Abstract
This study presents two new free-living marine nematodes, Haliplectus major sp. nov. and Haliplectus futianisus sp. nov., from a mangrove reservation in Shenzhen, China. Haliplectus major sp. nov. can be distinguished by its total body length of 1615–1998 µm; a cuticle annulated with eight rows of cuticle pores; amphidial fovea at 9–12 µm from the anterior end; a bipartite basal bulb with striated valve plates in both sexes; five to seven closely spaced precloacal supplements in males; a didelphic reproductive system in females, with the vulva at 49.64–52.37% of body length; and sexual dimorphism in tail shape, arcuate-conoid and 2.3–3.2 times anal body diameter long in males and elongate-conid and 3.5–4.2 times anal body diameter long in females. Haliplectus futianisus sp. nov. is characterized by a total body length of 574–655 µm; a cuticle annulated with eight rows of cuticle pores; amphidial fovea at 8–11 µm from the anterior end; a monopartite basal bulb with unstriated valve plates in both sexes; an absence of precloacal supplements in males; and a didelphic reproductive system in females, with the vulva at 42.33–45.63% of body length. The identification was confirmed by analyzing morphological characteristic and 18S rRNA gene sequences. The maximum likelihood (ML) topology was regarded as morphological evidence of H. major sp. nov. and H. futianisus sp. nov. being two distinct species.
1. Introduction
The free-living marine nematode is not only an important indicator species of the marine environment [1,2,3,4] but also a very important member of the mangrove wetland, where it plays an important role in nutrient cycling and carbon fixation [5,6,7]. Therefore, it is important to classify and identify this species accurately.
The genus Haliplectus was first identified by Cobb in 1913 with the type species H. pellucidus [8]. It is characterized by an annulated cuticle; the presence of body pores; subcephalic and cervical sensilla, with ocelli and deirid absent; inconspicuous cephalic and somatic setae; circular to unispiral amphids; stoma elongated into the corpus of the esophagus, narrow, with or without distinct cheilorhabdions, and terminating with joints in a small median esophageal bulb; the esophagus surrounding most of the stoma with the median bulb, isthmus and elongate, rather chromadoroid, posterior bulb, lining thickened; arcuate spicules and gubernaculum in males; ventral preanal supplementary organs present or absent, with two testes; and two reflexed, opposed ovaries in females [9,10]. So far, 28 species have been described in the genus Haliplectus and they are globally abundant organisms [10,11,12,13]. In particular, seven of them are mainly distributed in mangrove environments [10,14,15].
Compared to the Haliplectus species that have been identified through morphology, the existing genetic data are far from satisfactory. Currently, the sequences of nematodes are generally deposited at three databases: the National Center for Biotechnology Information (NCBI), the European Bioinformatics Institute (EMBL-EBI), and the Center for Information Biology and DNA Data Bank of Japan (DDBJ). Herein, the sequences of the genus Haliplectus were surveyed from the NCBI, which is the most used database. A total of 19 sequences, among which 5 are mitochondrial cytochrome c oxidase subunit I (COI) genes, only 1 belongs to the 28S rRNA gene, and the others are 18S rDNA sequences, were examined. Among them, three specific species were identified and could be matched to the ten sequences. The detailed information is as follows: Haliplectus floridanus has three vouchers, every voucher has both one COI gene sequence and one 18S rDNA sequence. Haliplectus dorsalis has two sequences, and one belongs to the COI gene fragment the other one is 18S rDNA. Haliplectus iranicus only has two sequences of 18S rDNA. In the other nine sequences, only one sequence has an exact species name, which is Haliplectus cf. dorsalis and it is different from Haliplectus dorsalis in the 18S rRNA gene, and the remaining eight sequences were not identified or shown to belong to a species (electronic database: https://www.ncbi.nlm.nih.gov/ (accessed on 22 June 2022)).
2. Materials and Methods
2.1. Sample Collection, Meiofauna Extraction, and Nematode Identification
Meiofaunal samples were collected from Futian mangrove mudflats in Shenzhen, China. Sediment sampling and nematode extraction were performed as described in our previous studies [16,17]. Some of the samples were fixed with 5% formalin for species identification. Measurements were taken in µm. Abbreviations are as follows: a = body length/maximum body diameter; b = body length/esophagus length; c = body length/tail length; abd = anal body diameter; cbd = corresponding body diameter; vbd = vulval body diameter; c’ = tail length/abd; V% = position of vulva as a percentage of body length from anterior end; that is, anterior end to vulva/body length. All holotypes and paratypes were deposited at the Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China.
