Assessing the Influence of Fumigation and Bacillus Subtilis-Based Biofungicide on the Microbiome of Chrysanthemum Rhizosphere
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Site
2.2. Soil Fumigant and Biofungicide
2.3. Experimental Design
2.4. Soil Sampling and DNA Extraction
2.5. PCR Amplification and Illumina Sequencing
2.6. Statistical and Bioinformatic Analyses
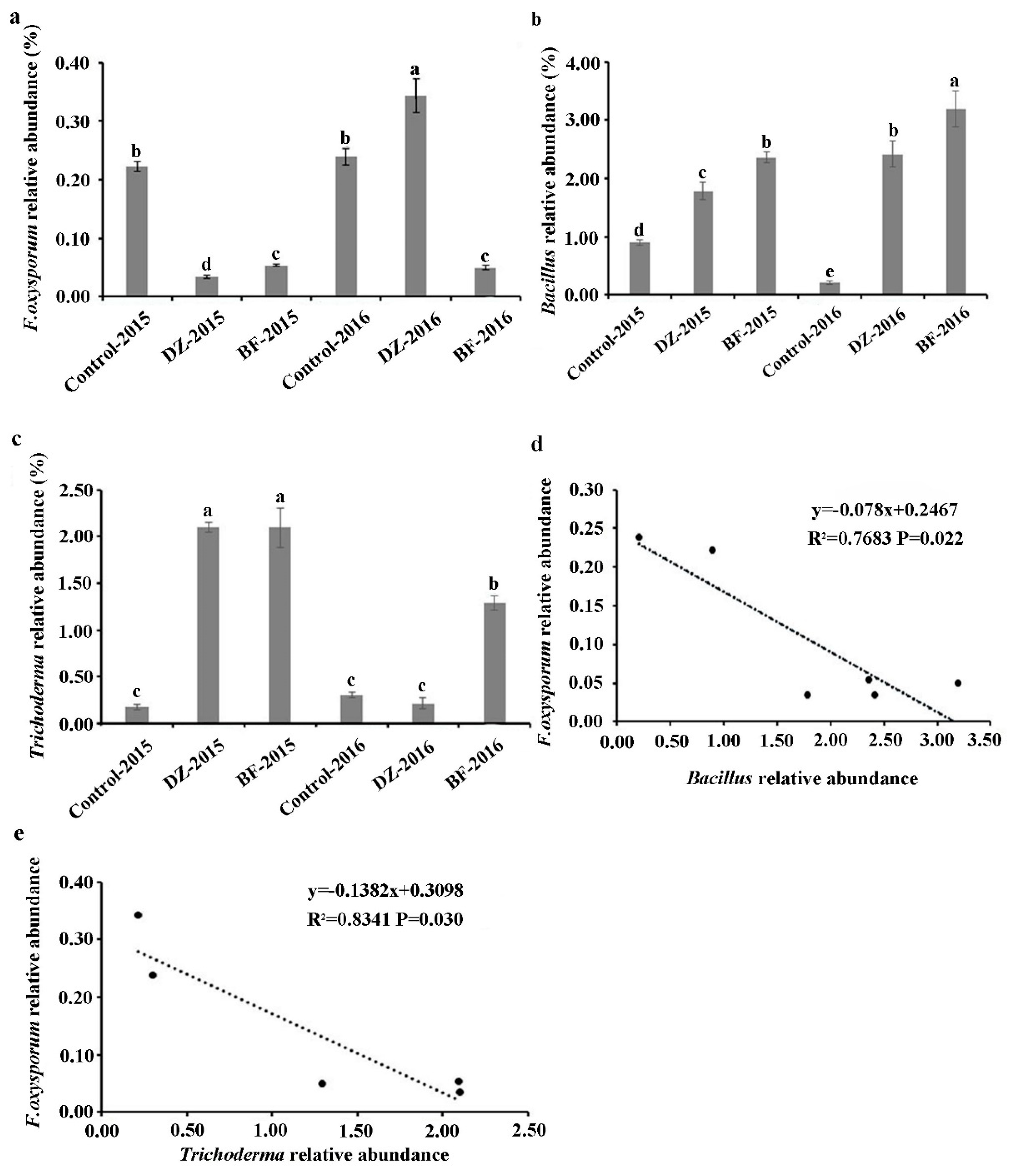
3. Results
3.1. Subsection Illumina Sequencing and Operational Taxonomic Unit (OTU) Classification
3.2. Alpha Diversity Analysis of the Soil Microbiome
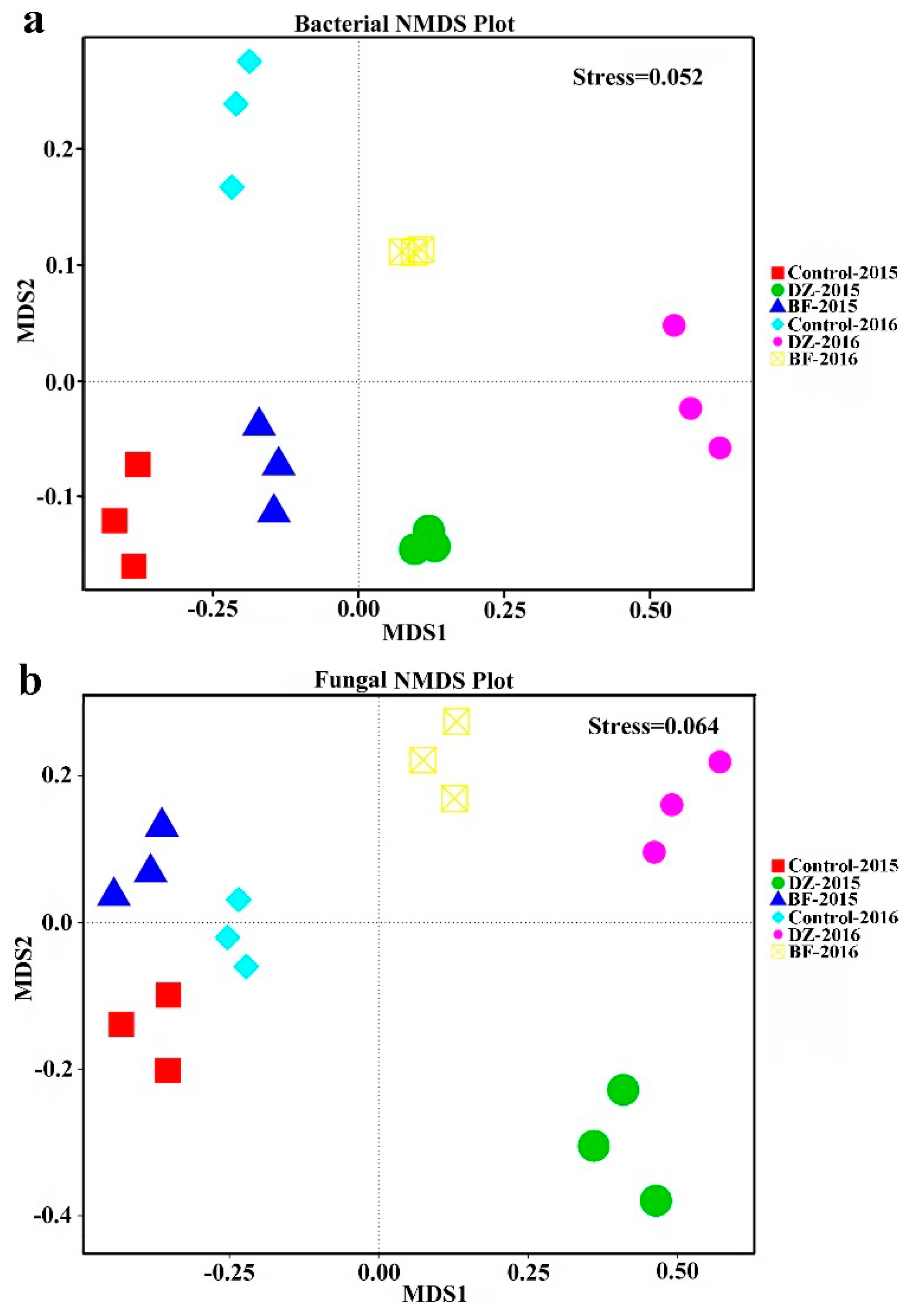
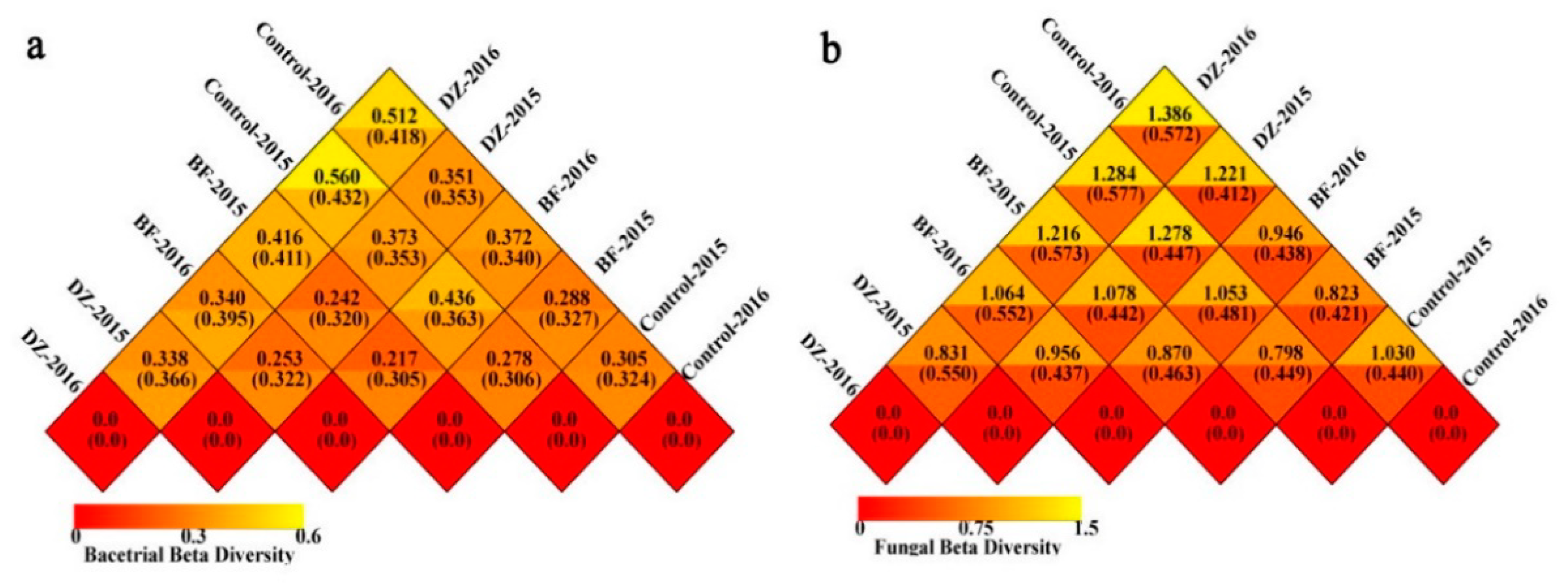
3.3. Beta Diversity Analysis of the Soil Microbiome
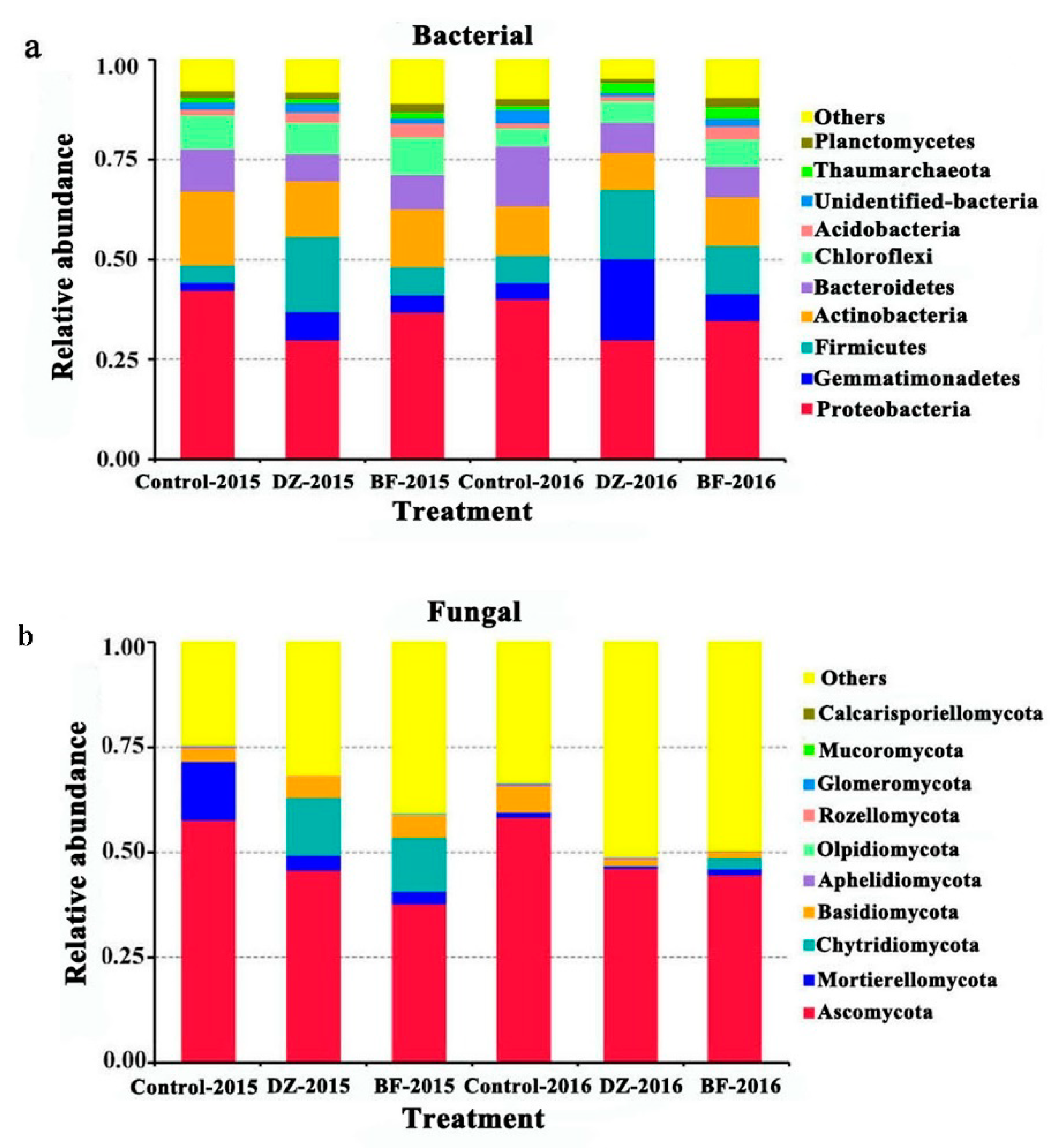
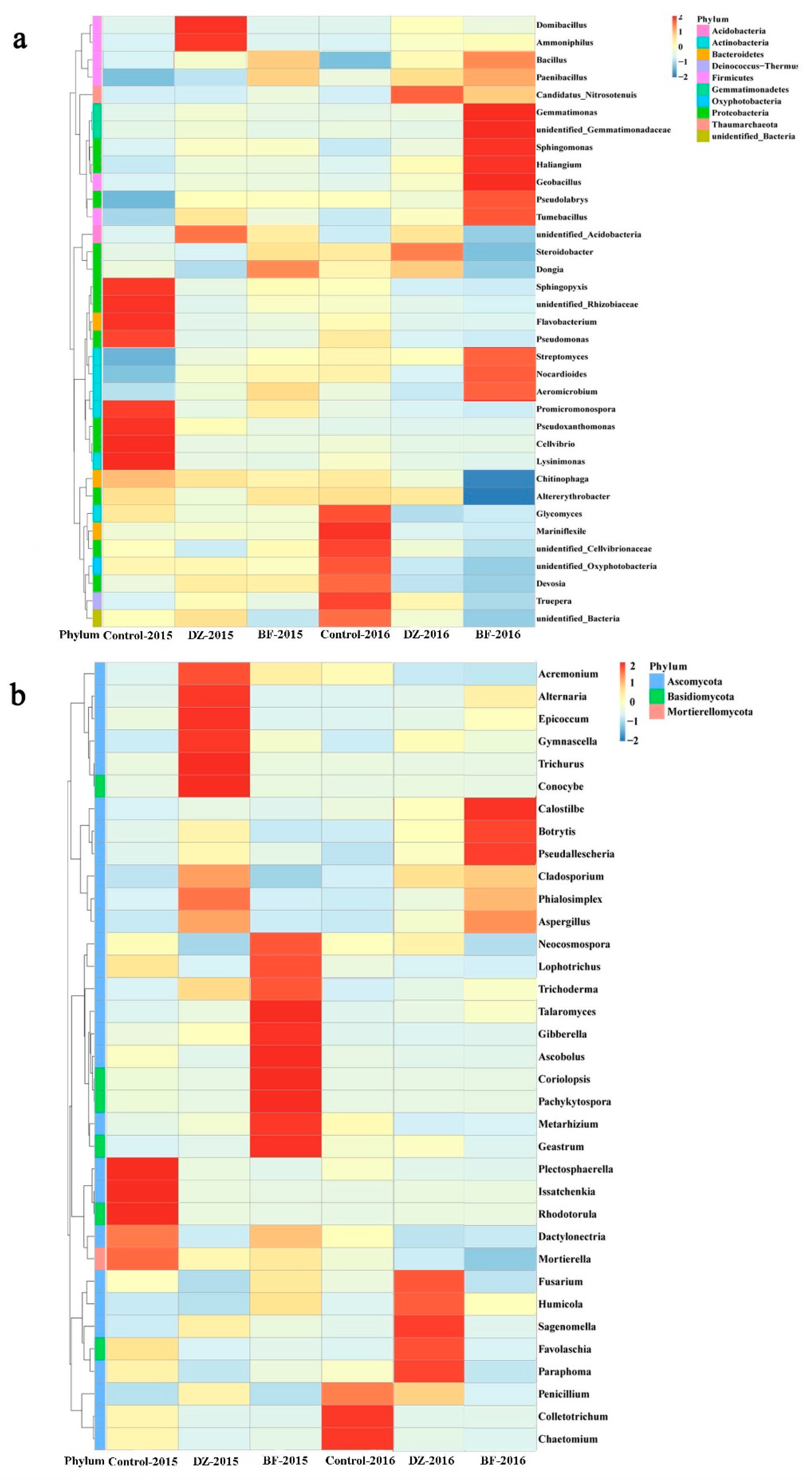
3.4. Taxonomic Composition of the Microbiome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wang, J.; Li, X.; Xing, S.; Ma, Z.; Hu, S.; Tu, C. Bio-organic fertilizer promotes plant growth and yield and improves soil microbial community in continuous monoculture system of Chrysanthemum morifolium cv. Chuju. Int. J. Agric. Biol. 2017, 19, 563–568. [Google Scholar] [CrossRef]
- Song, A.; Zhao, S.; Chen, S.; Jiang, J.; Chen, S.; Li, H.; Chen, Y.; Chen, X.; Fang, W.; Chen, F. The abundance and diversity of soil fungi in continuously monocropped chrysanthemum. Sci. World J. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Xue, C.; Taylor, P.W.J.; Ou, Y.; Wang, B.; Zhao, Y.; Ruan, Y.; Li, R.; Shen, Q. Soil pre-fumigation could effectively improve the disease suppressiveness of biofertilizer to banana Fusarium wilt disease by reshaping the soil microbiome. Biol. Fertil. Soils 2018, 54, 1–14. [Google Scholar] [CrossRef]
- Huang, B.; Li, J.; Wang, Q.; Guo, M.; Yan, D.; Fang, W.; Ren, Z.; Wang, Q.; Ouyang, C.; Li, Y. Effect of soil fumigants on degradation of abamectin and their combination synergistic effect to root-knot nematode. PLoS ONE 2018, 13, e0188245. [Google Scholar] [CrossRef]
