Abstract
In the crustacean immune system, leucine-rich repeat (LRR) is one of the major structures for recognizing pathogen-associated molecular patterns (PAMPs). LRR domain-containing proteins belong to the LRR family, which is a large group of proteins with more than 6000 genes in the database. They are involved in very diverse physiological functions, mainly by interacting with other proteins. In a previous study, the LvLRRm, a transmembrane protein containing only LRR domain, was identified in the white leg shrimp, Litopenaeus vannamei. Its versatile role in performing multiple immunomodulation activities has been reported. However, there is still a lack of research on its efficient function at the protein level. To investigate its interactions with other proteins, we applied a convenient method called the ‘Hybrid LRR technique’ to produce a recombinant LvLRRm. The LvLRRm and hagfish’s variable lymphocyte receptors (VLRs) fragments were fused to the conserved LxxLxLxxN motif while retaining the β-strand. In addition, we established interactions between hybrid proteins and the flagellin of Salmonella typhimurium by performing surface plasmon resonance (SPR) analysis. The results of the SPR analysis demonstrated notable affinity for both LvLRRm and hybrid proteins towards Salmonella flagellin. The designed LvLRRm hybrid proteins bring insight for universal applications without losing protein functions.
1. Introduction
The white leg shrimp, Litopenaeus vannamei (Boone, 1931), is a popular species worldwide in commercial farming. However, it is vulnerable to infectious threats such as bacterial and viral diseases [1,2,3]. Crustaceans lack a true adaptive immune response. Thus, their defense mechanisms depend entirely on an innate immune system that is activated upon recognition of pathogen-associated molecular patterns (PAMPs) from various microorganisms [4,5]. The innate immune system plays an important role as the first line of defense, protecting hosts against infections caused by invading pathogens through mechanisms that can be activated rapidly upon recognition of a foreign threat [6]. Pattern recognition receptors (PRRs) are key molecules that can trigger humoral and cellular immune responses and then generate a series of removal processes of harmful molecules [7]. They can also interact with matching PAMPs, including nucleic acids, proteins, polysaccharides, lipids, and lipoproteins, to induce intracellular signaling pathways through pathogen recognition [8].
Leucine-rich repeats (LRRs) are a pivotal structure in conformational complementarity in several classes of PRRs based on protein–protein interactions. They consist of an LRR motif in the center, an N-terminal (LRRNT), and a C-terminal (LRRCT) to protect a hydrophobic inner core [9,10]. Horseshoe-shaped LRRs typically have 20–30 amino acids. They are unusually rich in hydrophobic amino acid leucine and highly conserved modules that form domains from two or more tandem LRRs to generate solenoidal curves suitable for protein–protein interactions [9,11,12].
The LvLRRm is a newly reported PRR from Litopenaeus vannamei. It is an immune-related membrane protein that contains 12 LRR motifs, with a hallmark sequence of 11 residues, LxxLxLxxNxL (x is any amino acid), and is involved in the antibacterial defense process by regulating antibacterial innate immune signaling pathways [7]. Most immune responses in crustaceans are mediated by cellular receptors, and in the case of white leg shrimp, the LvLRRm gene was found to be involved in mitigating the mortality of white leg shrimp during bacterial infections [7,13].
Similar to other membrane proteins, LvLRRm has difficulties producing target proteins in soluble form, which can be a barrier to further research, such as protein overexpression and in vitro molecular experiments. These problems can sometimes be bypassed by removing nonessential domains because many proteins have multiple domains linked by flexible loops. In addition, the biological function of a protein is often dependent upon domains much smaller than the full-length protein [14,15,16]. Like the LRR repeat protein, α-solenoid proteins have been widely investigated. Examples include ankyrin repeat proteins, HEAT repeat proteins, armadillo repeat proteins (ArmRP), and tetratricopeptide repeat proteins (TPR) [17]. They are attractive targets for protein engineering and design for the protein’s stability and the specific recognition of other proteins. In particular, the ‘Hybrid LRR technique’, initially developed for the entire structure of toll-like receptor 4 (TLR4), can be applied to LRR proteins to enhance the expression and stability of the original protein [18]. This technique can be used to fuse truncated fragments of LvLRRm with LRRCT modules from other LRRs with variable lymphocyte receptor B (VLRB) as a hybrid partner [19,20]. This fusion strategy has been successfully used to design eight hybrid proteins, three of which are soluble.