Some of the samples were fixed with DESS [18] for DNA extraction. The slides were prepared for species identification as follows: (1) a paraffin ring was formed on the glass slide; (2) 2 μL DESS was dropped in the center of the paraffin ring; (3) the individual nematode was transferred into the DESS; (4) it was covered with a cover glass and heated until the paraffin melted; (5) some of the main features were identified and photographed; (6) after identification, the slide was placed on the heater and the cover glass removed; (7) the nematode was transferred to the tube for DNA extraction.
2.2. DNA Extraction and PCR Amplification
DNA was extracted from a single nematode following the steps for the TIANamp Marine Animals DNA Kit (DP324), which is produced by TIANGEN.
The 18S rDNA was selected for genetic analysis. The two pairs of primers used in this analysis were as follows:1096F (5′-GGTAATTCTGGAGCTAATAC-3′) and 1912R (5′-TTTACGGTCAGAACTAGGG-3′), 1813F (5′-CTGCGTGAGAGGTGAAAT-3′) and 2646R (5′-GCTACCTTGTTACGACTTTT-3′) [19]. The primer pair JB2F (5′-ATGTTTTGATTTTACCWGCWTTYGGTGT-3′) and JB5GEDR (5′-AGCACCTAAACTTAAAACATARTGRAARTG-3′) [20] was chosen to amplify COI gene sequences. PCR was conducted with 12.5 μL 2X Pro Taq Master Mix (dye plus), 0.5 μL forward and reverse primers (0.2–1.0 μM), 7.5 μL RNase free water, and 4 μL DNA to achieve a total volume of 25 μL. PCR cycling conditions from previous studies were used [19,20]. Following the PCR assay, 5 μL of each PCR product was confirmed with electrophoresis on a 1% agarose gel. The gel was strained with 0.5 μg/mL 4S Red Plus Nucleic Acid Stain and imaged with an ultraviolet transillumination system. All positive PCR amplicons were sequenced by Sangon Biotech (Shanghai, China) Co., Ltd.
2.3. Sequencing and Analyses
A total of 28 sequences were obtained from individual nematodes of H. major sp. nov. and H. futianisus sp. nov. in this study. All of the sequences were also deposited at GenBank. The reference sequences were selected together for phylogenetic analysis. Spirina antipodea was included as an outgroup for 18S rDNA sequence analysis, for which the same gene sequence [21] was amplified in this study. The sequence analysis was performed as described in our previous studies [7].
3. Results
3.1. Morphological Description of Haliplectus major sp. nov.
urn:lsid:zoobank.org:act:1A5A7745-5DBB-4E84-B71F-450384039E7F.
3.1.1. Type Material
Five males and three females were collected from station SZFT (22.52° N, 114.01° E) in July and November 2020. Holotype: ♂1 on slide number SZFT20207 3L113. Paratypes: ♂2 on slide number SZFT20207 1L111, ♂3 on slide number SZFT20201127 2M120, ♂4 on slide number SZFT20201127 1M115, ♂5 on slide number SZFT20201127 1M108, ♀1 on slide number SZFT20207 3L105, ♀2 on slide number SZFT20207 2H104, ♀3 on slide number SZFT20207 1L110.
3.1.2. Etymology
The species was named after its relatively large body size.
3.1.3. Type Locality and Habitat
All specimens were collected from a mangrove forest in the mangrove reservation in Shenzhen, China. The characteristics of surface sediments from the sampling stations are shown in Table 1.

Table 1.
Characteristics of surface sediments from sampling stations.
3.1.4. Measurements
Detailed measurement data are shown in Table 2.

Table 2.
Individual measurements for Haliplectus major sp. nov. (in µm).