- Jones, J.; Noling, J.; Rosskopf, E. Evaluation of various chemical treatments for potential as methyl bromide replacements for disinfestation of soilborne pests in polyethylene-mulched tomatoes. Proc. Fla. State Hortic. Soc. 2003, 116, 151–158. [Google Scholar]
- Li, R.; Shen, Z.; Sun, L.; Zhang, R.; Fu, L.; Deng, X.; Shen, Q. Novel soil fumigation method for suppressing cucumber Fusarium wilt disease associated with soil microflora alterations. Appl. Soil Ecol. 2016, 101, 28–36. [Google Scholar] [CrossRef]
- Zhao, S.; Chen, X.; Deng, S.; Dong, X.; Song, A.; Yao, J.; Fang, W.; Chen, F. The effects of fungicide, soil fumigant, bio-organic fertilizer and their combined application on chrysanthemum Fusarium wilt controlling, soil enzyme activities and microbial properties. Molecules 2016, 21, 526. [Google Scholar] [CrossRef]
- Du, N.; Shi, L.; Yuan, Y.; Sun, J.; Shu, S.; Guo, S. Isolation of a potential biocontrol agent Paenibacillus polymyxa NSY50 from vinegar waste compost and its induction of host defense responses against Fusarium wilt of cucumber. Microbiol. Res. 2017, 202, 1. [Google Scholar] [CrossRef]
- Jambhulkar, P.P.; Sharma, P.; Manokaran, R.; Lakshman, D.K.; Rokadia, P.; Jambhulkar, N. Assessing synergism of combined applications of Trichoderma harzianum and Pseudomonas fluorescens to control blast and bacterial leaf blight of rice. Eur. J. Plant Pathol. 2018, 152, 1–11. [Google Scholar] [CrossRef]
- Cao, Y.; Zhang, Z.; Ling, N.; Yuan, Y.; Zheng, X.; Shen, B.; Shen, Q. Bacillus subtilis SQR 9 can control Fusarium wilt in cucumber by colonizing plant roots. Biol. Fertil. Soils 2011, 47, 495–506. [Google Scholar] [CrossRef]
