Integrated Transcriptomic and Metabolomic Analyses Implicate Key Genes and Metabolic Pathways in Maize Lodging Resistance
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Measurement of Phenotypic and Physiological Parameters
2.3. Total RNA Extraction, Gene Expression Analysis and Gene Functional Annotation
2.4. Differential Gene Expression and Enrichment Analysis
2.5. Quantitative Reverse Transcription Polymerase Chain Reaction
2.6. Metabolite Extraction and Analysis
2.7. Transcriptome and Metabolome Analysis
2.8. Statistical Analysi
3. Results
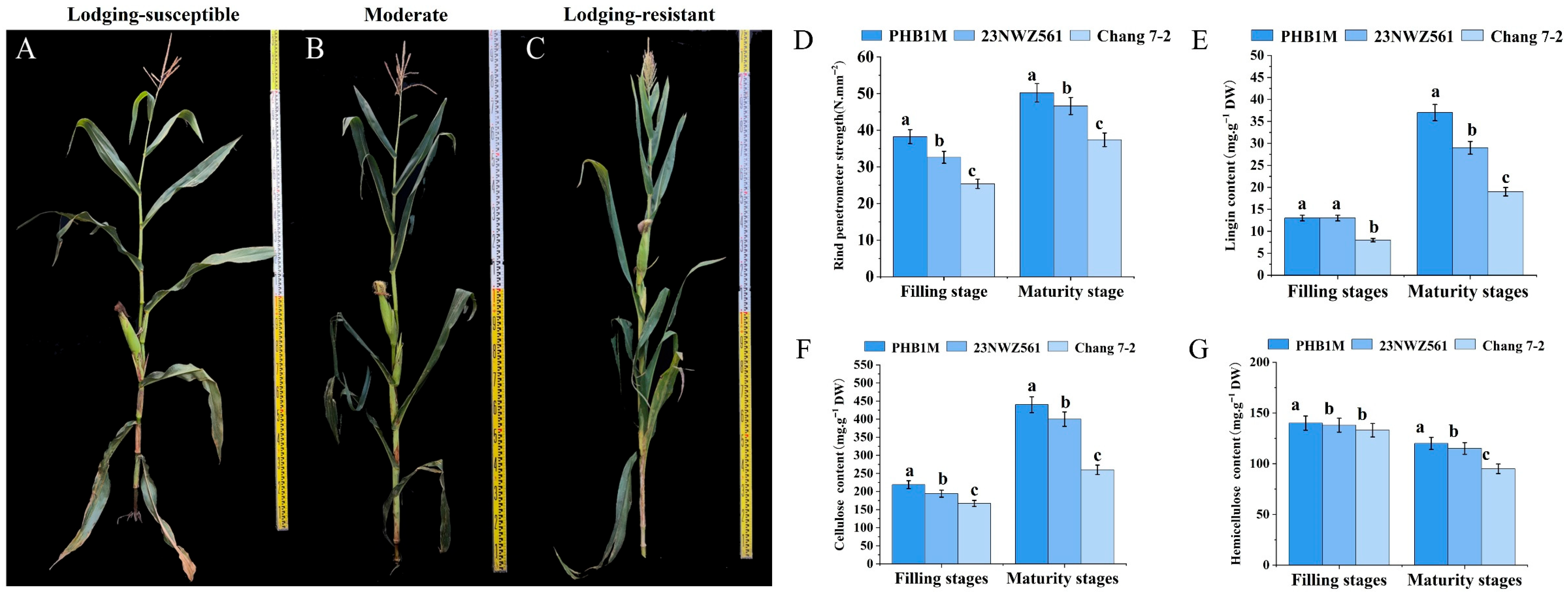