3.1.5. Description
Body elongate and cylindrical, slightly arcuate, c-shaped or irregularly curved upon fixation, tapering at both ends, body diameter at level of posterior esophagus end about 3.7–5.3 times cephalic region width. Head end convex-conoid. Cuticle strongly annulated, annuli 1–1.5 µm broad at neck region, 1.7–2.3 µm at midbody part, and about 1–1.4 µm at tail region. No distinct lips; lip region not set off. Cephalic region 6.8–8.2 µm. Labial and cephalic sensilla indistinct. Amphidial fovea unispiral to circular with posteriorly broken contour, 4–5 µm in diameter and situated at 10–12 µm from anterior end. Lateral fields not differentiated, with four rows of cuticular pores on each side. Buccal cavity rudimentary, cheilostom very weakly developed without teeth or denticles, formed as elongate tubular structure with slightly thickened cuticular lining that stretches into middle region of median esophageal bulb, 51–67 µm long. Esophagus muscular, 108–126 µm long, with a median bulb 16–19 µm long and 11–14 µm wide and a well-developed, distinct spherical muscular bipartite basal bulb 24–26 µm long, 23–32 µm wide. Esophageal median bulb and basal bulb connected by slightly narrower isthmus surrounded by nerve ring, basal bulb with striated valve plates. Nerve ring located at 65–71% of esophagus length. Cardia distinct, 19–22 µm long. Tail arcuate-conoid and 2.31–3.26 times anal body diameter long in males; elongate-conid and 3.48–4.21 times anal body diameter long in females. The caudal glands are packed in tandem series in anterior half of tail and have a distinct efferent duct-spinneret.
Male reproductive system diorchic, two testes arranged in tandem, outstretched. Spicules paired, arcuate, 35–42 µm long as arc and 31–36 µm long as chord, with distal ends pointed and gradually broadening to proximal, not cephalated. Gubernaculum slightly sclerotized, 14–18 µm long, lying almost parallel to distal portion of spicules. Five to seven closely spaced, bluntly conical precloacal supplements present, forming a ventral row located at 6–8 µm before anus. Four papillae on tail are partly ventral and partly submedian.
Female reproductive system didelphic. Two ovaries opposed and outstretched. Vulva at 49.64–52.37% of body length (Figure 1 and Figure 2).
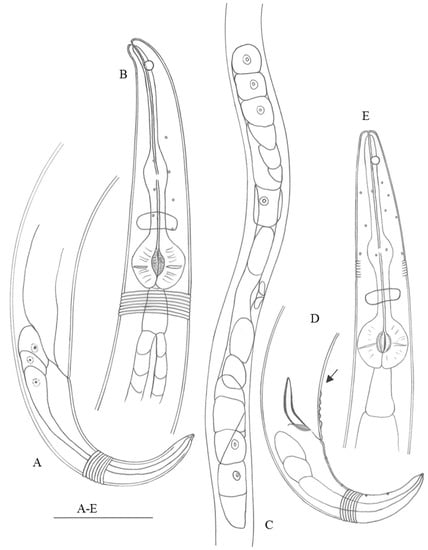
Figure 1.
Haliplectus major sp. nov. (A) lateral view of female tail region; (B) lateral view of female head end; (C) lateral view of female body part showing ovaries; (D) lateral view of male tail region (arrow indicating supplements); (E) lateral view of male head end. Scale bars: (A,B,D,E) = 50 µm; (C) = 125 µm.
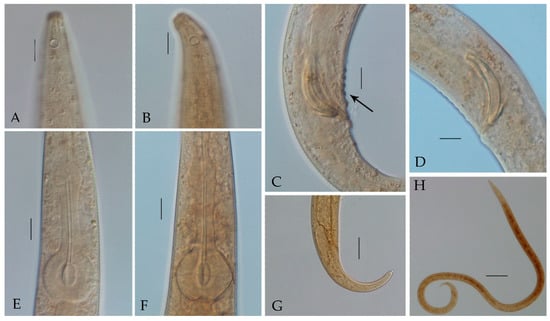
Figure 2.
Haliplectus major sp. nov. (A) lateral view of male head end showing amphidial fovea; (B) lateral view of female head end showing amphidial fovea; (C) lateral view of male supplements; (D) lateral view of male copulatory apparatus; (E) lateral view of male pharynx basal bulb; (F) lateral view of female pharynx basal bulb; (G) lateral view of female tail region; (H) lateral view of male total body. Scale bars: (A–F) = 10 μm; (G) = 25 μm; (H) = 100 μm.
3.1.6. Diagnosis
Haliplectus major sp. nov. is characterized by a total body length of 1615–1998 µm; annulated cuticle with eight rows of cuticle pores; amphidial fovea at 9–12 µm from the anterior end; bipartite basal bulb with striated valve plates in both sexes; five to seven closely spaced precloacal supplements in males; didelphic reproductive system in females, with the vulva at 49.64–52.37% of body length; and sexual dimorphism in tail shape, arcuate-conoid and 2.31–3.26 times anal body diameter long in males and elongate-conid and 3.48–4.21 times anal body diameter long in females.
