Quorum Sensing: Unravelling the Intricacies of Microbial Communication for Biofilm Formation, Biogeochemical Cycling, and Biotechnological Applications
Abstract
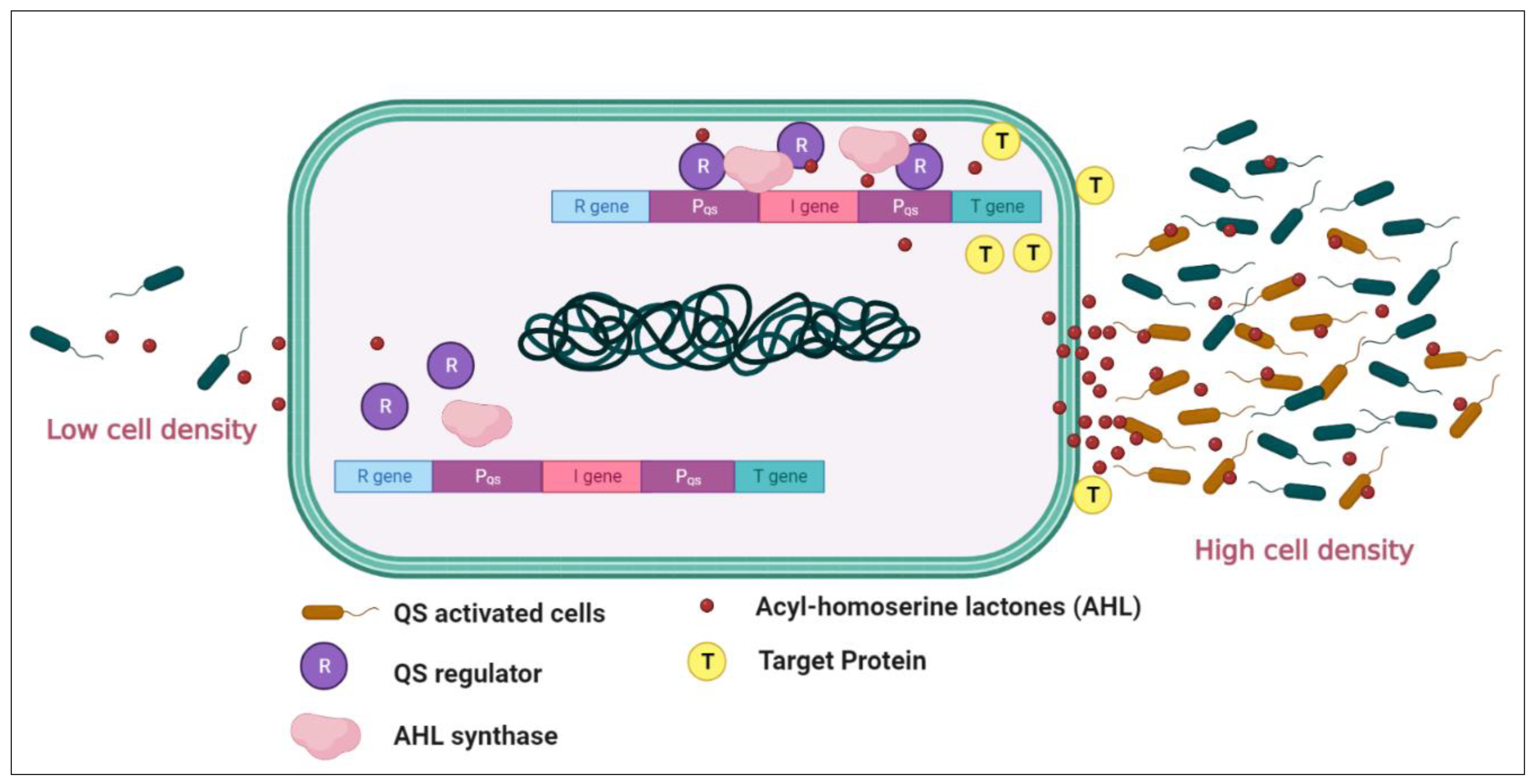
1. Introduction
2. Bacterial Quorum Sensing in Oceanic Settings
3. Biofilm Formation
4. Biogeochemical Cycling and Quorum Sensing
5. Methods of Monitoring Quorum Sensing
6. Biotechnological Applications
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sood, U.; Dhingra, G.G.; Anand, S.; Hira, P.; Kumar, R.; Kaur, J.; Verma, M.; Singhvi, N.; Lal, S.; Rawat, C.D.; et al. Microbial Journey: Mount Everest to Mars. Indian J. Microbiol. 2022, 62, 323–337. [Google Scholar] [CrossRef]
- Bassler, B.L.; Losick, R. Bacterially Speaking. Cell 2006, 125, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Nealson, K.H.; Platt, T.; Hastings, J.W. Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 1970, 104, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Nealson, K.H. Autoinduction of bacterial luciferase: Occurrence, mechanism and significance. Arch. Microbiol. 1977, 112, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Engebrecht, J.; Nealson, K.; Silverman, M. Bacterial bioluminescence: Isolation and genetic analysis of functions from Vibrio fischeri. Cell 1983, 32, 773–781. [Google Scholar] [CrossRef]
- Wang, D.; Bai, L.; Li, S.; Yan, W. Similarities and Differences in Quorum Sensing-Controlled Bioluminescence between Photobacterium phosphoreum T3 and Vibrio qinghaiensis sp.-Q67. Appl. Sci. 2022, 12, 2066. [Google Scholar] [CrossRef]
- Preda, V.G.; Săndulescu, O. Communication is the key: Biofilms, quorum sensing, formation and prevention. Discoveries 2019, 7, e10. [Google Scholar] [CrossRef] [PubMed]
- Shanker, E.; Federle, M.J. Quorum sensing regulation of competence and bacteriocins in Streptococcus pneumoniae and mutans. Genes 2017, 8, 15. [Google Scholar] [CrossRef]
- van Gestel, J.; Bareia, T.; Tenennbaum, B.; Dal Co, A.; Guler, P.; Aframian, N.; Puyesky, S.; Grinberg, I.; D’Souza, G.G.; Erez, Z.; et al. Short-range quorum sensing controls horizontal gene transfer at micron scale in bacterial communities. Nat. Commun. 2021, 12, 2324. [Google Scholar] [CrossRef]
- İnat, G.; Sırıken, B.; Başkan, C.; Erol, İ.; Yıldırım, T.; Çiftci, A. Quorum sensing systems and related virulence factors in Pseudomonas aeruginosa isolated from chicken meat and ground beef. Sci. Rep. 2021, 11, 15639. [Google Scholar] [CrossRef]
- Kai, K. Bacterial quorum sensing in symbiotic and pathogenic relationships with hosts. Biosci. Biotechnol. Biochem. 2018, 82, 363–371. [Google Scholar] [CrossRef]
- Albuquerque, P.; Casadevall, A. Quorum sensing in fungi—A review. Med. Mycol. 2012, 50, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Hull, C.M. Sporulation: How to survive on planet Earth (and beyond) HHS Public Access Author manuscript. Curr. Genet. 2017, 63, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Joshi, C.; Kothari, V.; Patel, P. Importance of selecting appropriate wavelength, while quantifying growth and production of quorum sensing regulated pigments in bacteria. Recent Pat. Biotechnol. 2016, 10, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Bramhachari, P.V.; Yugandhar, N.M.; Prathyusha, A.; Mohana Sheela, G.; Naravula, J.; Venkateswarlu, N. Quorum sensing regulated swarming motility and migratory behavior in bacteria. In Implication of Quorum Sensing System in Biofilm Formation and Virulence; Springer: Berlin/Heidelberg, Germany, 2018; pp. 49–66. [Google Scholar]
- Butrico, C.E.; Cassat, J.E. Quorum sensing and toxin production in Staphylococcus aureus osteomyelitis: Pathogenesis and paradox. Toxins 2020, 12, 516. [Google Scholar] [CrossRef]
- Armes, A.C.; Walton, J.L.; Buchan, A. Quorum Sensing and Antimicrobial Production Orchestrate Biofilm Dynamics in Multispecies Bacterial Communities. Microbiol. Spectr. 2022, 10, e02615-22. [Google Scholar] [CrossRef]
- Liu, X.; Bimerew, M.; Ma, Y.; Müller, H.; Ovadis, M.; Eberl, L.; Berg, G.; Chernin, L. Quorum-sensing signaling is required for production of the antibiotic pyrrolnitrin in a rhizospheric biocontrol strain of Serratia plymuthica. FEMS Microbiol. Lett. 2007, 270, 299–305. [Google Scholar] [CrossRef]
