Reproductive Characteristics of the Flat Oyster Ostrea denselamellosa (Bivalvia, Ostreidae) Found on the Southern Coast of South Korea
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Collection
2.2. DNA Extraction and DNA Sequencing
2.3. Sequence Alignment and Phylogenetic Analysis
2.4. Histological Analysis
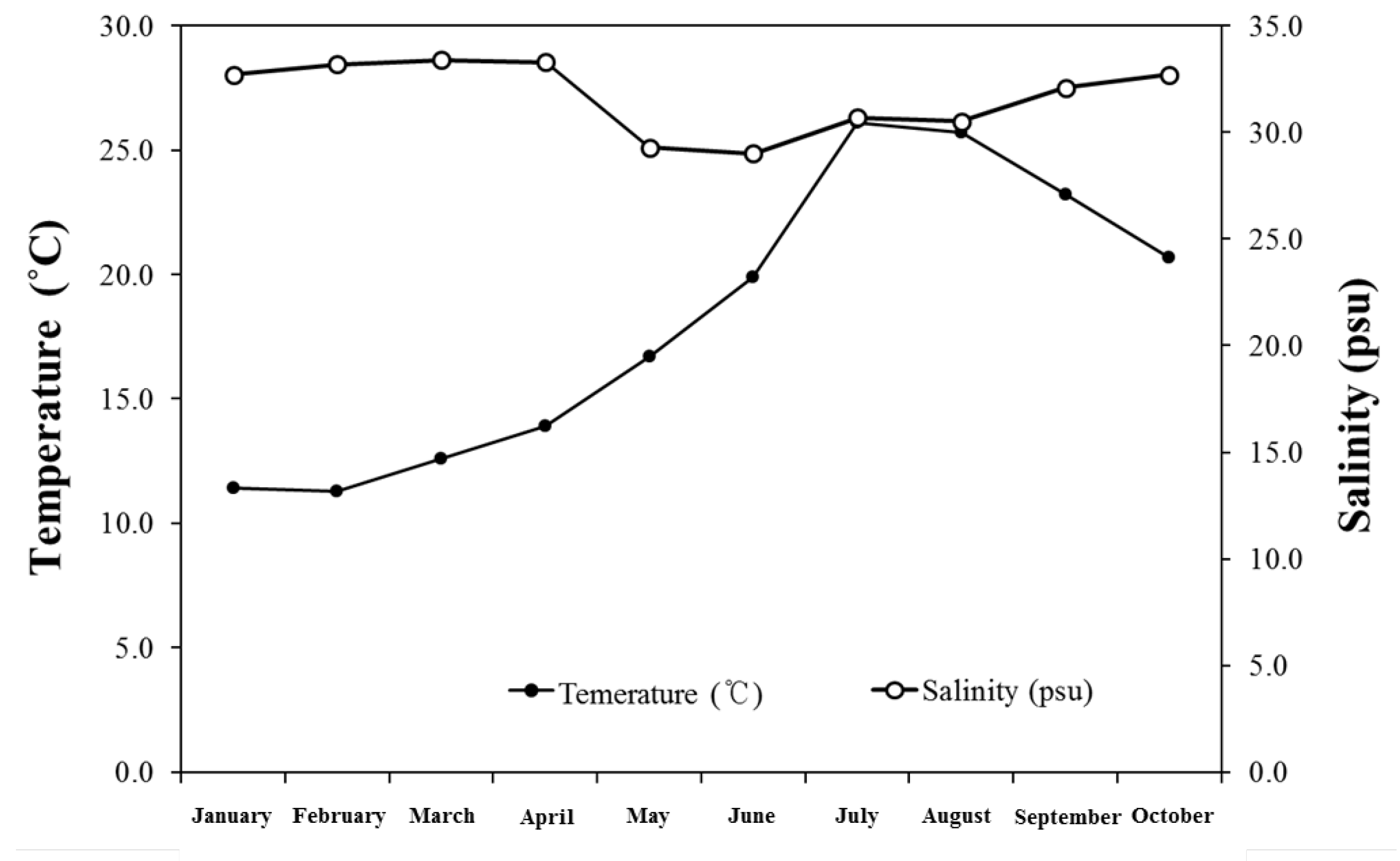
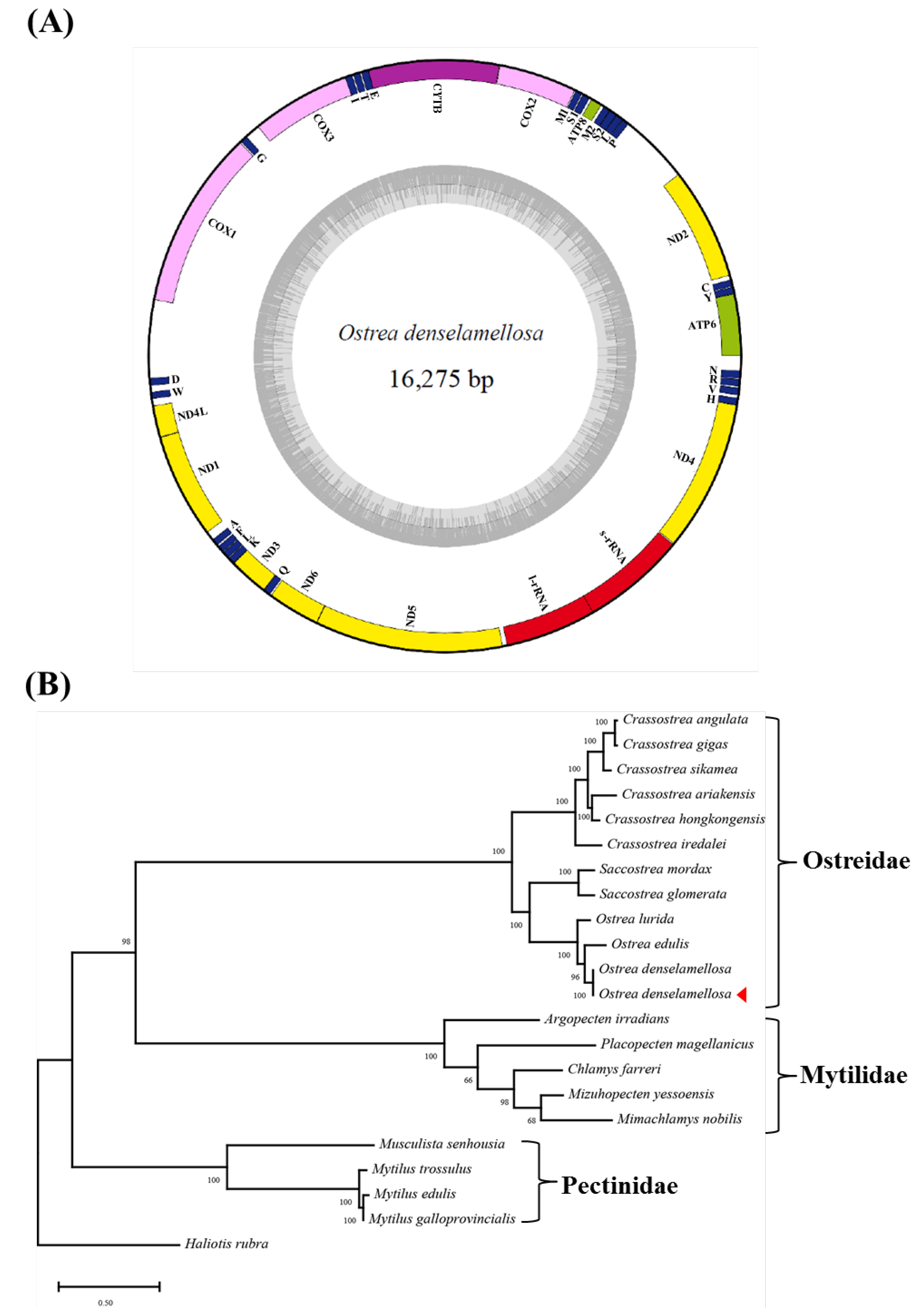
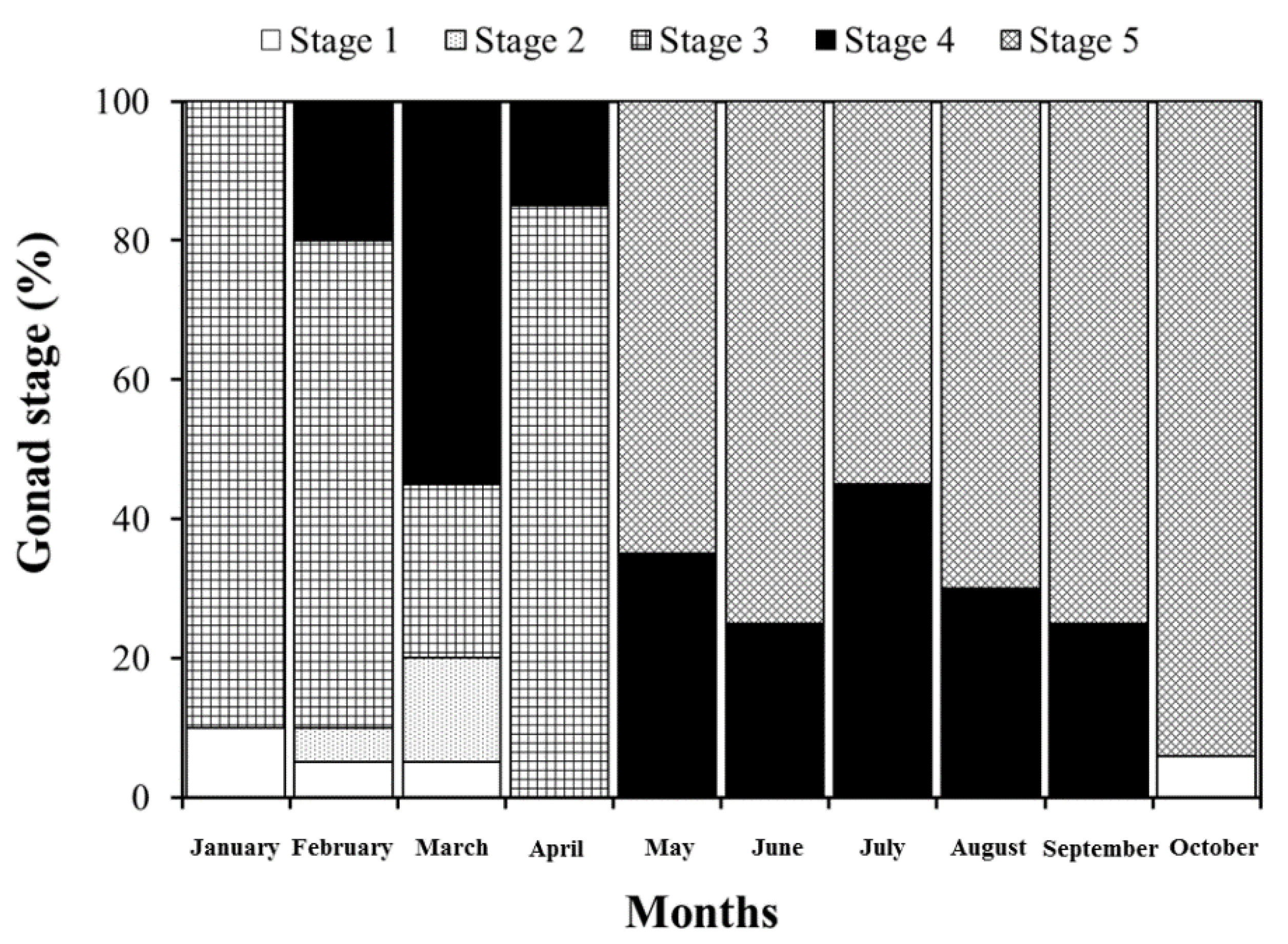
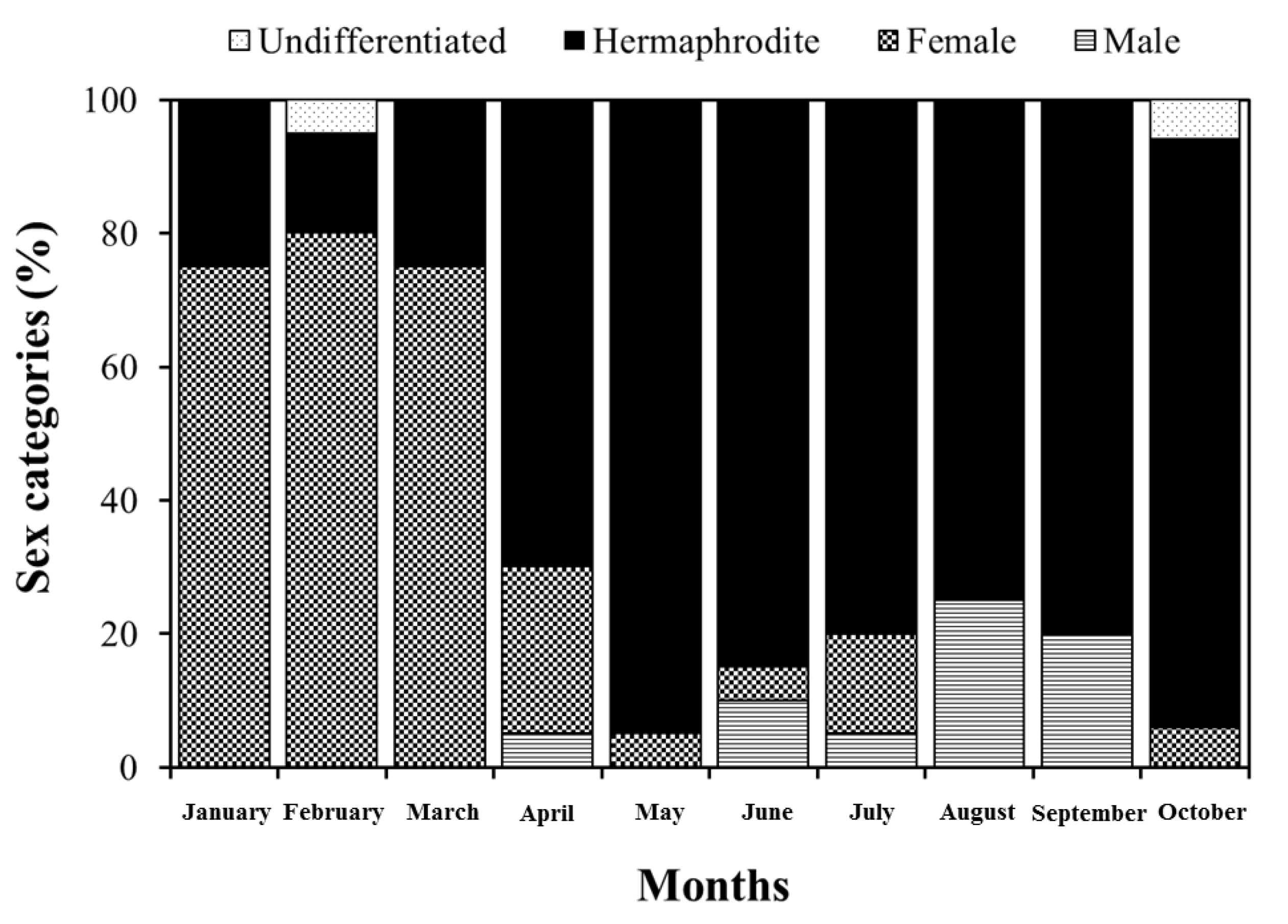
3. Results and Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smaal, A.C.; Ferreira, J.G.; Grant, J.; Petersen, J.K.; Strand, Ø. Goods and Services of Marine Bivalves; Springer Open: Gewerbestrasse, Switzerland, 2019; p. 591. [Google Scholar]
- FAO. Fishery Statistical Collections. Global Production, Global Capture Production and Global Aquaculture Production; FAO Fisheries and Aquaculture Department: Rome, Italy, 2020; Available online: http://www.fao.org/fishery/statistics/global-aquaculture-production/en (accessed on 24 March 2020).