- Barhate, B.G.; Musmade, N.A.; Nikhate, T.A. Management of Fusarium wilt of tomato by bioagents, fungicides and varietal resistance. Int. J. Plant Prot. 2015, 8, 49–52. [Google Scholar] [CrossRef]
- Schoeman, M.H.; Labuschagne, N.; Calitz, F.J. Efficacy of fungicides, plant resistance activators and biological control agents against guava wilt disease caused by Nalanthamala psidii. S. Afr. J. Plant Soil 2017, 34, 119–124. [Google Scholar] [CrossRef]
- Sharma, S.K.; Ramesh, A.; Sharma, M.P.; Joshi, O.P.; Govaerts, B.; Steenwerth, K.L.; Karlen, D.L. Microbial community structure and diversity as indicators for evaluating soil quality. In Biodiversity, Biofuels, Agroforestry and Conservation Agriculture; Springer: Berlin, Germany, 2010; Volume 5, pp. 317–358. [Google Scholar] [CrossRef]
- Shen, Z.; Wang, D.; Ruan, Y.; Xue, C.; Zhang, J.; Li, R.; Shen, Q. Deep 16S rRNA pyrosequencing reveals a bacterial community associated with banana Fusarium wilt disease suppression induced by bio-organic fertilizer application. PLoS ONE 2014, 9, e98420. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhao, S.; Zhang, K.; Zhao, J.; Jiang, J.; Chen, F.; Fang, W. Evaluation of soil-applied chemical fungicide and biofungicide for control of the Fusarium wilt of chrysanthemum and their effects on rhizosphere soil microbiota. Agriculture 2018, 8, 184. [Google Scholar] [CrossRef]
- Tanja, M.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996. [Google Scholar] [CrossRef]
- Qiong, W.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Kõljalg, U.; Larsson, K.H.; Abarenkov, K.; Nilsson, R.H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E. UNITE: A database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol. 2010, 166, 1063–1068. [Google Scholar] [CrossRef]
- Kandeler, E.; Marschner, P.; Tscherko, D.; Gahoonia, T.S.; Nielsen, N.E. Microbial community composition and functional diversity in the rhizosphere of maize. Plant Soil 2002, 238, 301–312. [Google Scholar] [CrossRef]