Through the LvLRRm, TLR-like proteins in crustacean species are predicted to play an important role in the defense against pathogens. However, the biochemical basis of ligand binding and the activation of the LvLRRm protein remains unclear. Here, we hypothesized that LvLRRm hybrid proteins would show biochemical functions similar to LvLRRm and tested their affinity with flagellin, a protein that composes the bacterial flagella.
2. Materials and Methods
2.1. Cell Culture, Recombinant Protein Expression, and Purification
Suspension Sf9 cells, a clonal isolate of Spodoptera frugiperda Sf21 cells (IPLB-Sf21-AE), were cultured in a 28 °C shaking incubator in Sf900™ II SFM (Gibco, Grand Island, NY, USA) supplemented with 5% fetal bovine serum (Gibco, Grand Island, NY, USA) and 1% antibiotic–antimycotic solution (GenDepot, Katy, TX, USA). Suspension High Five cells, an insect cell line that originated from ovarian cells of the cabbage looper (Trichoplusia ni), were cultured in a 28 °C shaking incubator in Express Five™ SFM (Gibco, Grand Island, NY, USA) supplemented with 9% L-glutamine (Gibco, Grand Island, NY, USA) and 1% antibiotic–antimycotic solution (GenDepot, Katy, TX, USA).
Hybrid proteins were cloned into BamHI and NotI sites of the pAcGP67 vector (BD Biosciences, Franklin Lakes, NJ, USA) by overlap PCR using primers listed in Table 1. The Fc domain of human IgG1 and the thrombin cleavage site were designed between NotI and the BglII sites of pAcGP67. The resulting C-terminal, Fc-tagged LvLRRm, and LvLRRm hybrid proteins were expressed in High Five insect cells using the baculovirus expression system with ProGreen™ (AB Vector, San Diego, CA, USA). All proteins were purified by protein A Sepharose (Amicogen, Seongnam, Korea) affinity chromatography in a buffer containing 20 mM Tris pH 8.0 and 200 mM NaCl. After cleavage by thrombin to remove the Fc tag, eluted proteins were further purified by Superdex® 200 10/300 GL (GE Healthcare, Louisville, KY, USA) size-exclusion chromatography (GE AKTA Prime Plus FPLC System). All proteins were concentrated for the assay and stored at −80 °C after being aliquoted.

Table 1.
Primer sets used in experiments.
The Salmonella typhimurium flagellin (stflagellin)-inserted vector was received from the Institute of Membrane Proteins (IMP) in Pohang, South Korea. Then, stflagellin residues F54–R451 were cloned into NcoI and XhoI sites of the pET28a vector containing a C-terminal hexa-His-tag and transformed into E. coli BL21 (DE3). Cells were grown at 30 °C in Luria–Bertani (LB) and expressed in a soluble fraction through the optimal induction condition (optical density (OD) 600 = 0.6~0.8) with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). The culture was allowed to grow for a period of 3~4 h at 37 °C.
Harvested cells were lysed in a high-pressure homogenizer (Genizer, Irvine, CA, USA) in a buffer containing 20 mM Tris pH 8.0, 200 mM NaCl, and 5 mM imidazole. The lysate from soluble proteins was centrifuged for 30 min at 17,000 rpm. The clarified lysate was loaded onto a Ni-NTA (Bioworks, Uppsala, Sweden) affinity chromatography resin, which was followed by step elution with 20 mM Tris pH 8.0, 200 mM NaCl, and 250 mM imidazole. It was then further purified by ion exchange and size-exclusion chromatography. The eluted stflagellin was concentrated without removing the hexa-his-tag and utilized in the assay.
2.2. SPR (Surface Plasmon Resonance) Experiments by Single-Cycle Kinetics
Binding kinetics of LvLRRm and LvLRRm hybrid proteins for stflagellin were determined with an iMSPR-Pro/A (icluebio, Seongnam, Korea) equipped with an NTA-Au chip (NTA functionalized monolayer surface, icluebio, Korea). Experiments were performed using the single-cycle kinetics (SCK) method. The 1xHBST (HEPES 100 mM, NaCl 150 mM, Tween 20 0.005%, and pH 7.4) and 1xHBST containing 5 mM NiCl2 were used as the reaction buffer and the running buffer, respectively. The stflagellin was immobilized on the surface of an NTA-Au chip by amine coupling using 2 ug/mL 1xHBST at a flow rate of 10 µL/mL. LvLRRm and its hybrid proteins, the analyte, were applied at five different concentrations (0.5, 1, 2, 4, and 8 µM) for 180 s at a flow rate of 30 µL/min following the 1xHBST running buffer. Analytes bound to sensor chips were dissociated by washing with 1xHBST running buffer for 180 s. Association () and dissociation () constants were both measured over 180 s intervals. The equilibrium dissociation constant () was calculated as the ratio of the off-rate to the on-rate (/). Kinetic parameter sensorgrams were determined with the global fitting function of the Biacore Insight Evaluation Software using a 1:1 binding model.