- Quayle, D.B. Pacific oyster culture in British Columbia. Can. J. Fish. Aquat. 1988, 218, 192. [Google Scholar]
- Beck, M.W.; Brumbaugh, R.D.; Airoldi, L.; Carranza, A.; Coen, L.D.; Crawford, C.; Defeo, O.; Edgar, G.J.; Hancock, B.; Kay, M.C.; et al. Oyster reefs at risk and recommendations for conservation, restoration, and management. Bioscience 2011, 61, 107–116. [Google Scholar] [CrossRef]
- Dumbauld, B.R.; Kauffman, B.E.; Trimble, A.C.; Ruesink, J.L. The Willipa Bay oyster reserves in Washington state: Fishery collapse, creating a sustainable replacement, and the potential for habitat conservation and restoration. J. Shellfish Res. 2011, 30, 71–83. [Google Scholar] [CrossRef]
- Nielsen, P.; Petersen, J.K. Flat oyster fishery management during a time with fluctuating population size. Aquat. Living Resour. 2019, 32, 22. [Google Scholar] [CrossRef]
- Min, D.-K.; Lee, J.-S.; Koh, D.-B.; Je, J.-G. Mollusks in Korea; Min Molluscan Research Institute: Seoul, Korea, 2004; p. 566. [Google Scholar]
- Noseworthy, R.G.; Lee, H.-J.; Choi, S.-D.; Choi, K.-S. Unique substrate preference of Ostrea denselamellosa Lischke, 1869 (Mollusca: Ostreidae) at Haechang Bay, on the south coast of Korea. Korean J. Malacol. 2016, 32, 31–36. [Google Scholar] [CrossRef][Green Version]
- Okutani, T. Marine Mollusks in Japan; Tokai University Press: Tokyo, Japan, 2000; p. 1173. [Google Scholar]
- Lam, K.; Morton, B. The oysters of Hong Kong (Bivalvia: Ostreidae and Gryphaeidae). Raffles Bull. Zool. 2004, 52, 11–28. [Google Scholar]
- Chen, L.; Li, Q.; Wang, Q.Z.; Kong, L.F.; Zheng, X.D. Techniques of artificial breeding of the oyster Ostrea denselamellosa. Period. Ocean. Univ. China 2011, 41, 43–46. [Google Scholar]
- Insua, A.; Thiriot-Quievreux, C. The characterization of Ostrea denselamellosa (Mollusca, Bivalvia) chromosomes: Karyotype, constitutive heterochromatin and nucleolus organizer regions. Aquaculture 1991, 97, 317–325. [Google Scholar] [CrossRef]
- Curole, J.P.; Kocher, T.D. Mitogenomics: Digging deeper with complete mitochondrial genomes. Trends Ecol. Evol. 1999, 14, 394–398. [Google Scholar] [CrossRef]
- Lin, C.P.; Danforth, B.N. How do insect nuclear and mitochondrial gene substitution patterns differ? Insights from Bayesian analyses of combined datasets. Mol. Phylogenetics Evol. 2004, 30, 686–702. [Google Scholar] [CrossRef]
- Elson, J.L.; Lightowlers, R.N. Mitochondrial DNA clonality in the dock: Can surveillance swing the case? Trends Genet. 2006, 22, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Kong, L.; Li, Q. Complete mitochondrial genome of Ostrea denselamellosa (Bivalvia, Ostreidae). Mitochondrial DNA Part A 2016, 27, 711–712. [Google Scholar] [CrossRef] [PubMed]
- Pemberton, A.J.; Noble, L.R.; Bishop, J.D.D. Frequency dependence in matings with water-borne sperm. J. Evol. Biol. 2003, 16, 304–316. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.M.; Qin, J.G.; Li, X. Gametogenesis, sex ratio and energy metabolism in Ostrea angasi: Implications for the reproductive strategy of spermcasting marine bivalves. J. Molluscan Stud. 2018, 84, 38–45. [Google Scholar] [CrossRef]