This new species is most morphologically similar to Haliplectus floridanus Cobb in Chitwood, 1956. Identification of H. floridanus was published by Chitwood in 1956 based on Cobb’s original notes with his sketches, attributing them to Cobb posthumously, and Gerlach redescribed it in 1957 [9,22]. Haliplectus major sp. nov. resembles H. floridanus Cobb in Chitwood, 1956 in its similar body size, the amphidial fovea spirals appearing as circles about one-third as wide as the corresponding portion of the head, the basal bulb being very distinctly developed and strongly transversely striated, and the number of precloacal supplements (six in H. floridanus) [9]. However, compared to the description by Gerlach (1957), it can be distinguished by the relatively short esophagus (b = 13.33–17.34 in males and 14.51–16.38 in females vs. b = 11.5, 13.6 in males and 13.3 in females in H. floridanus), the posterior location of the vulva (vulva at 49.71–52.37% of body length vs. 48% of body length in H. floridanus), the spicule being slightly longer (spicule length as arc = 35–42 µm vs. 33 and 34 µm in H. floridanus), the anterior location of the amphidial fovea (distance from anterior end to amphid/cephalic region width = 1.2–1.7 vs. 2 and 2.2 in H. floridanus), and the longer tail of the females (91–106 µm vs. 73–74 µm in H. floridanus) [22].
3.2. Morphological Description of Haliplectus futianisus sp. nov.
urn:lsid:zoobank.org:act:E4E94281-B30F-4208-9A28-38DC1A980D8E.
3.2.1. Type Material
Four males and three females were collected from station SZFT (22.52° N, 114.01° E) in July and November 2020. Holotype: ♂1 on slide number SZFT20201127 1H120. Paratypes: ♂2 on slide number SZFT20201127 1H109, ♂3 on slide number SZFT20201127 1H115, ♂4 on slide number SZFT20201127 1H104, ♀1 on slide number SZFT20207 1L106, ♀2 on slide number SZFT20201127 1H101, ♀3 on slide number SZFT20201127 1H115.
3.2.2. Type Locality and Habitat
All specimens were collected from a mangrove forest in the mangrove reservation in Shenzhen, China. The characteristics of surface sediments from the sampling stations are shown in Table 2.
3.2.3. Etymology
The species name refers to its first sampling location: the Futian mangrove in Shenzhen, China.
3.2.4. Measurements
Detailed measurement data are shown in Table 3.

Table 3.
Individual measurements for Haliplectus futianisus sp. nov. (in µm).
3.2.5. Description
Body elongate and cylindrical, slightly arcuate, tapering at both ends, body diameter at level of posterior esophagus end about 4.1–6.6 times cephalic region width. Head end markedly convex-conoid. Cuticle strongly annulated, annuli 1.1–1.3 µm broad at neck region, 1.2–1.5 µm at midbody part, and about 0.9–1.2 µm at tail region. No distinct lips; lip region not set off. Cephalic region 4.1–5.2 µm. Labial and cephalic sensilla indistinct. Amphidial fovea unispiral to circular with posteriorly broken contour, 3–4 µm in diameter and situated at 8–11 µm from anterior end. Lateral fields not differentiated, with four rows of cuticular pores on each side. Buccal cavity rudimentary, cheilostom very weakly developed without teeth or denticles, formed as elongate tubular structure with slightly thickened cuticular lining that stretches into middle region of median esophageal bulb, 41–43 µm long. Esophagus muscular, 83–90 µm long, with a median bulb 10–13 µm long and 7–9 µm wide and a well-developed, distinct spherical muscular monopartite basal bulb 17–19 µm long, 15–22 µm wide. Esophageal median bulb and basal bulb connected by slightly narrower isthmus surrounded by nerve ring, basal bulb with unstriated valve plates. Nerve ring located at 67–73% of esophagus length. Cardia distinct, 8–11 µm long. Tail conoid, 1.94–2.68 times anal body diameter long in males; 3.29–3.72 times anal body diameter long in females. The caudal glands are packed in tandem series in anterior half of tail and have a distinct efferent duct-spinneret.