- Sanchez, L.M.; Mannathan, S.; Lee, C.-K.; Merlo, M.E.; Meng, X.-F.; Nihira, T.; Takano, E.; Minnaard, A.J.; Dijkhuizen, L.; Petrusma, M. Identification and characterisation of enzymes involved in γ-butyrolactone biosynthesis in Streptomyces coelicolor. In Quorum Sensing in Streptomyces Coelicolor; University of Groningen: Groningen, The Netherlands, 2016; pp. 92–137. [Google Scholar]
- Pan, J.; Ren, D. Quorum sensing inhibitors: A patent overview. Expert Opin. Ther. Pat. 2009, 19, 1581–1601. [Google Scholar] [CrossRef]
- Carradori, S.; Di Giacomo, N.; Lobefalo, M.; Luisi, G.; Campestre, C.; Sisto, F. Biofilm and quorum sensing inhibitors: The road so far. Expert Opin. Ther. Pat. 2020, 30, 917–930. [Google Scholar] [CrossRef]
- Bassler, B.L. How bacteria talk to each other: Regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 1999, 2, 582–587. [Google Scholar] [CrossRef]
- Whiteley, M.; Diggle, S.P.; Greenberg, E.P. Progress in and promise of bacterial quorum sensing research. Nature 2017, 551, 313–320. [Google Scholar] [CrossRef]
- Tanet, L.; Tamburini, C.; Baumas, C.; Garel, M.; Simon, G.; Casalot, L. Bacterial bioluminescence: Light Emission in Photobacterium phosphoreum Is Not under Quorum-Sensing Control. Front. Microbiol. 2019, 10, 365. [Google Scholar] [CrossRef]
- Muras, A.; López-Pérez, M.; Mayer, C.; Parga, A.; Amaro-Blanco, J.; Otero, A. High prevalence of quorum-sensing and quorum-quenching activity among cultivable bacteria and metagenomic sequences in the Mediterranean Sea. Genes 2018, 9, 100. [Google Scholar] [CrossRef]
- Suresh, S.; Alva, P.P.; Premanath, R. Modulation of quorum sensing-associated virulence in bacteria: Carbohydrate as a key factor. Arch. Microbiol. 2021, 203, 1881–1890. [Google Scholar] [CrossRef]
- Pérez-Rodríguez, I.; Bolognini, M.; Ricci, J.; Bini, E.; Vetriani, C. From deep-sea volcanoes to human pathogens: A conserved quorum-sensing signal in Epsilonproteobacteria. ISME J. 2015, 9, 1222–1234. [Google Scholar] [CrossRef] [PubMed]
- Keller, L.; Surette, M.G. Communication in bacteria: An ecological and evolutionary perspective. Nat. Rev. Microbiol. 2006, 4, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Girard, L. Quorum sensing in Vibrio spp.: The complexity of multiple signalling molecules in marine and aquatic environments. Crit. Rev. Microbiol. 2019, 45, 451–471. [Google Scholar] [CrossRef] [PubMed]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef]
- Rehman, Z.U.; Leiknes, T.O. Quorum-quenching bacteria isolated from red sea sediments reduce biofilm formation by Pseudomonas aeruginosa. Front. Microbiol. 2018, 9, 1354. [Google Scholar] [CrossRef]
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2015, 6, 26–41. [Google Scholar] [CrossRef]
- Chen, X.; Schauder, S.; Potier, N.; Van Dorsselaer, A.; Pelczer, I.; Bassler, B.L.; Hughson, F.M. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 2002, 415, 545–549. [Google Scholar] [CrossRef]
- del Mar Cendra, M.; Torrents, E. Pseudomonas aeruginosa biofilms and their partners in crime. Biotechnol. Adv. 2021, 49, 107734. [Google Scholar] [CrossRef]
- Alain, K.; Zbinden, M.; Le Bris, N.; Lesongeur, F.; Quérellou, J.; Gaill, F.; Cambon-Bonavita, M.A. Early steps in microbial colonization processes at deep-sea hydrothermal vents. Environ. Microbiol. 2004, 6, 227–241. [Google Scholar] [CrossRef] [PubMed]
- Krupke, A.; Hmelo, L.R.; Ossolinski, J.E.; Mincer, T.J.; Van Mooy, B.A.S. Quorum sensing plays a complex role in regulating the enzyme hydrolysis activity of microbes associated with sinking particles in the ocean. Front. Mar. Sci. 2016, 3, 55. [Google Scholar] [CrossRef]
- Rumbaugh, K.P.; Sauer, K. Biofilm dispersion. Nat. Rev. Microbiol. 2020, 18, 571–586. [Google Scholar] [CrossRef] [PubMed]
- Di Donato, P.; Poli, A.; Taurisano, V.; Abbamondi, G.R.; Nicolaus, B.; Tommonaro, G. Recent advances in the study of marine microbial biofilm: From the involvement of quorum sensing in its production up to biotechnological application of the polysaccharide fractions. J. Mar. Sci. Eng. 2016, 4, 34. [Google Scholar] [CrossRef]
- Jamuna Bai, A.; Ravishankar Rai, V. Effect of small chain N acyl homoserine lactone quorum sensing signals on biofilms of food-borne pathogens. J. Food Sci. Technol. 2016, 53, 3609–3614. [Google Scholar] [CrossRef]
- Sakuragi, Y.; Kolter, R. Quorum-sensing regulation of the biofilm matrix genes (pel) of Pseudomonas aeruginosa. J. Bacteriol. 2007, 189, 5383–5386. [Google Scholar] [CrossRef]
- Friedman, L.; Kolter, R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 2004, 51, 675–690. [Google Scholar] [CrossRef]