- Lim, N.-L.; Lee, H.-M.; Jeung, H.D.; Noseworthy, R.G.; Jung, S.; Choi, K.-S. Early larval development and annual gametogenesis of the blooding oyster Ostrea circumpicta (Pilsbry, 1904) in the shallow subtidal benthic ecosystem in Jeju Island, off the south coast of Korea. Zool. Stud. 2019, 58, 29. [Google Scholar]
- Muranaka, M.S.; Lannan, J.E. Broodstock management of Crassostrea gigas: Environmental influences on broodstock conditioning. Aquaculture 1984, 39, 217–228. [Google Scholar] [CrossRef]
- Gosling, E. Bivalve Molluscs: Biology, Ecology and Culture. In Fishing News Books; John Wiley & Sons: Oxford, UK, 2003. [Google Scholar]
- Fabioux, C.; Huvet, A.; Le Souchu, P.; Le Pennec, M.; Pouvreau, S. Temperature and photoperiod drive Crassostrea gigas reproductive internal clock. Aquaculture 2005, 250, 458–470. [Google Scholar] [CrossRef]
- Bayne, B. Biology of Oysters; Academic Press: London, UK, 2017. [Google Scholar]
- Loosanoff, V.L.; Davis, H.C. Rearing of bivalve molluscs. Adv. Mar. Biol. 1963, 1, 1–136. [Google Scholar]
- Sastry, A.N. Temperature effects in reproduction of the Bay scallop, Aequipecten irradians Lamarck. Biol. Bull. 1966, 130, 118–134. [Google Scholar] [CrossRef]
- Xie, Q.; Burnell, G.M. A comparative study of the gametogenic cycles of the clams Tapes philippinarum (Adams and Reeve 1850) and Tapes decussatus (Linnaeus) on the south coast of Ireland. J. Shellfish Res. 1994, 13, 467–472. [Google Scholar]
- Meng, G.; Li, Y.; Yang, C.; Liu, S. MitoZ: A toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 2019, 47, e63. [Google Scholar] [CrossRef] [PubMed]
- del Moral, R.G. Laboratorio de Anatomía Patológica; McGraw-Hill-Interamericana de España: Madrid, Spain, 1993; pp. 405–425. [Google Scholar]
- Da Silva, P.M.; Fuentes, J.; Villalba, A. Differences in gametogenic cycle among strains of the European flat oyster Ostrea edulis and relationship between gametogenesis and bonamiosis. Aquaculture 2005, 287, 253–265. [Google Scholar] [CrossRef]
- Danic-Tchaleu, G.; Heurtebise, S.; Morga, B.; Lapègue, S. Complete mitochondrial DNA sequence of the European flat oyster Ostrea edulis confirms Ostreidae classification. BMC Res. Notes 2011, 4, 400. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Wu, X.; Li, L.; Yu, Z. Complete mitochondrial genome of the Olympia oyster Ostrea lurida (Bivalvia, Ostreidae). Mitochondrial DNA 2015, 26, 471–472. [Google Scholar] [CrossRef]
- MOF (Ministry of Oceans and Fisheries). The Development of Techniques to Increase Flat Oyster (Ostrea denselamellosa) Productivity for Its Industrial Application. In Research Report; Dong-Eui University: Busan, Korea, 1998; p. 215. [Google Scholar]
- Kang, C.-K.; Park, M.S.; Lee, P.-Y.; Choi, W.-J.; Lee, W.-C. Seasonal variation in condition, reproductive activity, and biochemical composition of the Pacific oyster Crassostrea gigas (Thunberg) in suspended culture in two coastal bays of Korea. J. Shellfish Res. 2000, 19, 771–778. [Google Scholar]
- Ngo, T.T.T.; Kang, S.-G.; Choi, K.-S. Seasonal changes in reproductive condition of the Pacific oysters, Crassostrea gigas (Thunburg) from suspended culture in Gosung Bay, Korea. Korea J. Environ. Biol. 2002, 20, 268–275. [Google Scholar]