3. Results
3.1. Design of LvLRRm Hybrid Proteins, LhV Proteins
The open reading frame (ORF) sequence of LvLRRm encodes 576 amino acid residues (accession number: MT180094). It contains a signal peptide (1M–95A), an LRR domain (L96–C496), and a transmembrane region (V511–L527). The LvLRRm of the LRR family proteins belongs to the ‘typical’ subfamily, with 22~24 amino acids in the LRR module. It is therefore predicted to have a characteristic horseshoe-shaped structure within concave and convex: LRRNT (L96–A127), 11 LRR modules (L128–D151, L152–H175, L176–K199, L200–Q223, L224–N247, L248–D271, L272–T295, L296–S319, L321–K344, L345–R368, and L369–M392)—linker (N393–E418)—12th LRR module (L419–I442), and LRRCT (I452–C496). The parallel β-sheet of the concave region contains the consensus sequence of “LxxLxLxxN”, a region suitable for protein engineering.
To increase the stability of the protein, we applied the known ‘Hybrid LRR Technique’ by removing the linker and replacing it with LRRCT from another protein. The technique generated a hybrid form of LvLRRm and a partner LRR family protein. Based on previous research, we chose a structurally compatible partner, such as the variable lymphocyte receptor B.61 (VLRB), which could mediate adaptive immune responses in jawless fish [21,22]. We designed the hybrid form of VLRB-fused LvLRRm and named these proteins ‘LhV no.’ (LvLRRm—hybrid VLRB, no. follows the designed order) (Table 2). Sheet and loop-based hybrids were designated as LhV1–4 and LhV5–8, respectively (Figure 1). We fused truncated fragments of LvLRRm with the LRRCT module from other LRRs in VLRB and yielded three soluble proteins of eight designed hybrid proteins. On size-exclusion chromatography, LhV6–8 were eluted as suitable fractions and showed band intensities in SDS-PAGE analysis (Figure 2A,B).

Table 2.
Sequential designs of LvLRRm hybrid proteins (LhVs).
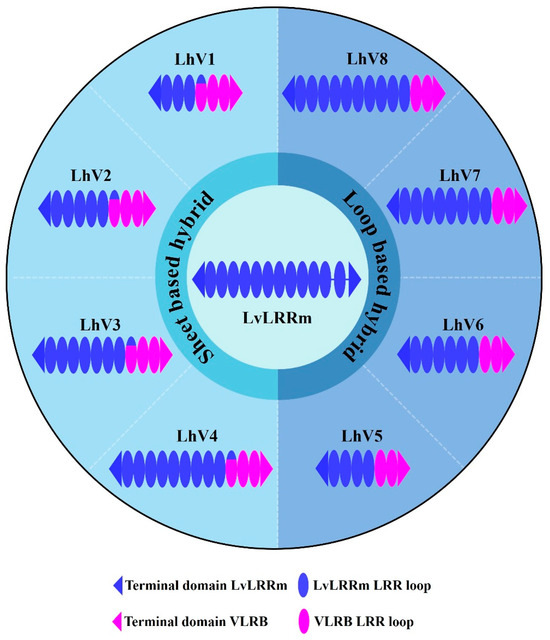
Figure 1.
Overall graphical schemes of LvLRRm and its hybrid proteins. An ellipse represents a loop in the LRR domain, and void gaps in LvLRRm reveal the linker domain. Navy and magenta represent LvLRRm and VLRB.61, respectively.

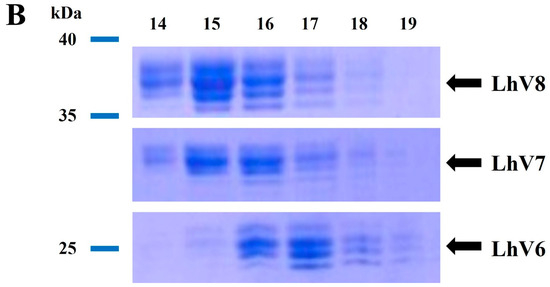
Figure 2.