- Garbeva, P.; Elsas, J.D.V.; Veen, J.A.V. Rhizosphere microbial community and its response to plant species and soil history. Plant Soil 2008, 302, 19–32. [Google Scholar] [CrossRef]
- Innes, L.; Hobbs, P.J.; Bardgett, R.D. The impacts of individual plant species on rhizosphere microbial communities in soils of different fertility. Biol. Fertil. Soils 2004, 40, 7–13. [Google Scholar] [CrossRef]
- Perla, G.M.V.; Lange, M.; Gleixner, G. The soil microbial community composition and soil microbial carbon uptake are more affected by soil type than by different vegetation types (C3 and C4 plants) and seasonal changes. Eur. Geosci. Union Gen. Assem. 2016, 18. [Google Scholar]
- Coppola, L.; Comitini, F.; Casucci, C.; Milanovic, V.; Monaci, E.; Marinozzi, M.; Taccari, M.; Ciani, M.; Vischetti, C. Fungicides degradation in an organic biomixture: Impact on microbial diversity. New Biotechnol. 2011, 29, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Yang, L.; Zhang, L.; Liu, C.; Han, M. Effects of cultivation ages and modes on microbial diversity in the rhizosphere soil of Panax ginseng. J. Ginseng Res. 2016, 40, 28–37. [Google Scholar] [CrossRef]
- Feld, L.; Hjelmsø, M.H.; Nielsen, M.S.; Jacobsen, A.D.; Rønn, R.; Ekelund, F.; Krogh, P.H.; Strobel, B.W.; Jacobsen, C.S. Pesticide side effects in an agricultural soil ecosystem as measured by amoA expression quantification and bacterial diversity changes. PLoS ONE 2015, 10, e0126080. [Google Scholar] [CrossRef]
- You, C.; Zhang, C.; Kong, F.; Feng, C.; Wang, J. Comparison of the effects of biocontrol agent Bacillus subtilis and fungicide metalaxyl–mancozeb on bacterial communities in tobacco rhizospheric soil. Ecol. Eng. 2016, 91, 119–125. [Google Scholar] [CrossRef]