- Park, M.S.; Lim, H.J.; Jo, Q.; Yoo, J.S.; Jeon, M.J. Assessment of reproductive health in the wild seed oysters, Crassostrea gigas, from two locations in Korea. J. Shellfish Res. 1999, 18, 445–450. [Google Scholar]
- Kim, B.-K.; Kang, D.-H.; Ko, D.-K.; Yang, H.-S.; Kim, D.-K.; Kang, C.-K.; Choi, K.-S. Annual reproductive cycle of the oyster, Saccostrea kegaki (Torigoe & Inaba 1981) on the southern coast of Jeju island, Korea. Invert. Reprod. Dev. 2010, 54, 19–26. [Google Scholar]
- Mazón-Suátergui, J.M.; Ruíz-García, M.C.; Chávez-Villalba, J.; Rodríguez-Jaramillo, C.; Saucedo, P.E. Analysis of growth and first reproduction of hatchery-reared juvenile Cortez oyster (Crassostrea corteziensis) in northwestern Mexico: Proposal of a minimal fishing size. Aquac. Res. 2011, 42, 1558–1568. [Google Scholar] [CrossRef]
- Kamphausen, S.L.; Jensen, A.; Hawkins, L. Unusually high proportion of males in a collapsing population of commercially fished oysters (Ostrea edulis) in the Solent, United Kingdom. J. Shellfish Res. 2011, 30, 217–222. [Google Scholar] [CrossRef]
- Acarli, S.; Lök, A.; Kirtik, A.; Acarli, D.; Serdar, S.; Kucukdermenci, A.; Yigitkurt, S.; Yildiz, H.; Saltan, A.N. Seasonal variation in reproductive activity and biochemical composition of flat oyster (Ostrea edulis) in the Homa Loagoon, Izmir Bay, Turkey. Sci. Mar. 2015, 79, 487–495. [Google Scholar] [CrossRef]
- Tamura, T. Shallow Sea Aquaculture; Series of Fisheries Science 2; Koseisha-koseikaku Inc.: Tokyo, Japan, 1960; p. 368. [Google Scholar]


| Tax On | Classification | Size (bp) | Accession No. | |
|---|---|---|---|---|
| Mollusca Bivalvia Pteriomorphia | Mytilus edulis | Mytiloida; Mytiloidea; Mytilidae | 16,740 | AY484747 |
| Mytilus galloprovincialis | Mytiloida; Mytiloidea; Mytilidae | 16,744 | AY497292 | |
| Mytilus trossulus | Mytiloida; Mytiloidea; Mytilidae | 18,652 | AY823625 | |
| Musculista senhousia | Mytiloida; Mytiloidea; Mytilidae | 20,612 | GU001954 | |
| Crassostrea angulata | Ostreoida; Ostreoidea; Ostreidae | 18,225 | EU672832 | |
| Crassostrea ariakensis | Ostreoida; Ostreoidea; Ostreidae | 18,414 | EU672835 | |
| Crassostrea gigas | Ostreoida; Ostreoidea; Ostreidae | 18,225 | EU672831 | |
| Crassostrea hongkongensis | Ostreoida; Ostreoidea; Ostreidae | 18,622 | EU672834 | |
| Crassostrea iredalei | Ostreoida; Ostreoidea; Ostreidae | 22,446 | FJ841967 | |
| Crassostrea sikamea | Ostreoida; Ostreoidea; Ostreidae | 18,243 | EU672833 | |
| Saccostrea mordax | Ostreoida; Ostreoidea; Ostreidae | 16,532 | FJ841968 | |
| Saccostrea glomerata | Ostreoida; Ostreoidea; Ostreidae | 16,281 | KU310918 | |
| Ostrea denselamellosa | Ostreoida; Ostreoidea; Ostreidae | 16,277 | HM015199 | |
| Ostrea edulis | Ostreoida; Ostreoidea; Ostreidae | 16,320 | JF274008 | |
| Ostrea lurida | Ostreoida; Ostreoidea; Ostreidae | 16,344 | KC768038 | |
| Argopecten irradians | Pectinoida; Pectinoidae; Pectinidae | 16,221 | EU023915 | |
| Chlamys farreri | Pectinoida; Pectinoidae; Pectinidae | 21,695 | EU715252 | |
| Mizuhopecten yessoensis | Pectinoida; Pectinoidae; Pectinidae | 20,414 | AB271769 | |