(A) Gel-filtration chromatography of the LvLRRm hybrid protein series. LvLRRm hybrid protein series were analyzed by Superdex 200 gel-filtration chromatography and calibrated with molecular weight standards. Apparent molecular weights of complexes calculated from elution volumes are shown. (B) SDS-PAGE analysis of the LvLRRm hybrid protein series. Purified constructors are arranged by fraction number in weight order.
3.2. Binding Kinetics Validation of LvLRRm and LhV6–8 to Stflagellin
We confirmed the binding of LhV6–8 to stflagellin by SPR analysis using recombinant stflagellin-immobilized NTA-Au chips. Analytes were then sequentially injected onto the target ligand in increasing concentrations (0.5 uM, 1 uM, 2 uM, 4 uM, and 8 uM) and dissociated in each trial. As expected, all hybrid proteins showed appropriate binding affinity between stflagellin and LhVs, indicating the potential for interaction as a binding partner.
LhV8–stflagellin (kon = 1.969 × 102 M−1 s−1; koff = 3.37 × 10−4 s−1; and KD = 1.69 × 10−6 M) and LhV7–stflagellin (kon = 1.48 × 103 M−1 s−1; koff = 6.63 × 10−4 s−1; and KD = 4.47 × 10−7 M) exhibited affinities high enough to recognize their partners between them, respectively (Figure 3A,B). LhV6–stflagellin (kon = 4.76 × 102 M−1 s−1; koff = 2.87 × 10−5 s−1; and KD = 5.99 × 10−8 M) showed a slightly lower dissociation rate with a robust binding affinity (Figure 3C). We also determined the binding kinetics between LvLRRm and stflagellin, although we used a small, purified amount, due to the unstable LvLRRm protein. An SPR sensorgram revealed that LvLRRm could bind to stflagellin with nanomolar affinity (kon = 2.66 × 10 M−1 s−1; koff = 4.36 × 10−4 s−1; and KD = 1.63 × 10−5 M) (Figure 3D).
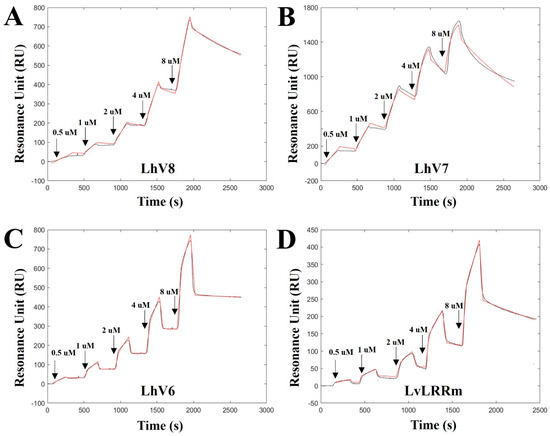
Figure 3.
The binding kinetics toward stflagellin determined by single-cycle kinetics SPR analysis for (A) LhV8, (B) LhV7, (C) LhV6, and (D) LvLRRm. The equilibrium dissociation constant () was calculated as the ratio of the off-rate to the on-rate (/). Kinetic parameters were determined with the global fitting function of the Biacore Insight Evaluation Software using a 1:1 binding model.
4. Discussion
The adaptive immune system evolved about 500 million years ago, and it is found mostly in vertebrates [22,23]. Furthermore, it is widely accepted that the innate immune response of invertebrates is sensitive and potent. However, some components and mechanisms are not pathogen specific [24]. Knockout of the LvLRRm gene has been shown to result in less expression of the LvLRRm protein in the cell membrane, leading to decreased survival for pathogen exposure and contributing significantly to the innate immune response [7].
For most proteins, it is not easy to perform various experiments because of their partially exposed hydrophobic surfaces, non-functional loops, and lack of stability [25,26]. In particular, LvLRRm was not stable for soluble expression due to the linker of unknown function. To advance our research on LvLRRm, we needed a technique to induce the overexpression of LvLRRm’s proteins while preserving the site responsible for recognizing external antigens in them.
