Transcriptome Analysis of Submergence Stress in Rice Provides Insights into the Molecular Mechanism of Rice Response to Flooding and the Roles of OsEXPB3 Under Submergence
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Submergence Stress Treatments and Sample Collection
2.3. RNA-Seq Analysis
2.4. Quantitative Real-Time PCR (qRT-PCR) Analysis
2.5. Phylogenetic, Gene Structure, and Promoter Cis-Element Analyses of the OsEXPB Gene Family
2.6. Gene Editing and Analysis of OsEXPB3
2.7. Determination of Endogenous Hormone Levels in Wild-Type and osexpb3 Mutants
2.8. Statistical Analysis
3. Results
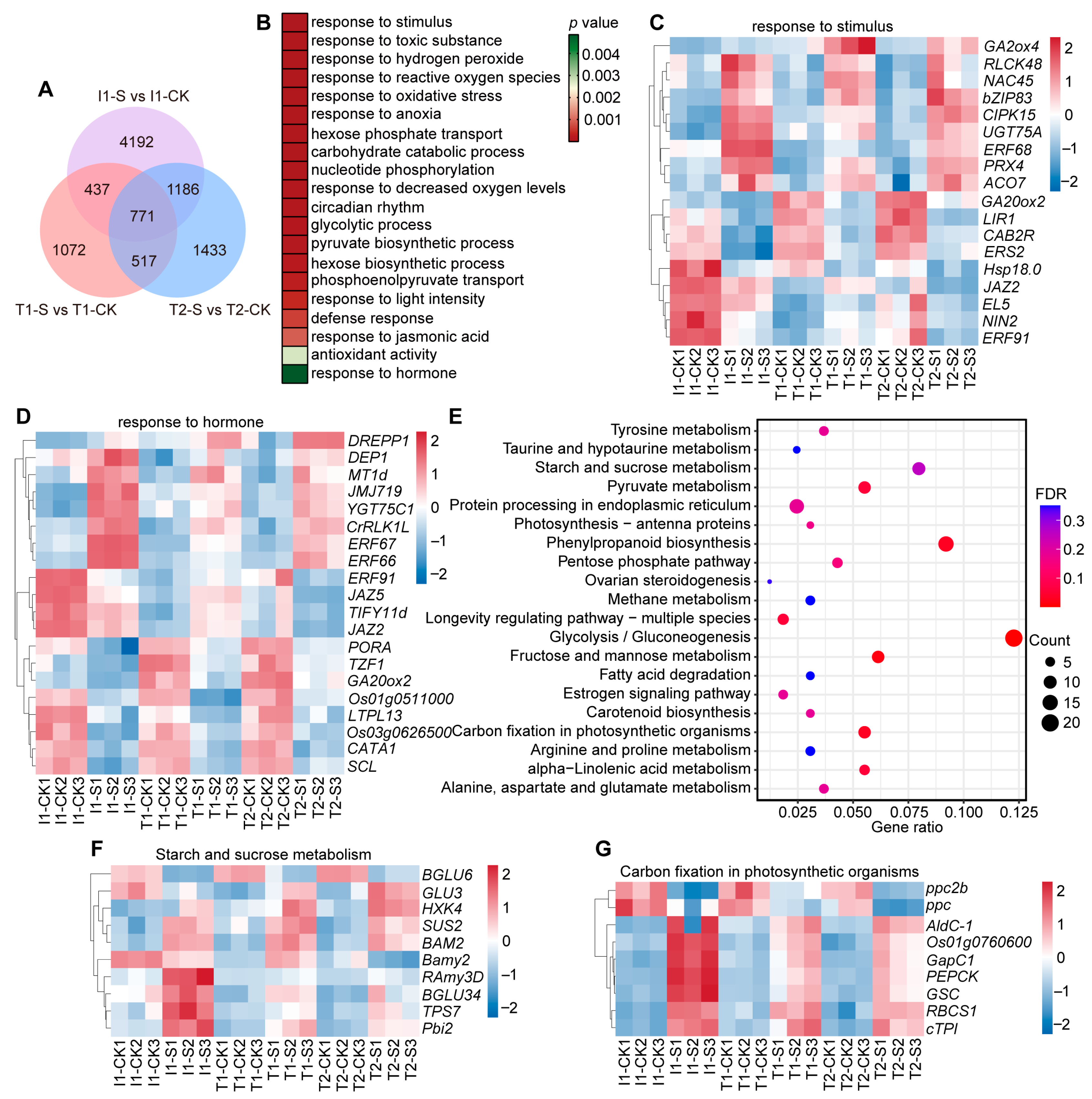
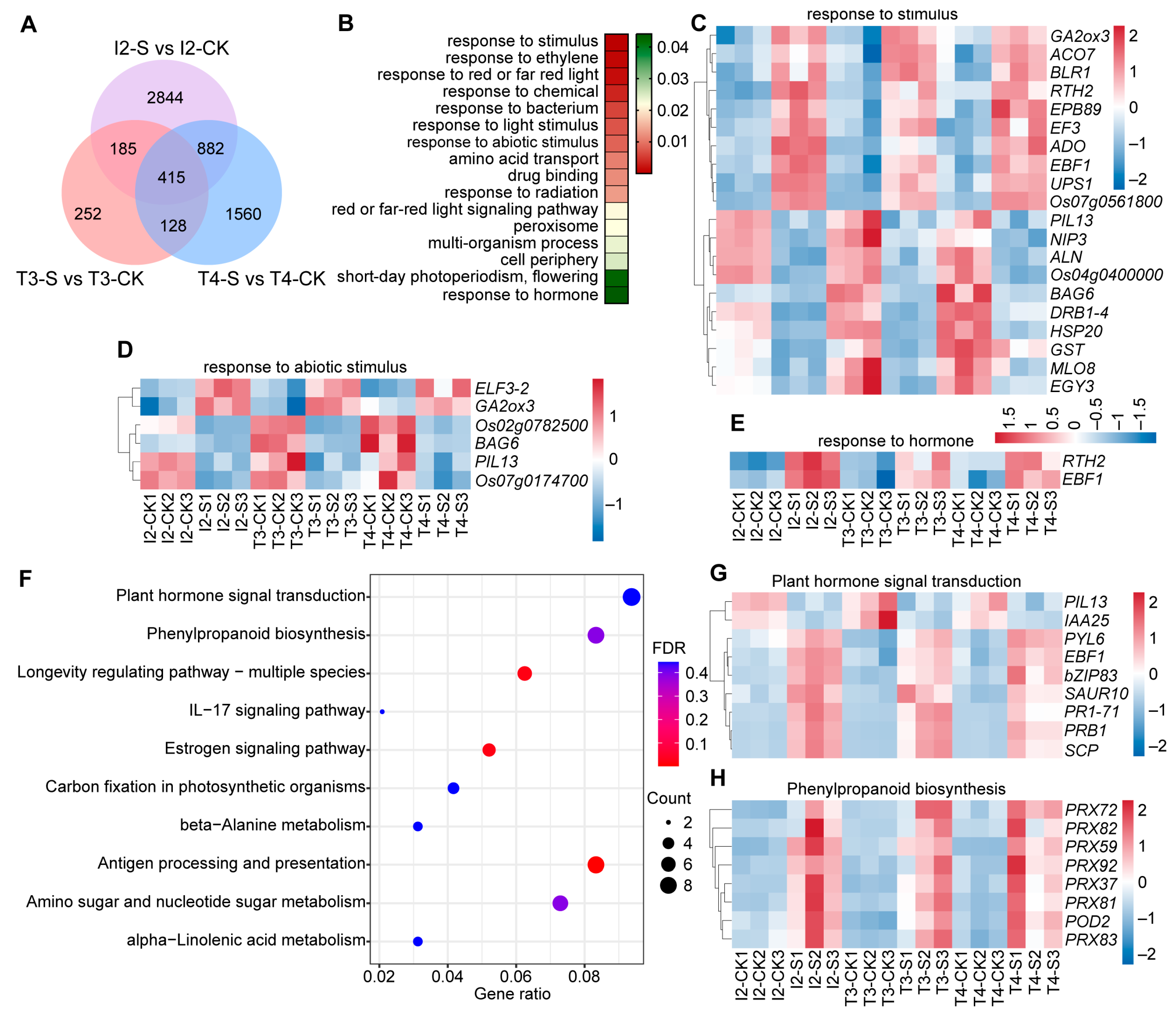
3.1. Analysis of DEGs in Japonica and Indica Rice in Response to Submergence Stress