- Ueda, A.; Wood, T.K. Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885). PLoS Pathog. 2009, 5, e1000483. [Google Scholar] [CrossRef]
- Diggle, S.P.; Winzer, K.; Chhabra, S.R.; Worrall, K.E.; Cámara, M.; Williams, P. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol. Microbiol. 2003, 50, 29–43. [Google Scholar] [CrossRef]
- Phippen, B.L.; Oliver, J.D. Clinical and environmental genotypes of Vibrio vulnificus display distinct, quorum-sensing-mediated, chitin detachment dynamics. Pathog. Dis. 2015, 73, ftv072. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef]
- Zhu, J.; Mekalanos, J.J. Quorum Sensing-Dependent Biofilms Enhance Colonization in Vibrio cholerae), the phenomenon by which bacteria monitor their cell population density through the extracellular accumulation of signaling molecules called autoinduc. Dev. Cell 2003, 5, 647–656. [Google Scholar] [CrossRef]
- Seyedsayamdost, M.R.; Carr, G.; Kolter, R.; Clardy, J. Roseobacticides: Small molecule modulators of an algal-bacterial symbiosis. J. Am. Chem. Soc. 2011, 133, 18343–18349. [Google Scholar] [CrossRef] [PubMed]
- Cude, W.N.; Mooney, J.; Tavanaei, A.A.; Hadden, M.K.; Frank, A.M.; Gulvik, C.A.; May, A.L.; Buchan, A. Production of the antimicrobial secondary metabolite indigoidine contributes to competitive surface colonization by the marine roseobacter Phaeobacter sp. strain Y4I. Appl. Environ. Microbiol. 2012, 78, 4771–4780. [Google Scholar] [CrossRef] [PubMed]
- Baysse, C.; Cullinane, M.; Dénervaud, V.; Burrowes, E.; Dow, J.M.; Morrissey, J.P.; Tam, L.; Trevors, J.T.; O’Gara, F. Modulation of quorum sensing in Pseudomonas aeruginosa through alteration of membrane properties. Microbiology 2005, 151, 2529–2542. [Google Scholar] [CrossRef]
- Eldar, A. Social conflict drives the evolutionary divergence of quorum sensing. Proc. Natl. Acad. Sci. USA. 2011, 108, 13635–13640. [Google Scholar] [CrossRef]
- Ball, A.S.; Chaparian, R.R.; Van Kessel, J.C. Quorum sensing gene regulation by LuxR/HapR master regulators in vibrios. J. Bacteriol. 2017, 199, 1110–1128. [Google Scholar] [CrossRef] [PubMed]
- Kumar, C.G.; Joo, H.-S.; Choi, J.-W.; Koo, Y.-M.; Chang, C.-S. Purification and characterization of an extracellular polysaccharide from haloalkalophilic Bacillus sp. I-450. Enzyme Microb. Technol. 2004, 34, 673–681. [Google Scholar] [CrossRef]
- Llamas, I.; Quesada, E.; Martínez-Cánovas, M.J.; Gronquist, M.; Eberhard, A.; Gonzalez, J.E. Quorum sensing in halophilic bacteria: Detection of N-acyl-homoserine lactones in the exopolysaccharide-producing species of Halomonas. Extremophiles 2005, 9, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Llamas, I.; Mata, J.A.; Tallon, R.; Bressollier, P.; Urdaci, M.C.; Quesada, E.; Béjar, V. Characterization of the exopolysaccharide produced by Salipiger mucosus A3T, a halophilic species belonging to the Alphaproteobacteria, isolated on the Spanish mediterranean seaboard. Mar. Drugs 2010, 8, 2240–2251. [Google Scholar] [CrossRef] [PubMed]
- Voordeckers, J.W.; Starovoytov, V.; Vetriani, C. Caminibacter mediatlanticus sp. nov., a thermophilic, chemolithoautotrophic, nitrate-ammonifying bacterium isolated from a deep-sea hydrothermal vent on the Mid-Atlantic Ridge. Int. J. Syst. Evol. Microbiol. 2005, 55, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Raguénès, G.H.C.; Peres, A.; Ruimy, R.; Pignet, P.; Christen, R.; Loaec, M.; Rougeaux, H.; Barbier, G.; Guezennec, J.G. Alteromonas infernus sp. nov., a new polysaccharide-producing bacterium isolated from a deep-sea hydrothermal vent. J. Appl. Microbiol. 1997, 82, 422–430. [Google Scholar] [CrossRef]
- Raguénès, G.; Christen, R.; Guezennec, J.; Pignet, P.; Barbier, G. Vibrio diabolicus sp. nov., a new polysaccharide-secreting organism isolated from a deep-sea hydrothermal vent polychaete annelid, Alvinella pompejana. Int. J. Syst. Evol. Microbiol. 1997, 47, 989–995. [Google Scholar]
- Floris, R.; Rizzo, C.; Lo Giudice, A. Biosurfactants from Marine Microorganisms. In Metabolomics—New Insights into Biology and Medicine; IntechOpen: London, UK, 2020. [Google Scholar] [CrossRef]
- Gutiérrez-Arnillas, E.; Rodríguez, A.; Sanromán, M.A.; Deive, F.J. New sources of halophilic lipases: Isolation of bacteria from Spanish and Turkish saltworks. Biochem. Eng. J. 2016, 109, 170–177. [Google Scholar] [CrossRef]