We obtained LhV proteins based on VLRs using LRR-module engineering. Three of eight characterized LvLRRm hybrid proteins were experimentally stable, folded, and successfully expressed at high levels of LhV 6–8 in an insect baculovirus expression system. Expressed LhV6–8 contained bindable loop arrangements and shape complementarity to patches of stflagellin in predicted structures performed by the I-TASSER tool (https://zhanggroup.org/I-TASSER/ accessed on 7 August 2023) (Figure 4). In the predicted structure, LhV6–8 exhibited a compact shape with repeated LRR loops, and the original C-terminal end of LvLRRm was successfully replaced by the hydrophobic inner core of VLR. Results showed that these developed hybrid proteins could be widely used to generate target-specific molecular binders for applications in biotechnology and crustacean immunology by a rational design.
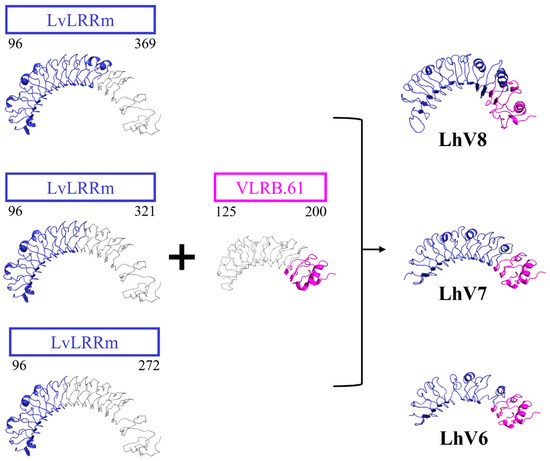
Figure 4.
The predicted structures of the LvLRRm hybrid proteins, LhVs. All the hybrid proteins were predicted to be horseshoe-shaped structures. LvLRRm (navy) formed different hybrids based on the number of loops, while VLRB (magenta) was joined together in the C-terminal direction. The non-colored region of each structure reveals truncated LRR loops.
Previous studies have shown that a family of type I transmembrane glycoproteins containing LRRs in their extracellular domains can recognize conserved patterns of pathogen-derived molecules, rather than the specificity of them [27,28,29]. PAMPs, well-known lipopolysaccharide (LPS), peptidoglycan (PGN), glucan (GLU), and polycytidine-polycitric acid (poly IC) are known to be involved in innate immune responses by mediating protein–ligand interactions [30]. Although PRRs can recognize a variety of molecules as ligands, flagellin was chosen in this study because its recombinant proteins could be easily engineered and produced to be suitable for SPR experiments.
Flagellin, a bacterial protein that forms the flagellar filament crucial for bacterial motility, serves as a pathogen invasion signal when bacteria invade the host. It can trigger an innate immune response through recognition by toll-like receptor 5 (TLR5) [31,32,33]. TLR5 is activated by a highly conserved sequence in flagellin that is essential for its function. It is sequestered in the protofilament of assembled flagella [34]. The interaction between flagellin and TLR5 appears to be direct. The binding site has been mapped to a single LRR in the receptor ectodomain [35]. In TLR5, two surfaces (loop1–4 and loop 7–9) could form the binding between hot spot residues (R89, L93, and E114) in D0–D1 domains of Bacillus subtilis flagellin through van der Waals interactions and a hydrogen bond [36]. Based on the SPR analysis results and the similar arrangement of LRR loop patterns, it cannot be ruled out that LvLRRm may recognize flagellin as a PRR like TLR5. Furthermore, judging from the remaining central domain LRR loops of the LhVs without truncation, there is a possibility that the hotspot for flagellin binding may be located in that region.
The hybrid LRR technique was first used to elucidate the structure of TLR4 by replacing the N- and C-terminal regions with VLRs, and it played a role in unraveling the structure of eritoran, an analog of LPS that binds to the TLR4–MD2 complex [15,18]. Also, it has been reported that the human TLR2 with the C-terminal end replaced by VLR had no issue recognizing tri-acylated lipopeptide [16]. Such biophysical recognition responses suggest that, as long as the antigen recognition site is preserved, other domains can be replaced. When loops are hybridized on a block-by-block basis while maintaining backbone-shape complementarity, it might be possible to create another repeat protein curvature that targets macromolecules [37]. In the present study, LhV6–8 were designed based on the number of central LRR loops, and the affinity for flagellin was confirmed in all hybrid proteins. Such diversity in LRRs suggests the potential for shape transformations in LvLRRm.