- Khezri, M.; Ahmadzadeh, M.; Jouzani, G.S.; Behboudi, K.; Ahangaran, A.; Mousivand, M.; Rahimian, H. Characterization of some biofilm-forming Bacillus subtilis strains and evaluation of their biocontrol potential against Fusarium culmorum. J. Plant Pathol. 2011, 93, 373–382. [Google Scholar] [CrossRef]
- Xiong, W.; Guo, S.; Jousset, A.; Zhao, Q.; Wu, H.; Li, R.; Kowalchuk, G.A.; Shen, Q. Bio-fertilizer application induces soil suppressiveness against Fusarium wilt disease by reshaping the soil microbiome. Soil Biol. Biochem. 2017, 114, 238–247. [Google Scholar] [CrossRef]
- Bending, G.D.; Rodríguez-Cruz, M.S.; Lincoln, S.D. Fungicide impacts on microbial communities in soils with contrasting management histories. Chemosphere 2007, 69, 82–88. [Google Scholar] [CrossRef]
- Jung, H.K.; Ryoo, J.C.; Kim, S.D. A Multi-microbial biofungicide for the biological control against several important plant pathogenic fungi. J. Korean Soc. Appl. Biol. Chem. 2005, 48, 40–47. [Google Scholar]
- Pishchik, V.N.; Vorobyev, N.I.; Moiseev, K.G.; Sviridova, O.V.; Surin, V.G. Influence of Bacillus subtilis on the physiological state of wheat and the microbial community of the soil under different rates of nitrogen fertilizers. Eurasian Soil Sci. 2015, 48, 77–84. [Google Scholar] [CrossRef]
- H, S.; Sarniguet, A.; Gazengel, K.; Moënne-Loccoz, Y.; Grundmann, G.L. Rhizosphere bacterial communities associated with disease suppressiveness stages of take-all decline in wheat monoculture. New Phytol. 2009, 184, 694–707. [Google Scholar] [CrossRef]
- Silvalacerda, G.R.; Santana, R.C.; Vicalvicosta, M.C.; Solidônio, E.G.; Sena, K.X.; Lima, G.M.; Araújo, J.M. Antimicrobial potential of actinobacteria isolated from the rhizosphere of the Caatinga biome plant Caesalpinia pyramidalis Tul. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef]