Male reproductive system diorchic, two testes arranged in tandem, outstretched. Spicules paired, arcuate, 24–25 µm long as arc and 22–23 µm long as chord, with distal ends pointed and gradually broadening to proximal, slightly cephalated. Gubernaculum slightly sclerotized, 8–9 µm long, lying parallel to distal portion of spicules. Precloacal supplements absent. In the holotype, it can be clearly seen that a row of cuticle pores range in a partly ventral and partly submedian manner.
Female reproductive system didelphic. Two ovaries opposed and outstretched. Vulva at 42.33–45.63% of body length (Figure 3 and Figure 4).
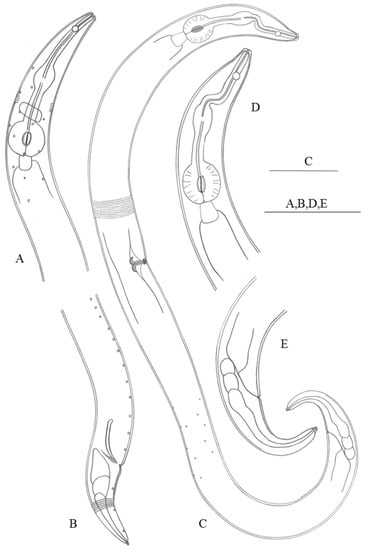
Figure 3.
Haliplectus futianisus sp. nov. (A) lateral view of male head end; (B) lateral view of male tail region; (C) lateral view of female total body; (D) lateral view of female head end; (E) lateral view of female tail region. Scale bars: (A–E) = 50 µm.

Figure 4.
Haliplectus futianisus sp. nov. (A) lateral view of male head end showing amphidial fovea; (B) lateral view of male copulatory apparatus; (C) lateral view of female vulva; (D) lateral view of female head end showing pharynx basal bulb; (E) lateral view of male tail region; (F) lateral view of female tail region; (G) lateral view of female body part showing ovaries; (H) lateral view of female total body. Scale bars: (A–F) = 10 μm; (G) = 25 μm; (H) = 50 μm.
3.2.6. Diagnosis
Haliplectus futianisus sp. nov. is characterized by a total body length of 574–655 µm; an annulated cuticle with eight rows of cuticle pores; amphidial fovea at 8–11 µm from the anterior end; a monopartite basal bulb with unstriated valve plates in both sexes; an absence of precloacal supplements in males; and a didelphic reproductive system in females, with the vulva at 42.33–45.63% of body length. It most closely resembles H. iranicus Gharahkhani, Pourjam, Leduk & Pedram, 2022 [10] in its similar body size and the unstriated valvar plates of the esophageal basal bulb. However, it can be easily distinguished by the plumper body (a = 18.86–25.31 vs. a = 28.1–33.7 in H. iranicus), the relatively anterior location of the amphidial fovea (distance from anterior end to amphid/cephalic region width = 1.8–2.7 vs. 2.5–3 in H. iranicus), the esophageal basal bulb monopartite (esophageal basal bulb bipartite in H. iranicus), the number of precloacal supplements (absent vs. three contiguous and one adcloacal in H. iranicus), and slightly longer spicule (spicule length as arc 24–25 µm vs. 18–21 µm in H. iranicus) [10].
3.3. Sequences Information and Cladogram Analyses
In the NCBI database, the 18S rDNA and COI genes have more complete resources than the 28S rDNA sequences [23]. Therefore, two fragments of these genes were chosen and respectively analyzed in this study. The Genbank accession numbers of the 18S rDNA are as follows: ON688994–ON689005 and ON689013–ON689024. The accession numbers of the COI gene are ON660976–ON660979. A 630 base pair (bp) sequence of 18S rDNA and a 310 bp sequence of the COI gene were used in this study. The maximum likelihood (ML) topology was used in this study (Figure 5 and Figure 6). Both the 18S rDNA and COI gene cladograms indicated morphological evidence of H. futianisus sp. nov. and H. major sp. nov. being distinct species. The interspecific and intrageneric thresholds of Kimura 2-parameter (K2P) distance divergence were as follows: 0.1–3.1% and 6.7–9.0% for 18S rDNA and 0–0.3% and 19.1%–29.5% for COI, respectively.
