | ns | ns |
| Precision CTRL | <0.001 | <0.001 | <0.001 | <0.001 | ns | ns | ns |
| Precision CD | <0.001 | <0.001 | <0.001 | 0.002 | ns | ns | ns |
| Precision ED | <0.001 | <0.001 | <0.001 | <0.001 | ns | ns | ns |
| Sections | |||||||
| (a) | Features | ALL | AI | MI | DI | ||
| Accuracy | ALL | b, ns | a, ns | c, B | c, B | ||
| QT | a, ns | a, ns | b, A | a, A | |||
| SQT | b, ns | a, ns | c, A | d, B | |||
| Precision CTRL | ALL | a, C | a, B | b, B | b, B | ||
| QT | ns, A | ns, A | ns, A | ns, A | |||
| SQT | a, B | a, B | b, B | b, B | |||
| Precision CD | ALL | b, C | a, ns | c, B | d, B | ||
| QT | a, A | a, ns | b, A | a, A | |||
| SQT | a, B | b, ns | b, A | c, B | |||
| Precision ED | ALL | a, B | a, ns | b, B | b, B | ||
| QT | b, A | b, ns | c, B | a, A | |||
| SQT | a, A | a, ns | ab, A | b, B | |||
| (b) | Accuracy | Precision CTRL | Precision CD | Precision ED | |||
| Algorithm | AB | ab | ab | ab | ab | ||
| DT | a | a | a | a | |||
| ES | c | b | abc | b | |||
| KNN | bc | ab | abc | b | |||
| LR | c | b | c | b | |||
| NB | ab | a | abc | ab | |||
| RF | bc | ab | abc | b | |||
| SVM | c | ab | bc | b | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliveira, J.; Barata, M.; Soares, F.; Pousão-Ferreira, P.; Oliva-Teles, A.; Couto, A. Machine Learning-Based Classification of Malnutrition Using Histological Biomarkers of Fish Intestine: Preliminary Data. J. Mar. Sci. Eng. 2024, 12, 2177. https://doi.org/10.3390/jmse12122177
Oliveira J, Barata M, Soares F, Pousão-Ferreira P, Oliva-Teles A, Couto A. Machine Learning-Based Classification of Malnutrition Using Histological Biomarkers of Fish Intestine: Preliminary Data. Journal of Marine Science and Engineering. 2024; 12(12):2177. https://doi.org/10.3390/jmse12122177
Chicago/Turabian StyleOliveira, Joana, Marisa Barata, Florbela Soares, Pedro Pousão-Ferreira, Aires Oliva-Teles, and Ana Couto. 2024. "Machine Learning-Based Classification of Malnutrition Using Histological Biomarkers of Fish Intestine: Preliminary Data" Journal of Marine Science and Engineering 12, no. 12: 2177. https://doi.org/10.3390/jmse12122177
APA StyleOliveira, J., Barata, M., Soares, F., Pousão-Ferreira, P., Oliva-Teles, A., & Couto, A. (2024). Machine Learning-Based Classification of Malnutrition Using Histological Biomarkers of Fish Intestine: Preliminary Data. Journal of Marine Science and Engineering, 12(12), 2177. https://doi.org/10.3390/jmse12122177