- Gutierrez, T.; Singleton, D.R.; Berry, D.; Yang, T.; Aitken, M.D.; Teske, A. Hydrocarbon-degrading bacteria enriched by the Deepwater Horizon oil spill identified by cultivation and DNA-SIP. ISME J. 2013, 7, 2091–2104. [Google Scholar] [CrossRef]
- Heng, S.-P.; Letchumanan, V.; Deng, C.-Y.; Ab Mutalib, N.-S.; Khan, T.M.; Chuah, L.-H.; Chan, K.-G.; Goh, B.-H.; Pusparajah, P.; Lee, L.-H. Vibrio vulnificus: An environmental and clinical burden. Front. Microbiol. 2017, 8, 997. [Google Scholar] [CrossRef]
- Zhou, Z.; St John, E.; Anantharaman, K.; Reysenbach, A.-L. Global patterns of diversity and metabolism of microbial communities in deep-sea hydrothermal vent deposits. Microbiome 2022, 10, 241. [Google Scholar] [CrossRef]
- Yang, Y.; Gao, Y.; Liu, Y.; Liu, B.; Wang, D.; Xu, Y.; Wei, Y. Pseudomonas marianensis sp. nov., a marine bacterium isolated from deep-sea sediments of the Mariana Trench. Arch. Microbiol. 2022, 204, 638. [Google Scholar] [CrossRef]
- Casillo, A.; Lanzetta, R.; Parrilli, M.; Corsaro, M.M. Exopolysaccharides from marine and marine extremophilic bacteria: Structures, properties, ecological roles and applications. Mar. Drugs 2018, 16, 69. [Google Scholar] [CrossRef]
- Raimundo, I.; Silva, R.; Meunier, L.; Valente, S.M.; Lago-Lestón, A.; Keller-Costa, T.; Costa, R. Functional metagenomics reveals differential chitin degradation and utilization features across free-living and host-associated marine microbiomes. Microbiome 2021, 9, 43. [Google Scholar] [CrossRef]
- Stout, L.M.; Joshi, S.R.; Kana, T.M.; Jaisi, D.P. Microbial activities and phosphorus cycling: An application of oxygen isotope ratios in phosphate. Geochim. Cosmochim. Acta 2014, 138, 101–116. [Google Scholar] [CrossRef]
- dos Santos, H.F.; Cury, J.C.; do Carmo, F.L.; dos Santos, A.L.; Tiedje, J.; van Elsas, J.D.; Rosado, A.S.; Peixoto, R.S. Mangrove Bacterial Diversity and the Impact of Oil Contamination Revealed by Pyrosequencing: Bacterial Proxies for Oil Pollution. PLoS ONE 2011, 6, e16943. [Google Scholar] [CrossRef]
- Visick, K.L.; Ruby, E.G. Vibrio fischeri and its host: It takes two to tango. Curr. Opin. Microbiol. 2006, 9, 632–638. [Google Scholar] [CrossRef]
- Weiland-Bräuer, N. Friends or foes—Microbial interactions in nature. Biology 2021, 10, 496. [Google Scholar] [CrossRef] [PubMed]
- Sievert, S.M.; Vetriani, C. Chemoautotrophy at deep-sea vents: Past, present, and future. Oceanography 2012, 25, 218–233. [Google Scholar] [CrossRef]
- Li, C.Y.; Mausz, M.A.; Murphy, A.; Zhang, N.; Chen, X.L.; Wang, S.Y.; Gao, C.; Aguilo-Ferretjans, M.M.; Silvano, E.; Lidbury, I.D.E.A.; et al. Ubiquitous occurrence of a dimethylsulfoniopropionate ABC transporter in abundant marine bacteria. ISME J. 2023, 17, 579–587. [Google Scholar] [CrossRef]
- Sánchez-Baracaldo, P.; Bianchini, G.; Wilson, J.D.; Knoll, A.H. Cyanobacteria and biogeochemical cycles through Earth history. Trends Microbiol. 2022, 30, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Dang, H.; Lovell, C.R. Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 2016, 80, 91–138. [Google Scholar] [CrossRef]
- Goffredi, S.K.; Johnson, S.B.; Vrijenhoek, R.C. Genetic diversity and potential function of microbial symbionts associated with newly discovered species of Osedax polychaete worms. Appl. Environ. Microbiol. 2007, 73, 2314–2323. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.Z.; Sasidharan, R.S.; Alghamdi, H.A.; Dang, H. Understanding Interaction Patterns within Deep-Sea Microbial Communities and Their Potential Applications. Mar. Drugs 2022, 20, 108. [Google Scholar] [CrossRef] [PubMed]
- Hualpa-Cutipa, E.; Andi, R.; Acosta, S.; Landa-Acuña, D.; Elena, M.; Salvatierra, S.; Reynaldo, J.; Quiroz, R. Omics Insights into Quorum Sensing and Biofilm Formation. In Omics for Environmental Engineering and Microbiology Systems; CRC Press: Boca Raton, FL, USA, 2022; pp. 143–157. [Google Scholar]
- Barbosa, A.; Miranda, S.; Azevedo, N.F.; Cerqueira, L.; Azevedo, A.S. Imaging biofilms using fluorescence in situ hybridization: Seeing is believing. Front. Cell. Infect. Microbiol. 2023, 13, 1195803. [Google Scholar] [CrossRef]
- Kai, K.; Furuyabu, K.; Tani, A.; Hayashi, H. Production of the quorum-sensing molecules N-acylhomoserine lactones by endobacteria associated with Mortierella alpina A-178. ChemBioChem 2012, 13, 1776–1784. [Google Scholar] [CrossRef]
- Li, J.; Liu, R.; Chen, Y.; Liu, S.; Chen, C.; Liu, T.; Yang, S.; Zhuang, Y.; Yang, R.; Cui, Y.; et al. Computer-Aided Rational Engineering of Signal Sensitivity of Quorum Sensing Protein LuxR in a Whole-Cell Biosensor. Front. Mol. Biosci. 2021, 8, 729350. [Google Scholar] [CrossRef]