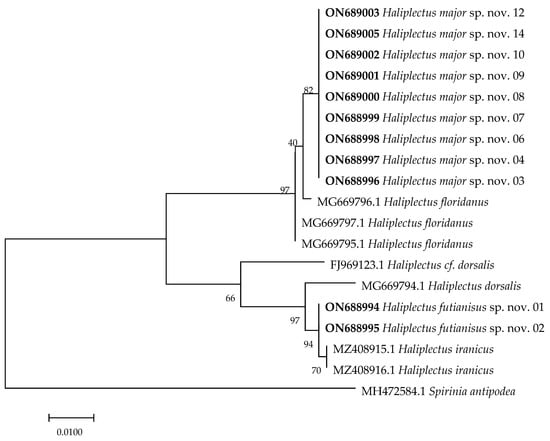
Figure 5.
Maximum likelihood tree based on a 630 bp region of the 18S rRNA gene. The sequences highlighted in bold are from this study. Numbers above/below the branches indicate branch support based on 1000 bootstrap replicates.
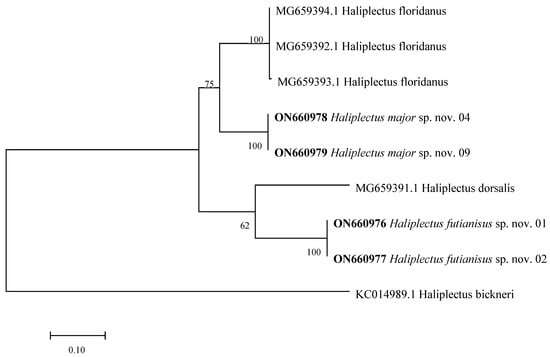
Figure 6.
Maximum likelihood tree based on a 310 bp region of the COI gene. The sequences highlighted in bold are from this study. Numbers above/below the branches indicate branch support based on 1000 bootstrap replicates.
4. Discussion
In the database (GenBank), as many as 42% of sequences are not assigned specific species names [7], which is why morphological characterization has an important role. Without morphological characterization, these sequences cannot be fully utilized due to the absence of species names. Therefore, it would greatly facilitate phylogenetic affinities to match species names to specific gene sequences. This study provides morphological diagnoses and sequences for two new species of the genus Haliplectus. The study also presents a key for the genus Haliplectus in mangroves worldwide.
4.1. The Key for the Genus Haliplectus Cobb, 1913 from Global Mangroves with Two New Species
| 1. Body length less than 1.6 mm·································································································2 |
| - Body length greater than 1.6 mm···································································H. major sp. nov. |
| 2. Esophagus basal bulb without striated valve plates····························································3 |
| - Esophagus basal bulb with striated valve plates·······················································H. minor |
| 3. Distance from anterior end to amphid/cephalic region width equal to 1··························4 |
| - Distance from anterior end to amphid/cephalic region width greater than 1····················5 |
| 4. Female: body length is 0.67–0.92 mm, c value is 16–23. Male: three contiguous supplements··········································································································H. pakistanensis |
| - Female: body length is 0.49–0.52 mm, c value is 13.2–14.6·····························H. monodelphis |
| 5. Distance from anterior end to amphid/cephalic region width equal to 2··························6 |
| - Distance from anterior end to amphid/cephalic region width not equal to 2·····················7 |
| 6. a value is 18.2–20.3, tail length is 33–35 μm···························································H. gracilis |
| - a value is 22.3–38.1, tail length is 34–44 μm, with supplements and caudal papilla········H. paradorsalis |
| 7. Spicule length less than 30 μm································································································8 |
| - Spicule length equal to 30 μm··················································································H. robustus |
| 8. No precloacal supplements····································································H. futianisus sp. nov. |
| - With precloacal supplements····················································································H. iranicus |
4.2. Molecular Results Discussion
Regarding phylogenetic analyses, the genes had suitable resolution for different taxonomic levels. The 18S rDNA sequences possess good resolution for identification at higher taxonomic rankings, such as to separate the allied genera, while COI is a useful gene for identification at the species level [20,24,25]. Gharahkhani et al. (2022) [10] constructed a consensus cladogram using 18S rDNA and suggested that the family Haliplectidae might be considered as incerta sedis within the class Chromadorea. However, the current molecular sequence database of marine nematodes remains incomplete, and misidentified or unidentified organisms are a huge problem [26]. Due to these reasons, the clarity of the systematics of the family Haliplectidae is limited. In the present work, we provided two different sequence fragments, 18S rDNA and COI, to establish the new species and enrich the database for further study.