- Trivedi, P.; Delgado-Baquerizo, M.; Trivedi, C.; Hamonts, K.; Anderson, I.C.; Singh, B.K. Keystone microbial taxa regulate the invasion of a fungal pathogen in agro-ecosystems. Soil Biol. Biochem. 2017, 111, 10–14. [Google Scholar] [CrossRef]
- Lu, S.W.; Kroken, S.; Lee, B.N.; Robbertse, B.; Churchill, A.C.; Yoder, O.C.; Turgeon, B.G. A novel class of gene controlling virulence in plant pathogenic ascomycete fungi. Proc. Natl. Acad. Sci. USA 2003, 100, 5980–5985. [Google Scholar] [CrossRef]
- Shen, Z.; Ruan, Y.; Chao, X.; Zhang, J.; Li, R.; Shen, Q. Rhizosphere microbial community manipulated by 2 years of consecutive biofertilizer application associated with banana Fusarium wilt disease suppression. Biol. Fertil. Soils 2015, 51, 1–10. [Google Scholar] [CrossRef]
- Edel-Herman, V.; Lecomte, C. Current Status of Fusarium oxysporum formae speciales and races. Phytopathology 2019, 109, 512–530. [Google Scholar] [CrossRef]
- Lee, S.H.; Ka, J.O.; Cho, J.C. Members of the phylum Acidobacteria are dominant and metabolically active in rhizosphere soil. FEMS Microbiol. Lett. 2008, 285, 263–269. [Google Scholar] [CrossRef]
- Zhang, F.; Huo, Y.; Cobb, A.B.; Luo, G.; Zhou, J.; Yang, G.; Wilson, G.W.T.; Zhang, Y. Trichoderma biofertilizer links to altered soil chemistry, altered microbial communities, and improved grassland biomass. Front. Microbiol. 2018, 9, 848. [Google Scholar] [CrossRef]
- Lozano-Tovar, M.D.; Garrido-Jurado, I.; Quesada-Moraga, E.; Raya-Ortega, M.C.; Trapero-Casas, A. Metarhizium brunneum and Beauveria bassiana release secondary metabolites with antagonistic activity against verticillium dahliae and phytophthora megasperma olive pathogens. Crop Prot. 2017, 100, 186–195. [Google Scholar] [CrossRef]
| Microbe | Treatment | Phylum (×104) | Class (×104) | Order (×104) | Family (×104) | Genus (×104) |
|---|---|---|---|---|---|---|