- Keleştemur, S.; Avci, E.; Çulha, M. Raman and surface-enhanced Raman scattering for biofilm characterization. Chemosensors 2018, 6, 5. [Google Scholar] [CrossRef]
- Manifold, B.; Fu, D. Quantitative Stimulated Raman Scattering Microscopy: Promises and Pitfalls. Annu. Rev. Anal. Chem. 2022, 15, 269–289. [Google Scholar] [CrossRef]
- Aljuhani, W.; Zhang, Y.; Wylie, M.P.; Xu, Y.; McCoy, C.P.; Bell, S.E.J. Probing the interaction of ex situ biofilms with plasmonic metal nanoparticles using surface-enhanced Raman spectroscopy. Analyst 2023, 148, 2002–2011. [Google Scholar] [CrossRef] [PubMed]
- Bodelón, G.; Montes-García, V.; López-Puente, V.; Hill, E.H.; Hamon, C.; Sanz-Ortiz, M.N.; Rodal-Cedeira, S.; Costas, C.; Celiksoy, S.; Pérez-Juste, I. Detection and imaging of quorum sensing in Pseudomonas aeruginosa biofilm communities by surface-enhanced resonance Raman scattering. Nat. Mater. 2016, 15, 1203–1211. [Google Scholar] [CrossRef]
- Prescott, R.D.; Decho, A.W. Flexibility and adaptability of quorum sensing in nature. Trends Microbiol. 2020, 28, 436–444. [Google Scholar] [CrossRef]
- Anetzberger, C.; Schell, U.; Jung, K. Single cell analysis of Vibrio harveyi uncovers functional heterogeneity in response to quorum sensing signals. BMC Microbiol. 2012, 12, 209. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Quan, C.; Liu, B.; Xu, Y.; Zhao, P.; Xiong, W.; Fan, S. Green fluorescent protein (GFP)-based overexpression screening and characterization of AgrC, a Receptor protein of quorum sensing in Staphylococcus aureus. Int. J. Mol. Sci. 2013, 14, 18470–18487. [Google Scholar] [CrossRef] [PubMed]
- Campagna, S.R.; Gooding, J.R.; May, A.L. Direct quantitation of the quorum sensing signal, autoinducer-2, in clinically relevant samples by liquid chromatography—tandem mass spectrometry. Anal. Chem. 2009, 81, 6374–6381. [Google Scholar] [CrossRef] [PubMed]
- May, A.L.; Eisenhauer, M.E.; Coulston, K.S.; Campagna, S.R. Detection and quantitation of bacterial acylhomoserine lactone quorum sensing molecules via liquid chromatography–isotope dilution tandem mass spectrometry. Anal. Chem. 2012, 84, 1243–1252. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.H.; Hegde, M.; Kim, J.; Wang, X.; Jayaraman, A.; Wood, T.K. Synthetic quorum-sensing circuit to control consortial biofilm formation and dispersal in a microfluidic device. Nat. Commun. 2012, 3, 613. [Google Scholar] [CrossRef]
- Burgess, J.G. New and emerging analytical techniques for marine biotechnology. Curr. Opin. Biotechnol. 2012, 23, 29–33. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, J.; Crisante, D.; Jaramillo Lopez, J.M.; Mahadevan, R. Dynamic cell programming with quorum sensing-controlled CRISPRi circuit. ACS Synth. Biol. 2020, 9, 1284–1291. [Google Scholar] [CrossRef]
- Dai, C.; Qu, Y.; Wu, W.; Li, S.; Chen, Z.; Lian, S.; Jing, J. QSP: An open sequence database for quorum sensing related gene analysis with an automatic annotation pipeline. Water Res. 2023, 235, 119814. [Google Scholar] [CrossRef]
- Barbarossa, M.V.; Kuttler, C.; Fekete, A.; Rothballer, M. A delay model for quorum sensing of Pseudomonas putida. Biosystems 2010, 102, 148–156. [Google Scholar] [CrossRef]
- Guo, J.; Liu, Y.; Chen, Y.; Li, J.; Ju, H. A multifunctional SERS sticky note for real-time quorum sensing tracing and inactivation of bacterial biofilms. Chem. Sci. 2018, 9, 5906–5911. [Google Scholar] [CrossRef]
- Senni, K.; Gueniche, F.; Changotade, S.; Septier, D.; Sinquin, C.; Ratiskol, J.; Lutomski, D.; Godeau, G.; Guezennec, J.; Colliec-Jouault, S. Unusual glycosaminoglycans from a deep sea hydrothermal bacterium improve fibrillar collagen structuring and fibroblast activities in engineered connective tissues. Mar. Drugs 2013, 11, 1351–1369. [Google Scholar] [CrossRef]
- Montgomery, K.; Charlesworth, J.C.; LeBard, R.; Visscher, P.T.; Burns, B.P. Quorum sensing in extreme environments. Life 2013, 3, 131–148. [Google Scholar] [CrossRef] [PubMed]
- Bzdrenga, J.; Daude, D.; Remy, B.; Jacquet, P.; Plener, L.; Elias, M.; Chabriere, E. Biotechnological applications of quorum quenching enzymes. Chem. Biol. Interact. 2017, 267, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Liao, N.Q.; Li, H.M. Conceivable bioremediation techniques based on quorum sensing. Appl. Mech. Mater. 2013, 295, 39–44. [Google Scholar] [CrossRef]