3.2. Functional Annotation and Enrichment Analysis of DEGs in Japonica Rice
3.3. Functional Annotation and Enrichment Analysis of DEGs in Indica Rice
3.4. Identification and Expression Analysis of Candidate Genes for Submergence Tolerance
3.5. Gene Structure and Promoter Cis-Element Analysis of OsEXPB Genes
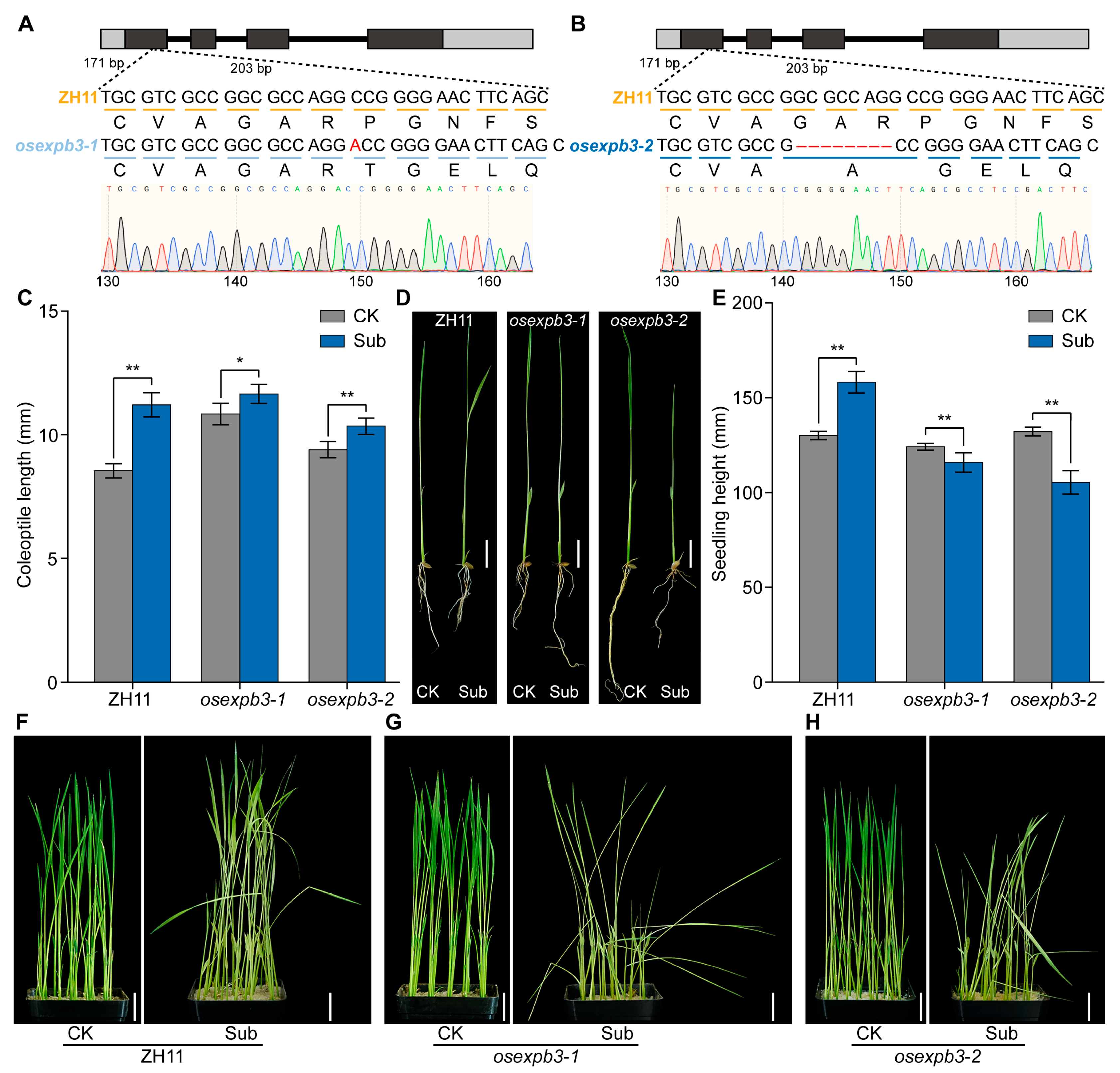
3.6. Comparison of Submergence Tolerance Between Wild-Type and osexpb3 Mutants
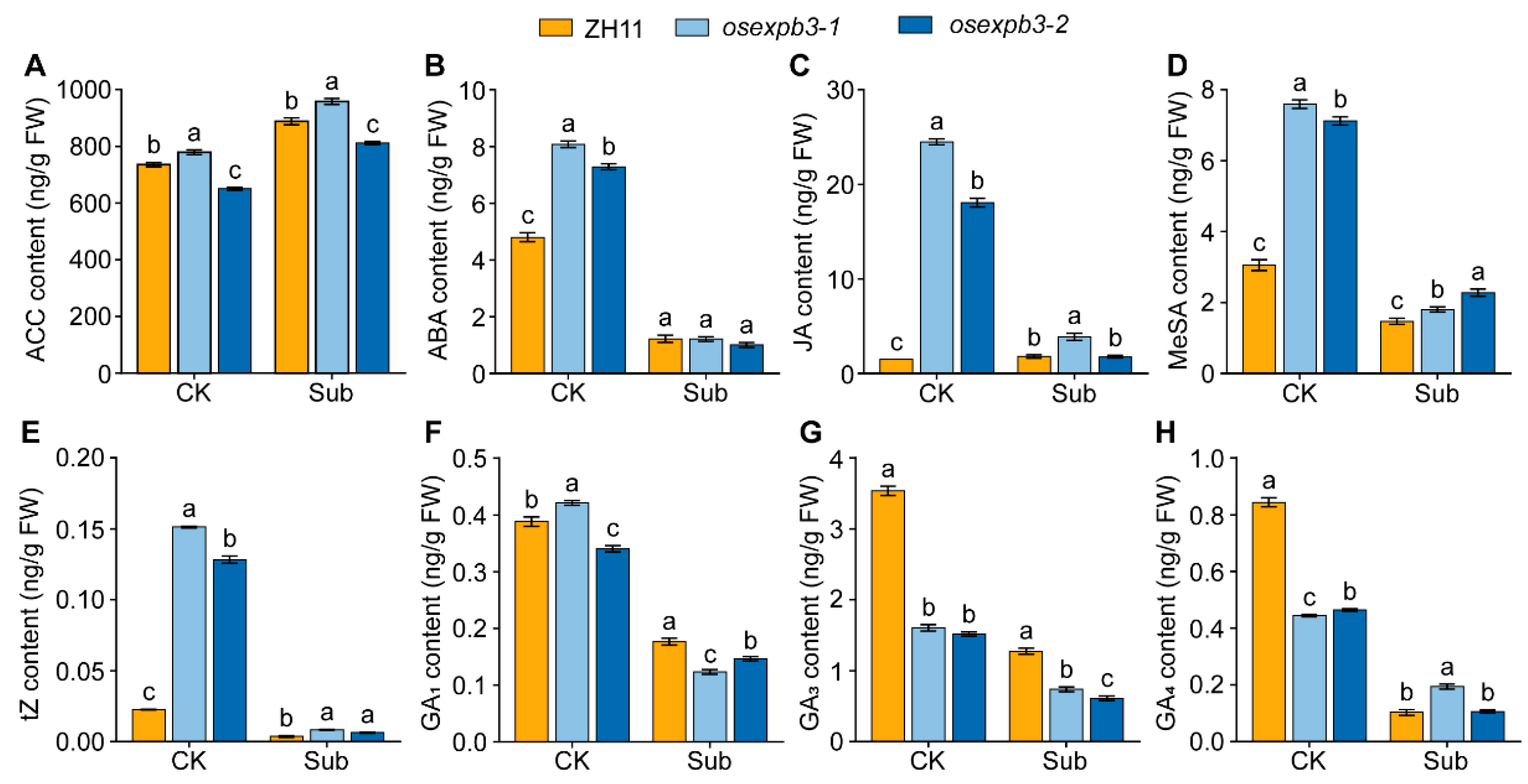
3.7. Comparison of Hormone Levels Between the Wild Type and osexpb3 Mutants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ETH | Ethylene |
| GA | Gibberellin |
| JA | Jasmonic acid |
| SA | Salicylic acid |
| ACC | 1-aminocyclopropane-1-carboxylic acid |