| Bacteria | Control-2015 | 6.16 ± 0.01 a | 5.88 ± 0.01 a | 5.38 ± 0.05 a | 4.94 ± 0.05 a | 3.54 ± 0.02 a |
| DZ-2015 | 5.99 ± 0.07 bc | 5.52 ± 0.03 c | 4.71 ± 0.04 d | 4.13 ± 0.03 d | 2.72 ± 0.03 d | |
| BF-2015 | 6.02 ± 0.01 b | 5.63 ± 0.03 bc | 5.06 ± 0.01 c | 4.40 ± 0.01 c | 3.01 ± 0.03 bc | |
| Control-2016 | 6.04 ± 0.01 b | 5.75 ± 0.07 b | 5.26 ± 0.05 ab | 4.72 ± 0.04 b | 3.21 ± 0.03 b | |
| DZ-2016 | 5.92 ± 0.03 c | 5.53 ± 0.04 c | 4.77 ± 0.04 d | 4.09 ± 0.04 d | 2.47 ± 0.02 e | |
| BF-2016 | 6.00 ± 0.02 bc | 5.71 ± 0.03 b | 5.16 ± 0.04 bc | 4.67 ± 0.04 b | 2.89 ± 0.03 cd | |
| Average | 6.02 | 5.67 | 5.06 | 4.49 | 2.97 | |
| Fungi | Control-2015 | 4.76 ± 0.09 a | 4.30 ± 0.05 b | 4.30 ± 0.04 b | 4.04 ± 0.04 a | 4.01 ± 0.04 a |
| DZ-2015 | 4.08 ± 0.18 b | 3.13 ± 0.12 c | 3.08 ± 0.13 c | 2.85 ± 0.07 b | 2.67 ± 0.21 b | |
| BF-2015 | 4.39 ± 0.22 ab | 3.09 ± 0.13 c | 3.12 ± 0.12 c | 2.96 ± 0.13 b | 2.91 ± 0.13 b | |
| Control-2016 | 4.58 ± 0.03 a | 4.70 ± 0.08 a | 4.70 ± 0.08 a | 4.08 ± 0.05 a | 4.00 ± 0.04 a | |
| DZ-2016 | 3.29 ± 0.11 c | 3.05 ± 0.14 c | 3.05 ± 0.14 c | 2.97 ± 0.16 b | 2.87 ± 0.16 b | |
| BF-2016 | 3.17 ± 0.05 c | 3.08 ± 0.07 c | 3.08 ± 0.07 c | 2.82 ± 0.07 b | 2.76 ± 0.08 b | |
| Average | 4.04 | 3.56 | 3.56 | 3.29 | 3.20 |
| Soil Microbiota | Treatment | Diversity (Shannon) | Richness (Chao1) | Faith’s PD | Evenness |
|---|---|---|---|---|---|
| Bacteria | Control-2015 | 9.625 ± 0.079 c | 4051.292 ± 28.873 b | 296.633 ± 1.903 b | 0.996 ± 0.000 b |
| DZ-2015 | 9.704 ± 0.033 c | 3915.980 ± 22.684 c | 277.514 ± 1.730 c | 0.996 ± 0.000 b | |
| BF-2015 | 10.216 ± 0.053 a | 4518.900 ± 42.726 a | 320.292 ± 8.509 a | 0.998 ± 0.000 a | |
| Control-2016 | 9.577 ± 0.040 c | 3953.088 ± 28.553 bc | 295.582 ± 4.951 b | 0.995 ± 0.000 c | |
| DZ-2016 | 8.814 ± 0.025 d | 3170.464 ± 29.194 d | 230.511 ± 0.851 d | 0.992 ± 0.000 d | |