- Sivasankar, P.; Poongodi, S.; Seedevi, P.; Sivakumar, M.; Murugan, T.; Loganathan, S. Bioremediation of wastewater through a quorum sensing triggered MFC: A sustainable measure for waste to energy concept. J. Environ. Manage 2019, 237, 84–93. [Google Scholar] [CrossRef]
- Smith, J.L.; Fratamico, P.M.; Novak, J.S. Quorum sensing: A primer for food microbiologists. J. Food Prot. 2004, 67, 1053–1070. [Google Scholar] [CrossRef] [PubMed]
- Tonkin, M.; Khan, S.; Wani, M.Y.; Ahmad, A. Quorum sensing-a stratagem for conquering multi-drug resistant pathogens. Curr. Pharm. Des. 2021, 27, 2835–2847. [Google Scholar] [CrossRef]
- Kameswaran, S.; Ramesh, B. Quenching and Quorum Sensing in Bacterial Bio-films. Res. Microbiol. 2023, 104085. [Google Scholar] [CrossRef] [PubMed]
- Abbamondi, G.R.; Tommonaro, G. Research progress and hopeful strategies of application of quorum sensing in food, agriculture and nanomedicine. Microorganisms 2022, 10, 1192. [Google Scholar] [CrossRef]
- Basu, S.; Gerchman, Y.; Collins, C.H.; Arnold, F.H.; Weiss, R. A synthetic multicellular system for programmed pattern formation. Nature 2005, 434, 1130–1134. [Google Scholar] [CrossRef]
- Kalia, V.C.; Patel, S.K.S.; Kang, Y.C.; Lee, J.-K. Quorum sensing inhibitors as antipathogens: Biotechnological applications. Biotechnol. Adv. 2019, 37, 68–90. [Google Scholar] [CrossRef] [PubMed]
- Shaaban, M.; Elgaml, A.; Habib, E.-S.E. Biotechnological applications of quorum sensing inhibition as novel therapeutic strategies for multidrug resistant pathogens. Microb. Pathog. 2019, 127, 138–143. [Google Scholar] [CrossRef] [PubMed]
| Bacteria | Activity Mediated by QS | Location of Occurrence | Signaling Molecules | QS System | Interaction within QS System | Reference |
|---|---|---|---|---|---|---|
| Staphylococcus aureus | Virulence factor production control Biofilm formation control through the agr system | Human body | Autoinducing peptides (AIPs) | Agr system | AIPs bind to the AgrC receptor, activating the AgrA response regulator, which regulates virulence factor production. | [31,32] |
| Bacillus cereus | Virulence factor control through the PlcR system | Various habitats, including soil and food | Cyclic peptide | PlcR system | The cyclic peptide acts as a quorum sensing signal, binding to PlcR and triggering virulence gene expression. | |
| Pseudomonas aeruginosa | Virulence factor control through LuxI/LuxR-type system | Various habitats, including soil, water, and plants | N-acyl homoserine lactones (AHLs) | LasR/RhlR system | AHLs bind to LasR and RhlR, initiating transcription of numerous genes, including those responsible for virulence factor production. | [33] |
| Vibrio fischerii | Light emission gene control through the LuxI system | Bobtail squid (Euprymna scolopes) in marine waters | N-acyl homoserine lactone (AHL) | LuxI/LuxR system | AHLs bind to LuxR, activating transcription of the luciferase operon, which is responsible for light production. | [25] |
| Vibrio diabolicus | Biofilm formation control | Polychaete annelid Alvinella pompejana deep-sea hydrothermal vent | Autoinducing peptides (AIPs) | Unknown QS system | Unknown | [34,35] |
| Escherichia coli | Regulation of biofilm formation, motility, and virulence | Various habitats, including the human gastrointestinal tract | Autoinducer-2 (AI-2) | LuxS/AI-2 system | AI-2 is internalized and binds to the LsrR repressor, leading to the de-repression of Lsr operon and other AI-2-responsive genes | [36,37] |
| Vibrio cholerae | Toxin production regulation and colonization of the human intestine | Hadel zones and human gastrointestinal tract | N-acyl homoserine lactones (AHLs) | LuxO/LuxR system | AHLs bind to LuxR, triggering the regulation of virulence genes, including the cholera toxin. | [38,39] |
| Streptococcus pneumoniae | Competence development and genetic transformation | Human respiratory tract | Peptides | ComDE system | The peptide pheromone binds to ComD, activating ComE which triggers genetic transformation and competence development | [40] |
| Acinetobacter baumannii | Biofilm formation and antibiotic resistance | Various environments, including hospitals and soil | Unknown | Unknown QS system | Unknown | [41,42] |
| Aliivibrio fischeri | Symbiotic colonization of the Hawaiian bobtail squid | Bobtail squid (Euprymna scolopes) in marine waters | N-acyl homoserine lactone (AHL) | LuxI/LuxR system | AHLs bind to LuxR, leading to changes in gene expression that allow symbiotic colonization of the bobtail squid. | [43] |