| MeSA | Methyl salicylate |
| tZ | Trans-zeatin |
| O2 | Oxygen |
| CO2 | Carbon dioxide |
| ATP | Adenosine triphosphate |
| EXP | Expansin |
| T-type | Tolerant type |
| I-type | Intolerant type |
| CK | Control group |
| Sub | Submergence |
| TPM | Transcripts Per Million |
| DEGs | Differentially expressed genes |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| CDS | Coding sequences |
| qRT-PCR | Quantitative Real-Time PCR |
References
- Chan, F.K.S.; Paszkowski, A.; Wang, Z.L.; Lu, X.H.; Mitchell, G.; Tran, D.D.; Warner, J.; Li, J.F.; Chen, Y.D.; Li, N.; et al. Building resilience in Asian mega-deltas. Nat. Rev. Earth Environ. 2024, 5, 522–537. [Google Scholar] [CrossRef]
- Han, J.C.; Zhang, Z.; Xu, J.L.; Chen, Y.; Jägermeyr, J.; Cao, J.; Luo, Y.C.; Cheng, F.; Zhuang, H.M.; Wu, H.Q.; et al. Threat of low-frequency high-intensity floods to global cropland and crop yields. Nat. Sustain. 2024, 7, 994–1006. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations (FAO). The Impact of Disasters on Agriculture and Food Security; FAO: Rome, Italy, 2023; Available online: https://www.fao.org/publications/fao-flagship-publications/the-impact-of-disasters-on-agriculture-and-food-security/en (accessed on 18 September 2025).
- International Rice Research Institute (IRRI). Impact of Flooding-Disaster on Agricultural Production; IRRI: Los Baños, Philippines, 2024; Available online: https://www.irri.org (accessed on 18 September 2025).