Among the processes used in our experiments, the single-cycle kinetics (SCK) method has limitations when it comes to observing molecule–molecule interactions. The SCK method involves a series of successive analyte injections at progressively higher concentrations onto the ligand surface, with no regeneration occurring at each concentration point [38]. Without regeneration, there is a possibility that while analytes continue to adhere to ligands during the association, the titer of ligand may decrease, compared to the previous association step. For this reason, the ligand surface may not be stable in each cycle. In another case of a problematic issue, the analyte may form such a tight complex with the ligand surface that no regeneration condition will disrupt the interaction without denaturing the ligand. This is commonly observed in high-affinity small-molecule/protein interactions [39]. To clarify these issues, structural studies of both LvLRRm and LvLRRm hybrid proteins regarding potential candidates for binding are necessary.
5. Conclusions
We developed hybrid proteins, the LhV6–8 series, by fusing LvLRRm with VLRB. LhV6–8 were well folded and stable in an aqueous solution. Furthermore, in the predicted structural model by I-TASER tools, VLRB successfully replaced the extended linker region between loops 11 and 12 of the original one, maintaining its compact loop arrangement. Given the affinity of the LvLRRm protein to flagellin observed in SPR analysis, we cannot rule out the possibility of an interaction between them. Nonetheless, given the limitations of the traditional SCK method and the lack of clear data supporting a 1:1 interaction between them, further investigation is still warranted. Hybrid LRR technology can be useful for structural studies, the characterization of LvLRRm, and the expression of recombinant proteins containing LRR modules.
Author Contributions
Conceptualization, T.K. and B.P.; methodology, J.C., W.L. and B.P.; validation, B.P.; formal analysis, W.L. and B.P.; investigation, T.K. and B.P.; resources, B.P.; data curation, J.C., T.K. and B.P.; writing—original draft, J.C. and B.P.; writing—review and editing, T.K. and B.P.; visualization, J.C.; supervision, T.K. and B.P.; project administration, T.K. and B.P.; funding acquisition, T.K. and B.P. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Korean Ministry of Oceans and Fisheries (No.20170411) and by the Technology Innovation Program (20019707, Cryo-EM-based vaccine discovery and optimization technology) funded by the Ministry of Trade, Industry and Energy (MOTIE, Korea), and KEIT. This work was also partially supported by the National Research Foundation of Korea (NRF) grants (No. 2015R1C1A1A01054639 and NRF2020R1A2C1005194).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained in the article.
Acknowledgments
We thank Jie-Oh Lee in POSTECH for providing us the Salmonella typhimurium flagellin gene.
Conflicts of Interest
The authors declare no conflict of interest.
References
- López-León, P.; Luna-González, A.; Escamilla-Montes, R.; del Carmen Flores-Miranda, M.; Fierro-Coronado, J.A.; Álvarez-Ruiz, P.; Diarte-Plata, G. Isolation and characterization of infectious Vibrio parahaemolyticus, the causative agent of AHPND, from the whiteleg shrimp (Litopenaeus vannamei). Lat. Am. J. Aquat. Res. 2016, 44, 470–479. [Google Scholar] [CrossRef]
- Lomelí-Ortega, C.O.; Martínez-Díaz, S.F. Phage therapy against Vibrio parahaemolyticus infection in the whiteleg shrimp (Litopenaeus vannamei) larvae. Aquaculture 2014, 434, 208–211. [Google Scholar] [CrossRef]