The nucleotide divergence of the genes could be used as a diagnostic tool for the discrimination of nematode species [20,26]. Derycke et al. (2010) [20] have suggested that the intraspecific threshold is approximately 0.05 K2P distance for the COI gene. However, these data are incomplete for nematodes at present, and an intraspecific threshold is yet to be defined. Therefore, further study on the divergence of different species is necessary.
Author Contributions
Conceptualization, data curation, writing—original draft preparation, and writing—review and editing, R.Z., Y.S. and Y.G.; methodology and graphic plotting, R.Z. and Y.C.; funding acquisition, Y.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Natural Science Foundation of Fujian Province, grant number 2022J01324, and the National Natural Science Foundation of China, grant number 31772416.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors thank Zhu Huilan and Futian-CityU Mangrove Research and Development Centre for great assistance in the sampling.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Avó, A.P.; Daniell, T.J.; Roy, N.; Solange, O.; Jordana, B.; Adão, H. DNA barcoding and morphological identification of benthic nematodes assemblages of estuarine intertidal sediments: Advances in molecular tools for biodiversity assessment. Front. Mar. Sci. 2017, 4, 66. [Google Scholar] [CrossRef]
- Gielings, R.; Fais, M.; Fontaneto, D.; Creer, S.; Costa, F.O.; Renema, W.; Macher, J. DNA metabarcoding methods for the study of marine benthic meiofauna: A review. Front. Mar. Sci. 2021, 8, 730063. [Google Scholar]
- Leduc, D.; Zhao, Z.Q. Morphological and molecular characterisation of new Acanthopharynx and Desmodora species (Nematoda: Desmodorinae) from intertidal sediments of New Zealand. Nematology 2016, 18, 905–924. [Google Scholar] [CrossRef]
- Tytgat, B.; Nguyen, D.T.; Nguyen TX, P.; Pham, T.M.; Long, P.K.; Vanreusel, A.; Derycke, S. Monitoring of marine nematode communities through 18S rRNA metabarcoding as a sensitive alternative to morphology. Ecol. Indic. 2019, 107, 105554. [Google Scholar] [CrossRef]
- Bhadury, P.; Austen, M.C.; Bilton, D.T.; Lambshead PJ, D.; Rogers, A.D.; Smerdon, G.R. Evaluation of combined morphological and molecular techniques for marine nematode (Terschellingia spp.) identification. Mar. Biol. 2008, 154, 509–518. [Google Scholar] [CrossRef]
- Nisa, R.U.; Tantray, A.Y.; Shah, A.A. Shift from morphological to recent advanced molecular approaches for the identification of nematodes. Genomics 2022, 114, 110295. [Google Scholar] [CrossRef]
- Shih, Y.-J.; Chen, Y.-Z.; Guo, Y.-Q. A new species of free-living marine nematode (Ptycholaimellus: Chromadoridae: Chromadorida: Nematoda) from mangrove wetlands in China. Zool. Stud. 2022, 61, 20. [Google Scholar]
- Cobb, N.A. New Nematode genera found inhabiting fresh water and nonbrackish soils. J. Wash. Acad. Sci. 1913, 3, 432–444. [Google Scholar]
- Chitwood, B.G. A revision of the genus Haliplectus Cobb, 1913. Proc. Helminthol. Soc. Wash. 1956, 23, 78–87. [Google Scholar]
- Gharahkhani, A.; Pourjam, E.; Leduc, D.; Pedram, M. The nematode genus Haliplectus Cobb, 1913 (Chromadorea: Haliplectidae): Phylogenetic relationships, description of a new species from the Persian Gulf, southern Iran, and a tabular key to valid species. Nematology 2022, 1–17. Available online: https://brill.com/view/journals/nemy/24/6/article-p639_4.xml?ebody=abstract%2Fexcerpt (accessed on 28 September 2022).