- Winkel, A.; Colmer, T.D..; Ismail, A.M.; Pedersen, O. Internal aeration of paddy field rice (Oryza sativa) during complete submergence---importance of light and floodwater O2. New Phytol. 2013, 197, 1193–1203. [Google Scholar] [CrossRef] [PubMed]
- Renziehausen, T.; Frings, S.; Schmidt-Schippers, R. ‘Against all floods’: Plant adaptation to flooding stress and combined abiotic stresses. Plant J. 2024, 117, 1836–1855. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, G.; Cui, Z.; Kong, X.; Yu, X.; Gui, R.; Han, Y.; Li, Z.; Lang, H.; Hua, Y.; et al. Regain flood adaptation in rice through a 14-3-3 protein OsGF14h. Nat. Commun. 2022, 13, 5664. [Google Scholar] [CrossRef]
- Yamauchi, T.; Nakazono, M. Modeling-based age-dependent analysis reveals the net patterns of ethylene-dependent and -independent aerenchyma formation in rice and maize roots. Plant Sci. 2022, 321, 111340. [Google Scholar] [CrossRef]
- Reynoso, M.A.; Borowsky, A.T.; Pauluzzi, G.C.; Yeung, E.; Zhang, J.; Formentin, E.; Velasco, J.; Cabanlit, S.; Duvenjian, C.; Prior, M.J.; et al. Gene regulatory networks shape developmental plasticity of root cell types under water extremes in rice. Dev. Cell 2022, 57, 1177–1192.e6. [Google Scholar] [CrossRef]
- Xu, K.; Xu, X.; Fukao, T.; Canlas, P.; Maghirang-Rodriguez, R.; Heuer, S.; Ismail, A.M.; Bailey-Serres, J.; Ronald, P.C.; Mackill, D.J. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 2006, 442, 705–708. [Google Scholar] [CrossRef]
- Hussain, W.; Anumalla, M.; Ismail, A.M.; Walia, H.; Singh, V.K.; Kohli, A.; Bhosale, S.; Bhardwaj, H. Revisiting FR13A for submergence tolerance: Beyond the SUB1A gene. J. Exp. Bot. 2024, 75, 5477–5483. [Google Scholar] [CrossRef]
- Kuroha, T.; Nagai, K.; Gamuyao, R.; Wang, D.R.; Furuta, T.; Nakamori, M.; Kitaoka, T.; Adachi, K.; Minami, A.; Mori, Y.; et al. Ethylene-gibberellin signaling underlies adaptation of rice to periodic flooding. Science 2018, 361, 181–186. [Google Scholar] [CrossRef]
- Nagai, K.; Mori, Y.; Ishikawa, S.; Furuta, T.; Gamuyao, R.; Niimi, Y.; Hobo, T.; Fukuda, M.; Kojima, M.; Takebayashi, Y.; et al. Antagonistic regulation of the gibberellic acid response during stem growth in rice. Nature 2020, 584, 109–114. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef]
- Waadt, R.; Seller, C.A.; Hsu, P.K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef] [PubMed]
- Voesenek, L.A.C.J.; Bailey-Serres, J. Flood adaptive traits and processes: An overview. New Phytol. 2015, 206, 57–73. [Google Scholar] [CrossRef] [PubMed]
- Fukao, T.; Bailey-Serres, J. Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc. Natl. Acad. Sci. USA 2008, 105, 16814–16819. [Google Scholar] [CrossRef]
- Sharma, N.; Dang, T.M.; Singh, N.; Ruzicic, S.; Mueller-Roeber, B.; Baumann, U.; Heuer, S. Allelic variants of OsSUB1A cause differential expression of transcription factor genes in response to submergence in rice. Rice 2018, 11, 2. [Google Scholar] [CrossRef]
- Hattori, Y.; Nagai, K.; Furukawa, S.; Song, X.J.; Kawano, R.; Sakakibara, H.; Wu, J.; Matsumoto, T.; Yoshimura, A.; Kitano, H.; et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 2009, 460, 1026–1030. [Google Scholar] [CrossRef]
- Saika, H.; Okamoto, M.; Miyoshi, K.; Kushiro, T.; Shinoda, S.; Jikumaru, Y.; Fujimoto, M.; Arikawa, T.; Takahashi, H.; Ando, M.; et al. Ethylene promotes submergence-induced expression of OsABA8ox1, a gene that encodes ABA 8′-hydroxylase in rice. Plant Cell Physiol. 2007, 48, 287–298. [Google Scholar] [CrossRef]