- Wang, J.; Huang, Y.; Xu, K.; Zhang, X.; Sun, H.; Fan, L.; Yan, M. White spot syndrome virus (WSSV) infection impacts intestinal microbiota composition and function in Litopenaeus vannamei. Fish Shellfish Immunol. 2019, 84, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, L.; Alpuche, J.; Maldonado, G.; Agundis, C.; Pereyra-Morales, A.; Zenteno, E. Review: Immunity mechanisms in crustaceans. Innate Immun. 2009, 15, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Soderhall, I. Crustacean hematopoiesis and the astakine cytokines. Blood 2011, 117, 6417–6424. [Google Scholar] [CrossRef] [PubMed]
- Tran, N.T.; Kong, T.; Zhang, M.; Li, S. Pattern recognition receptors and their roles on the innate immune system of mud crab (Scylla paramamosain). Dev. Comp. Immunol. 2020, 102, 103469. [Google Scholar] [CrossRef]
- Zhang, H.; Li, S.; Wang, F.; Xiang, J.; Li, F. Identification and functional study of an LRR domain containing membrane protein in Litopenaeus vannamei. Dev. Comp. Immunol. 2020, 109, 103713. [Google Scholar] [CrossRef] [PubMed]
- Habib, Y.J.; Zhang, Z. The involvement of crustaceans toll-like receptors in pathogen recognition. Fish Shellfish Immunol. 2020, 102, 169–176. [Google Scholar] [CrossRef]
- Park, H.; Huxley-Jones, J.; Boot-Handford, R.P.; Bishop, P.N.; Attwood, T.K.; Bella, J. LRRCE: A leucine-rich repeat cysteine capping motif unique to the chordate lineage. BMC Genom. 2008, 9, 599. [Google Scholar] [CrossRef]
- Wen, D.; Wildes, C.P.; Silvian, L.; Walus, L.; Mi, S.; Lee, D.H.; Meier, W.; Pepinsky, R.B. Disulfide structure of the leucine-rich repeat C-terminal cap and C-terminal stalk region of Nogo-66 receptor. Biochemistry 2005, 44, 16491–16501. [Google Scholar] [CrossRef]
- Kajava, A.V.; Kobe, B. Assessment of the ability to model proteins with leucine-rich repeats in light of the latest structural information. Protein Sci. 2002, 11, 1082–1090. [Google Scholar] [CrossRef]
- Wang, X.W.; Gao, J.; Xu, Y.H.; Xu, J.D.; Fan, Z.X.; Zhao, X.F.; Wang, J.X. Novel Pattern Recognition Receptor Protects Shrimp by Preventing Bacterial Colonization and Promoting Phagocytosis. J. Immunol. 2017, 198, 3045–3057. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.C.; Xu, J.Y.; Liu, H.P. Cellular entry of white spot syndrome virus and antiviral immunity mediated by cellular receptors in crustaceans. Fish Shellfish Immunol. 2019, 93, 580–588. [Google Scholar] [CrossRef]
- Jin, M.S.; Lee, J.O. Application of hybrid LRR technique to protein crystallization. BMB Rep. 2008, 41, 353–357. [Google Scholar] [CrossRef]
- Park, B.S.; Song, D.H.; Kim, H.M.; Choi, B.S.; Lee, H.; Lee, J.O. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 2009, 458, 1191–1195. [Google Scholar] [CrossRef]
- Jin, M.S.; Kim, S.E.; Heo, J.Y.; Lee, M.E.; Kim, H.M.; Paik, S.G.; Lee, H.; Lee, J.O. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell 2007, 130, 1071–1082. [Google Scholar] [CrossRef]
- Speltz, E.B.; Brown, R.S.H.; Hajare, H.S.; Schlieker, C.; Regan, L. A designed repeat protein as an affinity capture reagent. Biochem. Soc. Trans. 2015, 43, 874–880. [Google Scholar] [CrossRef]
- Kim, H.M.; Park, B.S.; Kim, J.I.; Kim, S.E.; Lee, J.; Oh, S.C.; Enkhbayar, P.; Matsushima, N.; Lee, H.; Yoo, O.J.; et al. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell 2007, 130, 906–917. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.M.; Oh, S.C.; Lim, K.J.; Kasamatsu, J.; Heo, J.Y.; Park, B.S.; Lee, H.; Yoo, O.J.; Kasahara, M.; Lee, J.O. Structural diversity of the hagfish variable lymphocyte receptors. J. Biol. Chem. 2007, 282, 6726–6732. [Google Scholar] [CrossRef]
- Pancer, Z.; Saha, N.R.; Kasamatsu, J.; Suzuki, T.; Amemiya, C.T.; Kasahara, M.; Cooper, M.D. Variable lymphocyte receptors in hagfish. Proc. Natl. Acad. Sci. USA 2005, 102, 9224–9229. [Google Scholar] [CrossRef]