| Photobacterium phosphoreum | Bioluminescence control | Deep-sea waters | Autoinducer-2 | LuxS/AI-2 system | AI-2 molecules are synthesized and used to modulate bioluminescence in a cell-density-dependent manner | [23] |
| Sulfitobacter sp. | Production of extracellular enzymes and biofilm formation | Marine environments, including surface water and sediments | Unknown | Unknown QS system | Unknown. Likely involves signaling molecules that regulate the production of extracellular enzymes and biofilm formation. | [44] |
| Ruegeria sp. | Biofilm formation and production of extracellular enzymes | Marine environments, including coastal seawater and sediment | N-acyl homoserine lactone (AHL) | LuxI/LuxR system | AHLs bind to LuxR, leading to changes in gene expression, including biofilm formation and the production of extracellular enzymes | [45] |
| Shewanella oneidensis | Regulation of biofilm formation and metal oxide reduction | Marine and freshwater environments | Unknown | Unknown QS system | Unknown | [46,47] |
| Colwellia psychrerythraea | Cold adaptation and biofilm formation | Cold marine environments | Unknown | Unknown QS system | Unknown | [48,49] |
| Psychrobacter sp. | Production of extracellular enzymes and biofilm formation | marine sediments | N-acylhomoserine lactones (AHL) | LuxI/LuxR system QS system | AHLs bind to LuxR, leading to changes in gene expression, including the production of extracellular enzymes and biofilm formation. | [50] |
| Marinobacter sp. | Biofilm formation and quorum quenching | Marine environments, including deep-sea sediments | Unknown | Unknown QS system | Unknown | [51] |
| Sulfurovum lithotrophicum | Quorum sensing in deep-sea vent bacteria | Deep-sea hydrothermal vents | Autoinducer-2 (AI-2) | LuxS/AI-2 system | AI-2 is synthesized and interacts within the LuxS/AI-2 system to regulate quorum sensing in these deep-sea vent bacteria. | [26,52] |
| Caminibacter mediatlanticus | Quorum sensing in deep-sea vent bacteria | Deep-sea hydrothermal vents | Autoinducer-2 (AI-2) | LuxS/AI-2 system | AI-2 is produced and interacts within the LuxS/AI-2 system to regulate quorum sensing, thus enabling these deep-sea vent bacteria to adapt and thrive in their extreme environment. | [26,52] |
| Thiomicrospira sp. | Symbiotic interactions and biofilm formation | Deep-sea hydrothermal vents | Unknown | Unknown QS system | Unknown | [53] |
| Location | Bacteria | Biofilm Composition | Functions | Reference | Quorum Sensing Involvement |
|---|---|---|---|---|---|
| Antarctic Waters | Marine bacteria | Charged uronic acid moieties and sulfate groups | Plays a role in cold adaptation | [38] | QS system(s) involved |
| Korean yellow sea | Bacillus sp. I450 | Neutral sugars and uronic acids | Exhibits antimicrobial activity | [38,52] | QS system(s) involved |
| Solar Saltern and Spanish Mediterranean Seaboard | Halomonas maura & Salipiger mucosus | Sulfated polysaccharide with high uronic acid content and fucose-rich polysaccharides | Adapted to high salinity environments and high capacity for binding cations | [53,54] | QS system(s) involved |
| Deep-sea hydrothermal vent | Caminibacter mediatlanticus | Sulfated polysaccharide high in glucosamine | Thrives in high-temperature hydrothermal vents | [27,55] | QS system(s) involved |
| Deep-sea hydrothermal vent | Alteromonas infernus | Lacking lipopolysaccharides (LPS) | Exhibits unique adaptations to extreme conditions, including high-pressure and high-temperature environments | [56] | QS system(s) involved |
| Host in deep-sea hydrothermal vent | Vibrio diabolcus | Large amounts of uronic acid and no sulfate groups | Forms symbiotic relationship with the host organism, the polychaete annelid Alvinella pompejana, in deep-sea hydrothermal vent ecosystems | [57] | QS system(s) involved |
| Antarctic Ocean | Psychrobacter sp. | Extracellular polysaccharides | Exhibits cold-adapted enzymatic activity and plays a role in biofilm formation under low temperatures | [58] | Not specified |
| Deep-sea sediments | Marinobacter sp. | Exopolysaccharides | Involved in sediment stabilization and biogeochemical cycling in deep-sea sediments | [59,60] | Not specified |