- Kretzschmar, T.; Pelayo, M.A.; Trijatmiko, K.R.; Gabunada, L.F.; Alam, R.; Jimenez, R.; Mendioro, M.S.; Slamet-Loedin, I.H.; Sreenivasulu, N.; Bailey-Serres, J.; et al. A trehalose-6-phosphate phosphatase enhances anaerobic germination tolerance in rice. Nat. Plants 2015, 1, 15124. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Diffuse Growth of Plant Cell Walls. Plant Physiol. 2018, 176, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Brasileiro, A.C.M.; Lacorte, C.; Pereira, B.M.; Oliveira, T.N.; Ferreira, D.S.; Mota, A.P.Z.; Saraiva, M.A.P.; Araujo, A.C.G.; Silva, L.P.; Guimaraes, P.M. Ectopic expression of an expansin-like B gene from wild Arachis enhances tolerance to both abiotic and biotic stresses. Plant J. 2021, 107, 1681–1696. [Google Scholar] [CrossRef]
- Wang, Z.; Cao, J.; Lin, N.; Li, J.; Wang, Y.; Liu, W.; Yao, W.; Li, Y. Origin, Evolution, and Diversification of the Expansin Family in Plants. Int. J. Mol. Sci. 2024, 25, 11814. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Microbial Expansins. Annu. Rev. Microbiol. 2017, 71, 479–497. [Google Scholar] [CrossRef]
- Wang, S.; Yang, H.; Mei, J.; Liu, X.; Wen, Z.; Zhang, L.; Xu, Z.; Zhang, B.; Zhou, Y. Rice Homeobox Protein KNAT7 Integrates the Pathways Regulating Cell Expansion and Wall Stiffness. Plant Physiol. 2019, 181, 669–682. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Voesenek, L.A. Flooding stress: Acclimations and genetic diversity. Annu. Rev. Plant Biol. 2008, 59, 313–339. [Google Scholar] [CrossRef]
- Liu, S.Q.; Ai, H.; Duan, S.M.; Li, R.N.; Owusu, E.A.; Lv, L.L.; Sun, Z.L.; Xu, D.C.; Zhang, M.Z.; Zhou, A.F.; et al. Integrated analysis of laboratory and field experiments to screen rice germplasm for submergence tolerance across various growth stages. Field Crops Res. 2026, 336, 110218. [Google Scholar] [CrossRef]
- de Sena Brandine, G.; Smith, A.D. Falco: High-speed FastQC emulation for quality control of sequencing data. F1000Research 2019, 8, 1874. [Google Scholar] [CrossRef] [PubMed]
- Gill, N.; Dhillon, B. RNA-seq Data Analysis for Differential Expression. Methods Mol. Biol. 2022, 2391, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, S.; Li, W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics 2012, 28, 2184–2185. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Pertea, G.; Pertea, M. GFF Utilities: GffRead and GffCompare. F1000Research 2020, 9, 304. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids. Res. 2012, 40, D109–D114. [Google Scholar] [CrossRef]
- Ding, R.; Qu, Y.; Wu, C.H.; Vijay-Shanker, K. Automatic gene annotation using GO terms from cellular component domain. BMC Med. Inform. Decis. Mak. 2018, 18, 119. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Vij, S.; Giri, J.; Dansana, P.K.; Kapoor, S.; Tyagi, A.K. The receptor-like cytoplasmic kinase (OsRLCK) gene family in rice: Organization, phylogenetic relationship, and expression during development and stress. Mol. Plant 2008, 1, 732–750. [Google Scholar] [CrossRef] [PubMed]
- Nijhawan, A.; Jain, M.; Tyagi, A.K.; Khurana, J.P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008, 146, 333–350. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Hu, H.; Ren, H.; Kong, Y.; Lin, H.; Guo, J.; Wang, L.; He, Y.; Ding, X.; Grabsztunowicz, M.; et al. LIGHT-INDUCED RICE1 Regulates Light-Dependent Attachment of LEAF-TYPE FERREDOXIN-NADP+ OXIDOREDUCTASE to the Thylakoid Membrane in Rice and Arabidopsis. Plant Cell 2016, 28, 712–728. [Google Scholar] [CrossRef] [PubMed]
- Tseng, T.S.; Rajendran, S.K.; Liu, Y.H.; Wu, S.J.; Lu, C.A.; Yeh, C.H. Overexpression of OsHsp 18.0 in rice enhanced tolerance to heavy metal stress. Plant Cell Rep. 2023, 42, 1841–1843. [Google Scholar] [CrossRef]
- He, Y.; Sun, S.; Zhao, J.; Huang, Z.; Peng, L.; Huang, C.; Tang, Z.; Huang, Q.; Wang, Z. UDP-glucosyltransferase OsUGT75A promotes submergence tolerance during rice seed germination. Nat. Commun. 2023, 14, 2296. [Google Scholar] [CrossRef] [PubMed]