| BF-2016 | 9.979 ± 0.015 b | 4020.658 ± 9.112 bc | 294.688 ± 0.200 b | 0.997 ± 0.000 a | |
| Fungi | Control-2015 | 6.000 ± 0.314 ab | 811.333 ± 29.525 ab | 184.107 ± 15.443 a | 0.952 ± 0.018 a |
| DZ-2015 | 5.720 ± 0.271 bc | 751.791 ± 34.465 bc | 186.743 ± 6.017 a | 0.933 ± 0.010 a | |
| BF-2015 | 6.551 ± 0.115 a | 863.942 ± 11.664 a | 217.206 ± 5.406 a | 0.968 ± 0.003 a | |
| Control-2016 | 6.404 ± 0.080 ab | 787.092 ± 13.298 ab | 192.826 ± 5.303 a | 0.959 ± 0.003 a | |
| DZ-2016 | 6.229 ± 0.122 ab | 800.751 ± 18.828 ab | 183.855 ± 6.544 a | 0.963 ± 0.004 a | |
| BF-2016 | 5.041 ± 0.335 c | 702.647 ± 36.261 c | 192.406 ± 18.111 a | 0.882 ± 0.023 b |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.; Zhao, J.; Jiang, J.; Chen, S.; Guan, Z.; Chen, F.; Fang, W.; Zhao, S. Assessing the Influence of Fumigation and Bacillus Subtilis-Based Biofungicide on the Microbiome of Chrysanthemum Rhizosphere. Agriculture 2019, 9, 255. https://doi.org/10.3390/agriculture9120255
Chen H, Zhao J, Jiang J, Chen S, Guan Z, Chen F, Fang W, Zhao S. Assessing the Influence of Fumigation and Bacillus Subtilis-Based Biofungicide on the Microbiome of Chrysanthemum Rhizosphere. Agriculture. 2019; 9(12):255. https://doi.org/10.3390/agriculture9120255
Chicago/Turabian StyleChen, Huijie, Jiamiao Zhao, Jing Jiang, Sumei Chen, Zhiyong Guan, Fadi Chen, Weimin Fang, and Shuang Zhao. 2019. "Assessing the Influence of Fumigation and Bacillus Subtilis-Based Biofungicide on the Microbiome of Chrysanthemum Rhizosphere" Agriculture 9, no. 12: 255. https://doi.org/10.3390/agriculture9120255
APA StyleChen, H., Zhao, J., Jiang, J., Chen, S., Guan, Z., Chen, F., Fang, W., & Zhao, S. (2019). Assessing the Influence of Fumigation and Bacillus Subtilis-Based Biofungicide on the Microbiome of Chrysanthemum Rhizosphere. Agriculture, 9(12), 255. https://doi.org/10.3390/agriculture9120255