| Placopecten magellanicus | Pectinoida; Pectinoidae; Pectinidae | 32,115 | DQ088274 | |
| Mimachlamys nobilis | Pectinoida; Pectinoidae; Pectinidae | 17,963 | FJ415225 | |
| Gastropoda Vetigastropoda | Haliotis rubra | Haliotoidea; Haliotidae | 16,907 | NC_005940 |
| Gene | Location | Size (bp) | Start Codon | Stop Codon | Intergenic Region * |
|---|---|---|---|---|---|
| ATP6 | 1–550 | 550 | GTG | CGT | 0 |
| trnY | 551–613 | 63 | - | - | 2 |
| trnC | 616–678 | 62 | - | - | 36 |
| ND2 | 716–1714 | 999 | ATG | TAA | 37 |
| trnP | 2336–2399 | 64 | - | - | 621 |
| trnL | 2401–2467 | 67 | - | - | 1 |
| trnS1 | 2468–2537 | 70 | - | - | 0 |
| trnM1 | 2538–2600 | 63 | - | - | 0 |
| ATP8 | 2625–2726 | 102 | ATA | TAA | 24 |
| trnS2 | 2745–2814 | 70 | 18 | ||
| trnM2 | 2821–2883 | 63 | 6 | ||
| COX2 | 2889–3585 | 696 | ATG | TAG | 1 |
| CYTB | 3587–4747 | 1161 | ATA | TAA | 1 |
| trnE | 4745–4812 | 67 | - | - | 8 |
| trnT | 4821–4885 | 65 | - | - | 8 |
| trnI | 4894–4960 | 67 | - | - | 18 |
| COX3 | 4941–5840 | 900 | ATG | TAA | 130 |
| trnG | 5971–6037 | 67 | - | - | 8 |
| COX1 | 6046–7641 | 1596 | ATG | TAA | 688 |
| trnD | 8330–8397 | 68 | - | - | 55 |
| trnW | 8453–8515 | 63 | - | - | 60 |
| ND4L | 8576–8857 | 282 | GTG | TAA | 1 |
| ND1 | 8859–9791 | 933 | ATG | TAA | 78 |
| trnA | 9870–9934 | 65 | - | - | 12 |
| trnF | 9947–10,013 | 67 | - | - | 2 |
| trnL | 10,016–10,081 | 66 | - | - | 2 |
| trnK | 10,084–10,148 | 65 | - | - | −1 |
| ND3 | 10,148–10,501 | 354 | ATG | TAG | 0 |
| trnQ | 10,502–10,567 | 66 | - | - | −1 |
| ND6 | 10,579–11,046 | 468 | ATG | TAA | −1 |
| ND5 | 11,046–12,716 | 1671 | ATG | TAA | 37 |
| l-rRNA | 12,754–14,221 | 1468 | - | - | −660 |
| s-rRNA | 13,561–14,488 | 928 | - | - | 0 |
| ND4 | 14,495–15,844 | 1350 | ATG | TAA | 24 |
| trnH | 15,845–15,907 | 63 | - | - | 24 |
| trnV | 15,932–15,999 | 68 | - | - | 24 |
| trnR | 16,008–16,074 | 67 | - | - | 4 |
| trnN | 16,079–16,147 | 69 | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, J.; Kim, H.-J.; Oh, S.-Y.; Choi, Y.-U. Reproductive Characteristics of the Flat Oyster Ostrea denselamellosa (Bivalvia, Ostreidae) Found on the Southern Coast of South Korea. J. Mar. Sci. Eng. 2022, 10, 1326. https://doi.org/10.3390/jmse10091326
Han J, Kim H-J, Oh S-Y, Choi Y-U. Reproductive Characteristics of the Flat Oyster Ostrea denselamellosa (Bivalvia, Ostreidae) Found on the Southern Coast of South Korea. Journal of Marine Science and Engineering. 2022; 10(9):1326. https://doi.org/10.3390/jmse10091326
Chicago/Turabian StyleHan, Jeonghoon, Han-Jun Kim, Sung-Yong Oh, and Young-Ung Choi. 2022. "Reproductive Characteristics of the Flat Oyster Ostrea denselamellosa (Bivalvia, Ostreidae) Found on the Southern Coast of South Korea" Journal of Marine Science and Engineering 10, no. 9: 1326. https://doi.org/10.3390/jmse10091326
APA StyleHan, J., Kim, H.-J., Oh, S.-Y., & Choi, Y.-U. (2022). Reproductive Characteristics of the Flat Oyster Ostrea denselamellosa (Bivalvia, Ostreidae) Found on the Southern Coast of South Korea. Journal of Marine Science and Engineering, 10(9), 1326. https://doi.org/10.3390/jmse10091326