- Ye, N.H.; Wang, F.Z.; Shi, L.; Chen, M.X.; Cao, Y.Y.; Zhu, F.Y.; Wu, Y.Z.; Xie, L.J.; Liu, T.Y.; Su, Z.Z.; et al. Natural variation in the promoter of rice calcineurin B-like protein10 (OsCBL10) affects flooding tolerance during seed germination among rice subspecies. Plant J. 2018, 94, 612–625. [Google Scholar] [CrossRef]
- Ye, H.; Du, H.; Tang, N.; Li, X.; Xiong, L. Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol. Biol. 2009, 71, 291–305. [Google Scholar] [CrossRef]
- Hsiao, P.Y.; Zeng, C.Y.; Shih, M.C. Group VII ethylene response factors forming distinct regulatory loops mediate submergence responses. Plant Physiol. 2024, 194, 1745–1763. [Google Scholar] [CrossRef]
- Iwai, T.; Miyasaka, A.; Seo, S.; Ohashi, Y. Contribution of ethylene biosynthesis for resistance to blast fungus infection in young rice plants. Plant Physiol. 2006, 142, 1202–1215. [Google Scholar] [CrossRef] [PubMed]
- Todaka, D.; Nakashima, K.; Maruyama, K.; Kidokoro, S.; Osakabe, Y.; Ito, Y.; Matsukura, S.; Fujita, Y.; Yoshiwara, K.; Ohme-Takagi, M.; et al. Rice phytochrome-interacting factor-like protein OsPIL1 functions as a key regulator of internode elongation and induces a morphological response to drought stress. Proc. Natl. Acad. Sci. USA 2012, 109, 15947–15952. [Google Scholar] [CrossRef]
- Wei, J.; Chen, J.; Zhang, Z.; Ban, Y.; Guo, J.; Dong, L.; Feng, Z. Toxicity and Glutathione S-Transferase-Catalyzed Metabolism of R-/S-Metolachlor in Rice. J. Agric. Food Chem. 2024, 72, 25001–25014. [Google Scholar] [CrossRef] [PubMed]
- Ning, Y.; Shi, X.; Wang, R.; Fan, J.; Park, C.H.; Zhang, C.; Zhang, T.; Ouyang, X.; Li, S.; Wang, G.L. OsELF3-2, an Ortholog of Arabidopsis ELF3, Interacts with the E3 Ligase APIP6 and Negatively Regulates Immunity against Magnaporthe oryzae in Rice. Mol. Plant 2015, 8, 1679–1682. [Google Scholar] [CrossRef]
- Tong, H.; Xiao, Y.; Liu, D.; Gao, S.; Liu, L.; Yin, Y.; Jin, Y.; Qian, Q.; Chu, C. Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell 2014, 26, 4376–4393. [Google Scholar] [CrossRef]
- Ma, F.; Yang, X.; Shi, Z.; Miao, X. Novel crosstalk between ethylene- and jasmonic acid-pathway responses to a piercing-sucking insect in rice. New Phytol. 2020, 225, 474–487. [Google Scholar] [CrossRef]
- Zhao, Y.; Cao, X.; Zhong, W.; Zhou, S.; Li, Z.; An, H.; Liu, X.; Wu, R.; Bohora, S.; Wu, Y.; et al. A viral protein orchestrates rice ethylene signaling to coordinate viral infection and insect vector-mediated transmission. Mol. Plant 2022, 15, 689–705. [Google Scholar] [CrossRef]
- Jain, M.; Kaur, N.; Garg, R.; Thakur, J.K.; Tyagi, A.K.; Khurana, J.P. Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct. Integr. Genom. 2006, 6, 47–59. [Google Scholar] [CrossRef]
- Tian, X.; Wang, Z.; Li, X.; Lv, T.; Liu, H.; Wang, L.; Niu, H.; Bu, Q. Characterization and Functional Analysis of Pyrabactin Resistance-Like Abscisic Acid Receptor Family in Rice. Rice 2015, 8, 28. [Google Scholar] [CrossRef]
- Huang, X.; Lu, Z.; Zhai, L.; Li, N.; Yan, H. The Small Auxin-Up RNA SAUR10 Is Involved in the Promotion of Seedling Growth in Rice. Plants 2023, 12, 3880. [Google Scholar] [CrossRef]
- Tognolli, M.; Penel, C.; Greppin, H.; Simon, P. Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 2002, 288, 129–138. [Google Scholar] [CrossRef]
- Lee, Y.; Choi, D. Biochemical properties and localization of the beta-expansin OsEXPB3 in rice (Oryza sativa L.). Mol. Cells 2005, 20, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Polko, J.K.; Kieber, J.J. 1-Aminocyclopropane 1-Carboxylic Acid and Its Emerging Role as an Ethylene-Independent Growth Regulator. Front. Plant Sci. 2019, 10, 1602. [Google Scholar] [CrossRef] [PubMed]