- Guo, P.; Gartland, L.; Li, J.X.; Hirano, M.; Alder, M.; Herrin, B.; Sides, J.; Kasahara, M.; Cooper, M. Characterization of adaptive immune receptors in hagfish. J. Immunol. 2011, 186, 170.13. [Google Scholar] [CrossRef]
- Boehm, T.; Hirano, M.; Holland, S.J.; Das, S.; Schorpp, M.; Cooper, M.D. Evolution of Alternative Adaptive Immune Systems in Vertebrates. Annu. Rev. Immunol. 2018, 36, 19–42. [Google Scholar] [CrossRef]
- Usharauli, D. Chronic infection and the origin of adaptive immune system. Med. Hypotheses 2010, 75, 241–243. [Google Scholar] [CrossRef] [PubMed]
- Hauton, C. The scope of the crustacean immune system for disease control. J. Invertebr. Pathol. 2012, 110, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, E.P.; Beis, K.; Cameron, A.D.; Iwata, S. Overcoming the challenges of membrane protein crystallography. Curr. Opin. Struct. Biol. 2008, 18, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Errasti-Murugarren, E.; Bartoccioni, P.; Palacin, M. Membrane Protein Stabilization Strategies for Structural and Functional Studies. Membranes 2021, 11, 155. [Google Scholar] [CrossRef] [PubMed]
- Nurnberger, T.; Brunner, F.; Kemmerling, B.; Piater, L. Innate immunity in plants and animals: Striking similarities and obvious differences. Immunol. Rev. 2004, 198, 249–266. [Google Scholar] [CrossRef]
- Coscia, M.R.; Giacomelli, S.; Oreste, U. Toll-like receptors: An overview from invertebrates to vertebrates. ISJ Invertebr. Surviv. J. 2011, 8, 210–226. [Google Scholar]
- Bella, J.; Hindle, K.L.; McEwan, P.A.; Lovell, S.C. The leucine-rich repeat structure. Cell. Mol. Life Sci. 2008, 65, 2307–2333. [Google Scholar] [CrossRef]
- Shen, X.; Wang, Y.; Hu, J.; Bao, Z.; Wang, M. Molecular characterization of an LRR-only protein gene in Pacific white shrimp Litopenaeus vannamei: Sequence feature, expression pattern, and protein activity. Fish Shellfish Immunol. 2022, 129, 199–206. [Google Scholar] [CrossRef]
- Yoon, S.I.; Kurnasov, O.; Natarajan, V.; Hong, M.; Gudkov, A.V.; Osterman, A.L.; Wilson, I.A. Structural basis of TLR5-flagellin recognition and signaling. Science 2012, 335, 859–864. [Google Scholar] [CrossRef]
- Hayashi, F.; Smith, K.D.; Ozinsky, A.; Hawn, T.R.; Yi, E.C.; Goodlett, D.R.; Eng, J.K.; Akira, S.; Underhill, D.M.; Aderem, A. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 2001, 410, 1099–1103. [Google Scholar] [CrossRef]
- Jacchieri, S.G.; Torquato, R.; Brentani, R.R. Structural study of binding of flagellin by Toll-like receptor 5. J. Bacteriol. 2003, 185, 4243–4247. [Google Scholar] [CrossRef]
- Buchanan, S.G.; Gay, N.J. Structural and functional diversity in the leucine-rich repeat family of proteins. Prog. Biophys. Mol. Biol. 1996, 65, 1–44. [Google Scholar] [CrossRef] [PubMed]
- Mizel, S.B.; West, A.P.; Hantgan, R.R. Identification of a sequence in human toll-like receptor 5 required for the binding of Gram-negative flagellin. J. Biol. Chem. 2003, 278, 23624–23629. [Google Scholar] [CrossRef] [PubMed]
- Song, W.S.; Jeon, Y.J.; Namgung, B.; Hong, M.; Yoon, S.I. A conserved TLR5 binding and activation hot spot on flagellin. Sci. Rep. 2017, 7, 40878. [Google Scholar] [CrossRef] [PubMed]
- Park, K.; Shen, B.W.; Parmeggiani, F.; Huang, P.S.; Stoddard, B.L.; Baker, D. Control of repeat-protein curvature by computational protein design. Nat. Struct. Mol. Biol. 2015, 22, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Bi, C.; Li, Z.; Podariu, M.; Hage, D.S. Analytical methods for kinetic studies of biological interactions: A review. J. Pharm. Biomed. Anal. 2015, 113, 163–180. [Google Scholar] [CrossRef]
- Karlsson, R.; Katsamba, P.S.; Nordin, H.; Pol, E.; Myszka, D.G. Analyzing a kinetic titration series using affinity biosensors. Anal. Biochem. 2006, 349, 136–147. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).