- Gee, J.M.; Somerfield, P.J. Do mangrove diversity and leaf litter decay promote meiofaunal diversity? J. Exp. Mar. Biol. Ecol. 1997, 218, 13–33. [Google Scholar]
- Netto, S.A.; Gallucci, F. Meiofauna and macrofauna communities in a mangrove from the Island of Santa Catarina, South Brazil. Hydrobiologia 2003, 505, 159–170. [Google Scholar] [CrossRef]
- Zhou, H. Effects of leaf litter addition on meiofaunal colonization of azoic sediments in a subtropical mangrove in Hong Kong. J. Exp. Mar. Biol. Ecol. 2001, 256, 99–121. [Google Scholar] [CrossRef]
- Zhu, H.; Liu, M.; Zhou, Y.; Guo, Y.Q. Meiofauna community structure and marine nematode (a new record) in Futian mangrove wetland of Shenzhen Bay, Guangdong Province. Chin. J. Ecol. 2020, 39, 1806–1812. (In Chinese) [Google Scholar]
- Shahina, F.; Siddiqi, M.R.; Nasira, K. Six new species of Haliplectus Cobb 1913 (Nematoda: Haliplectidae) from mangroves of Karachi, Pakistan. Int. J. Nematol. 2014, 24, 68–80. [Google Scholar]
- Yang, P.; Liu, M.; Guo, Y. One new species and one new record species of Sphaerolaimus (Nematode, Sphaerolaimidae) in offshore wetlands, China. J. Oceanol. Limnol. 2021, 39, 683–692. [Google Scholar] [CrossRef]
- Guo, Y.; Chang, Y.; Chen, Y.; Yongxiang, L.; Aiyuan, L. Description of a marine nematode Hopperia sinensis sp. nov. (Comesomatidae) from mangrove forests of Quanzhou, China, with a pictorial key to Hopperia species. J. Ocean Univ. China 2015, 14, 1111–1115. [Google Scholar]
- Yoder, M.; De Ley, I.T.; King, I.W.; Mundo-Ocampo, M.; Mann, J.; Blaxter, M.; Poiras, L.; De Ley, P. DESS: A versatile solution for preserving morphology and extractable DNA of nematodes. Nematology 2006, 8, 367–376. [Google Scholar]
- Holterman, M.; van der Wurff, A.; van den Elsen, S.; van Megen, H.; Bongers, T.; Holovachov, O.; Bakker, J.; Helder, J. Phylum-wide wnalysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown clades. Mol. Biol. Evol. 2006, 23, 1792–1800. [Google Scholar]
- Derycke, S.; Vanaverbeke, J.; Rigaux, A.; Backeljau, T.; Moens, T.; Roopnarine, P. Exploring the use of Cytochrome Oxidase c Subunit 1 (COI) for DNA Barcoding of free-living marine nematodes. PLoS ONE 2010, 5, e13716. [Google Scholar] [CrossRef]
- Leduc, D.; Zhao, Z.Q. Morphological and molecular characterisation of Spirinia antipodea Leduc n. sp. (Nematoda: Desmodoridae), a cryptic species related to S. parasitifera, from the coast of New Zealand. Nematology 2019, 21, 91–105. [Google Scholar]
- Gerlach, S.A. Marine nematoden aus dem mangrove-gebiet von cananéia (Brasilianische Meeres-Nematoden III). Abh. Math. -Nat. Kl. 1957, 5, 129–176. [Google Scholar]
- De Ley, P.; De Ley, I.T.; Morris, K.; Abebe, E.; Mundo-Ocampo, M.; Yoder, M.; Heras, J.; Waumann, D.; Rocha-Olivares, A.; Jay Burr, A.H.; et al. An integrated approach to fast and informative morphological vouchering of nematodes for applications in molecular barcoding. Phil. Trans. R. Soci. B 2005, 360, 1945–1958. [Google Scholar]
- Derycke, S.; Remerie, T.; Vierstraete, A.; Backeljau, T.; Vanfleteren, J.; Vincx, M.; Moens, T. Mitochondrial DNA variation and cryptic speciation within the free-living marine nematode Pellioditis marina. Mar. Ecol. Prog. Ser. 2005, 300, 91–103. [Google Scholar] [CrossRef]
- Derycke, S.; Backeljau, T.; Vlaeminck, C.; Vierstraete, A.; Vanfleteren, J.; Vincx, M.; Moens, T. Spatiotemporal analysis of population genetic structure in Geomonhystera disjuncta (Nematoda, Monhysteridae) reveals high levels of molecular diversity. Mar. Biol. 2007, 151, 1799–1812. [Google Scholar] [CrossRef]
- Pitusi, V.; Søreide, J.E.; Hassett, B.T.; Marquardt, M.; Andreasen, M.H. The occurrence of Nematoda in coastal sea ice on Svalbard (European Arctic) determined with the 18S small subunit rRNA gene. Polar Biol. 2021, 44, 1153–1162. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).