- Yim, H.K.; Lim, M.N.; Lee, S.E.; Lim, J.; Lee, Y.; Hwang, Y.S. Hexokinase-mediated sugar signaling controls expression of the calcineurin B-like interacting protein kinase 15 gene and is perturbed by oxidative phosphorylation inhibition. J. Plant Physiol. 2012, 169, 1551–1558. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Zheng, Z.; Bao, Y.; Zhao, X.; Lv, J.; Tang, C.; Wang, N.; Liang, Z.; Li, H.; Xiang, J.; et al. Identification and Regulation of Hypoxia-Tolerant and Germination-Related Genes in Rice. Int. J. Mol. Sci. 2024, 25, 2177. [Google Scholar] [CrossRef]
- Minami, A.; Yano, K.; Gamuyao, R.; Nagai, K.; Kuroha, T.; Ayano, M.; Nakamori, M.; Koike, M.; Kondo, Y.; Niimi, Y.; et al. Time-Course Transcriptomics Analysis Reveals Key Responses of Submerged Deepwater Rice to Flooding. Plant Physiol. 2018, 176, 3081–3102. [Google Scholar] [CrossRef]
- Loreti, E.; Perata, P. The Many Facets of Hypoxia in Plants. Plants 2020, 9, 745. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.C.; Lee, W.J.; Zeng, C.Y.; Chou, M.Y.; Lin, T.J.; Lin, C.S.; Ho, M.C.; Shih, M.C. SUB1A-1 anchors a regulatory cascade for epigenetic and transcriptional controls of submergence tolerance in rice. Pnas Nexus 2023, 2, 229. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhu, K.; Xia, H.; Chen, L.; Chen, K. Comparative proteomic analysis of indica and japonica rice varieties. Genet. Mol. Biol. 2014, 37, 652–661. [Google Scholar] [CrossRef] [PubMed]
- Mickelbart, M.V.; Hasegawa, P.M.; Bailey-Serres, J. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat. Rev. Genet. 2015, 16, 237–251. [Google Scholar] [CrossRef]
- Long, W.; He, Q.; Wang, Y.; Wang, Y.; Wang, J.; Yuan, Z.; Wang, M.; Chen, W.; Luo, L.; Luo, L.; et al. Genome evolution and diversity of wild and cultivated rice species. Nat. Commun. 2024, 15, 9994. [Google Scholar] [CrossRef]
- Singh, P.; Sinha, A.K. A Positive Feedback Loop Governed by SUB1A1 Interaction with MITOGEN-ACTIVATED PROTEIN KINASE3 Imparts Submergence Tolerance in Rice. Plant Cell 2016, 28, 1127–1143. [Google Scholar] [CrossRef]
- Locke, A.M.; Barding, G.A., Jr.; Sathnur, S.; Larive, C.K.; Bailey-Serres, J. Rice SUB1A constrains remodelling of the transcriptome and metabolome during submergence to facilitate post-submergence recovery. Plant Cell Environ. 2018, 41, 721–736. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Sun, Z.; Lv, L.; Huang, X.; Fan, H.; Li, M.; Shi, B.; Gao, Y.; Ai, H.; Xu, D.; et al. Transcriptome Analysis of Submergence Stress in Rice Provides Insights into the Molecular Mechanism of Rice Response to Flooding and the Roles of OsEXPB3 Under Submergence. Agriculture 2025, 15, 2556. https://doi.org/10.3390/agriculture15242556
Liu S, Sun Z, Lv L, Huang X, Fan H, Li M, Shi B, Gao Y, Ai H, Xu D, et al. Transcriptome Analysis of Submergence Stress in Rice Provides Insights into the Molecular Mechanism of Rice Response to Flooding and the Roles of OsEXPB3 Under Submergence. Agriculture. 2025; 15(24):2556. https://doi.org/10.3390/agriculture15242556
Chicago/Turabian StyleLiu, Shengqin, Zhanglun Sun, Liangliang Lv, Xinyu Huang, Huailin Fan, Mengya Li, Boxin Shi, Ya Gao, Hao Ai, Dachao Xu, and et al. 2025. "Transcriptome Analysis of Submergence Stress in Rice Provides Insights into the Molecular Mechanism of Rice Response to Flooding and the Roles of OsEXPB3 Under Submergence" Agriculture 15, no. 24: 2556. https://doi.org/10.3390/agriculture15242556
APA StyleLiu, S., Sun, Z., Lv, L., Huang, X., Fan, H., Li, M., Shi, B., Gao, Y., Ai, H., Xu, D., Feng, T., & Huang, X. (2025). Transcriptome Analysis of Submergence Stress in Rice Provides Insights into the Molecular Mechanism of Rice Response to Flooding and the Roles of OsEXPB3 Under Submergence. Agriculture, 15(24), 2556. https://doi.org/10.3390/agriculture15242556