| Gulf of Mexico | Vibrio vulnificus | Alginate and extracellular DNA | Forms biofilms on oyster shells and contributes to oyster pathogenesis | [61] | QS system(s) involved |
| Deep-sea hydrothermal vent | Thiomicrospira sp. | Extracellular sulfur and polysaccharides | Capable of sulfur oxidation and plays a role in ecosystem functioning in deep-sea hydrothermal vents | [62] | Not specified |
| Mariana Trench | Pseudomonas sp. | Exopolysaccharides | Exhibits unique adaptations to extreme pressures and low nutrient availability in the Mariana Trench | [63,64] | QS system(s) involved |
| Methods | Advantages | Disadvantages | References |
|---|---|---|---|
| Time-series methods | Capture dynamics of QS over time, good for deep-sea environments. | Sample collection at multiple time points can be logistically challenging. | [93] |
| Molecular sequence-based methods (16S and 18S rRNA gene sequencing) | Widely used, can provide broad overview of microbial community. | PCR biases and mismatches, inability to decode metabolic pathways. | [25] |
| Omics approaches (Metagenomics, Metatranscriptomics, Metaproteomics) | Provide comprehensive view of microbial community, identify QS-related genes and regulatory networks. | Require high-throughput sequencing and sophisticated data analysis, potential for missing rare species or transient events. | [76] |
| FISH | Visualize and identify specific microbial populations, reveal spatial distribution of QS-active cells. | Difficult to correlate with metabolic activity or function. | [77,78] |
| Bioengineered Biosensors | Real-time analysis, designed to detect specific QS signaling molecules. | Stability and functionality can be affected by marine environment conditions. | [79] |
| Raman Microscopy | Non-destructive analysis of individual cells, insights into cellular metabolism. | Complex data interpretation: Equipment can be expensive. | [82] |
| Single-cell genomics | Provides specific insights into QS activity at the single-cell level. | High sequencing cost: Data analysis can be complex. | [85] |
| Isotope tracing | Tracks specific metabolites and their utilization by QS-active cells, insights into metabolic activities. | Technically challenging, requires specialized equipment and expertise. | [94] |
| Fluorescent Protein Tagging | Visualization of protein localization and expression dynamics in real-time. | May not work for all proteins, can affect protein function. | [86] |
| Mass Spectrometry | Allows to identify and quantify small molecules, including QS signaling molecules. | Requires sophisticated equipment and expertise, can be expensive. | [87] |
| Tandem Mass Tag (TMT) Spectrometry | Allows for relative quantification of proteins in multiple samples simultaneously. | Complex data analysis, requires access to specialized equipment. | [88] |
| Microfluidic Devices | Allow for precise control over environment and high-resolution imaging. | Fabrication can be complex, handling and analysis require specialized knowledge. | [89] |
| CRISPRi (CRISPR interference) | Gene knockdown to study the function of specific genes in QS. | Requires genetic manipulation, off-target effects. | [91] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Israel, E.; Ramganesh, S.; Abia, A.L.K.; Chikere, C.B. Quorum Sensing: Unravelling the Intricacies of Microbial Communication for Biofilm Formation, Biogeochemical Cycling, and Biotechnological Applications. J. Mar. Sci. Eng. 2023, 11, 1586. https://doi.org/10.3390/jmse11081586
Israel E, Ramganesh S, Abia ALK, Chikere CB. Quorum Sensing: Unravelling the Intricacies of Microbial Communication for Biofilm Formation, Biogeochemical Cycling, and Biotechnological Applications. Journal of Marine Science and Engineering. 2023; 11(8):1586. https://doi.org/10.3390/jmse11081586
Chicago/Turabian StyleIsrael, Edamkue, Selvarajan Ramganesh, Akebe Luther King Abia, and Chioma Blaise Chikere. 2023. "Quorum Sensing: Unravelling the Intricacies of Microbial Communication for Biofilm Formation, Biogeochemical Cycling, and Biotechnological Applications" Journal of Marine Science and Engineering 11, no. 8: 1586. https://doi.org/10.3390/jmse11081586
APA StyleIsrael, E., Ramganesh, S., Abia, A. L. K., & Chikere, C. B. (2023). Quorum Sensing: Unravelling the Intricacies of Microbial Communication for Biofilm Formation, Biogeochemical Cycling, and Biotechnological Applications. Journal of Marine Science and Engineering, 11(8), 1586. https://doi.org/10.3390/jmse11081586