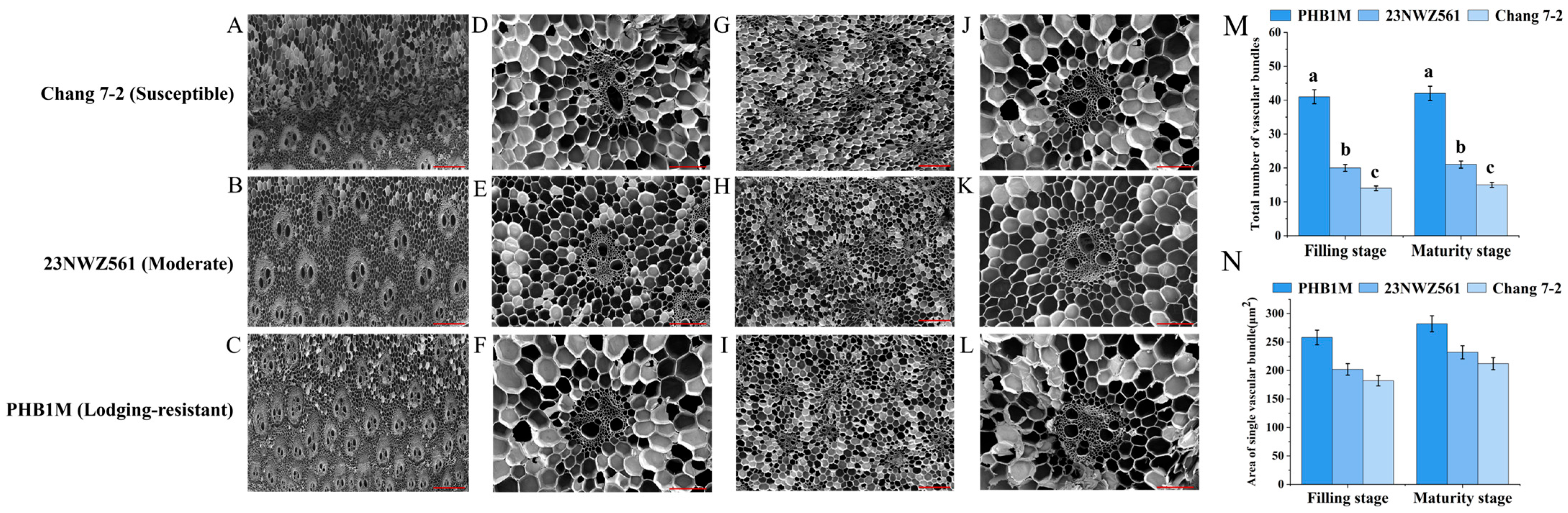
3.1. Phenotypic and Physiological Parameters of the Three Maize Varieties
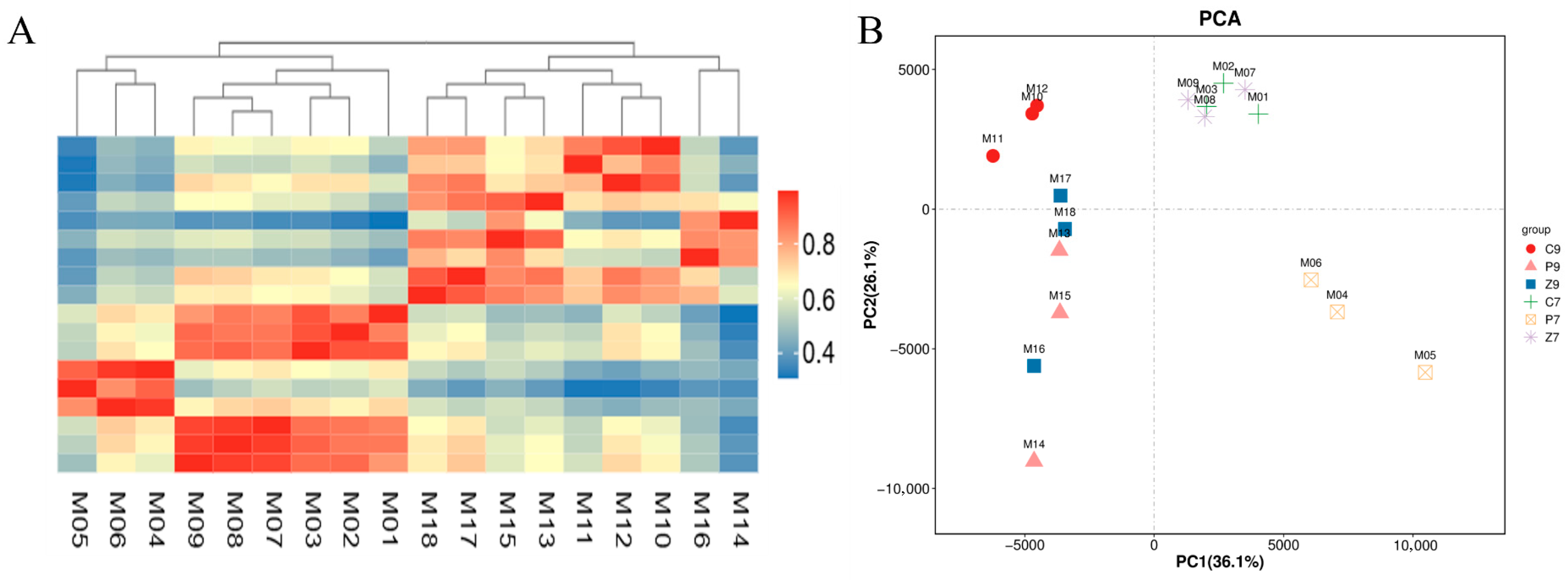
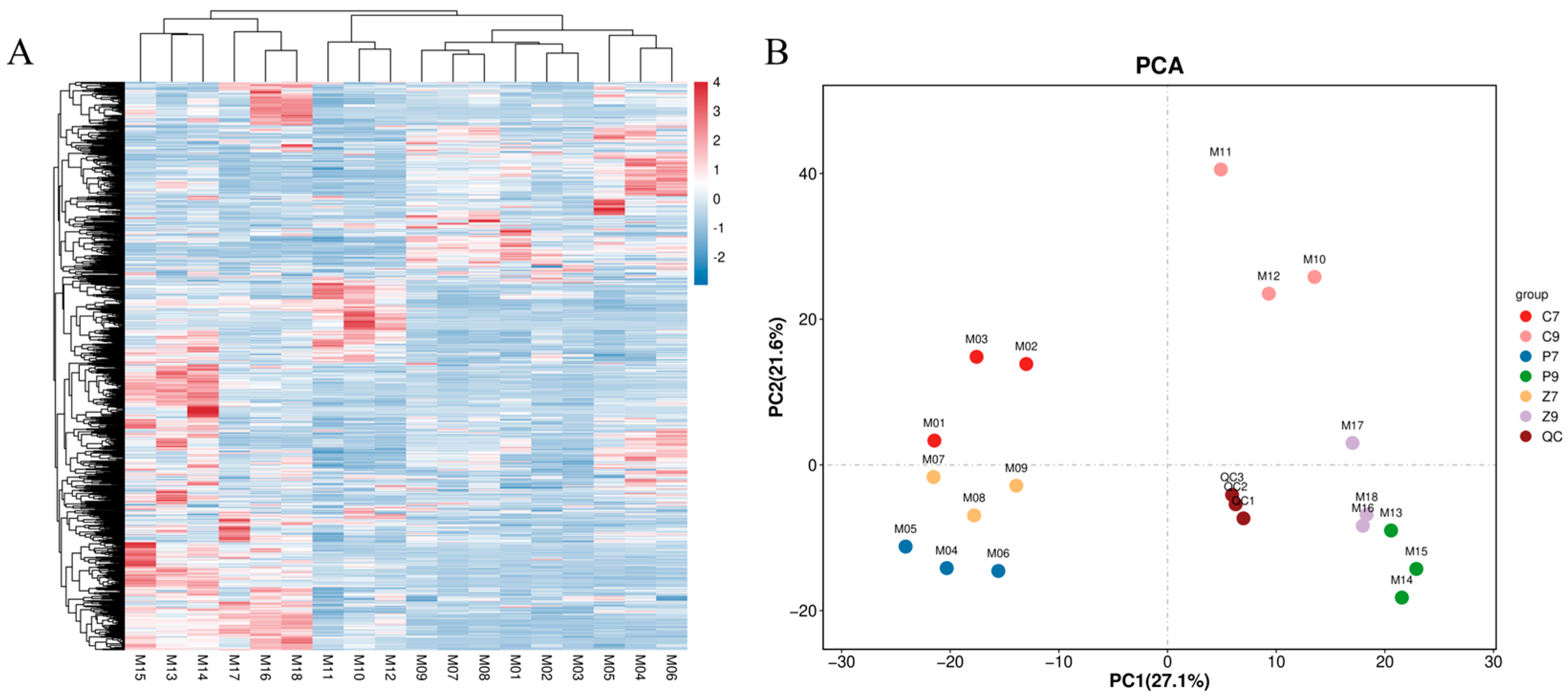
3.2. Transcriptomic Analysis of Stalk Internodes in Three Maize Varieties at Filling and Maturity Stages
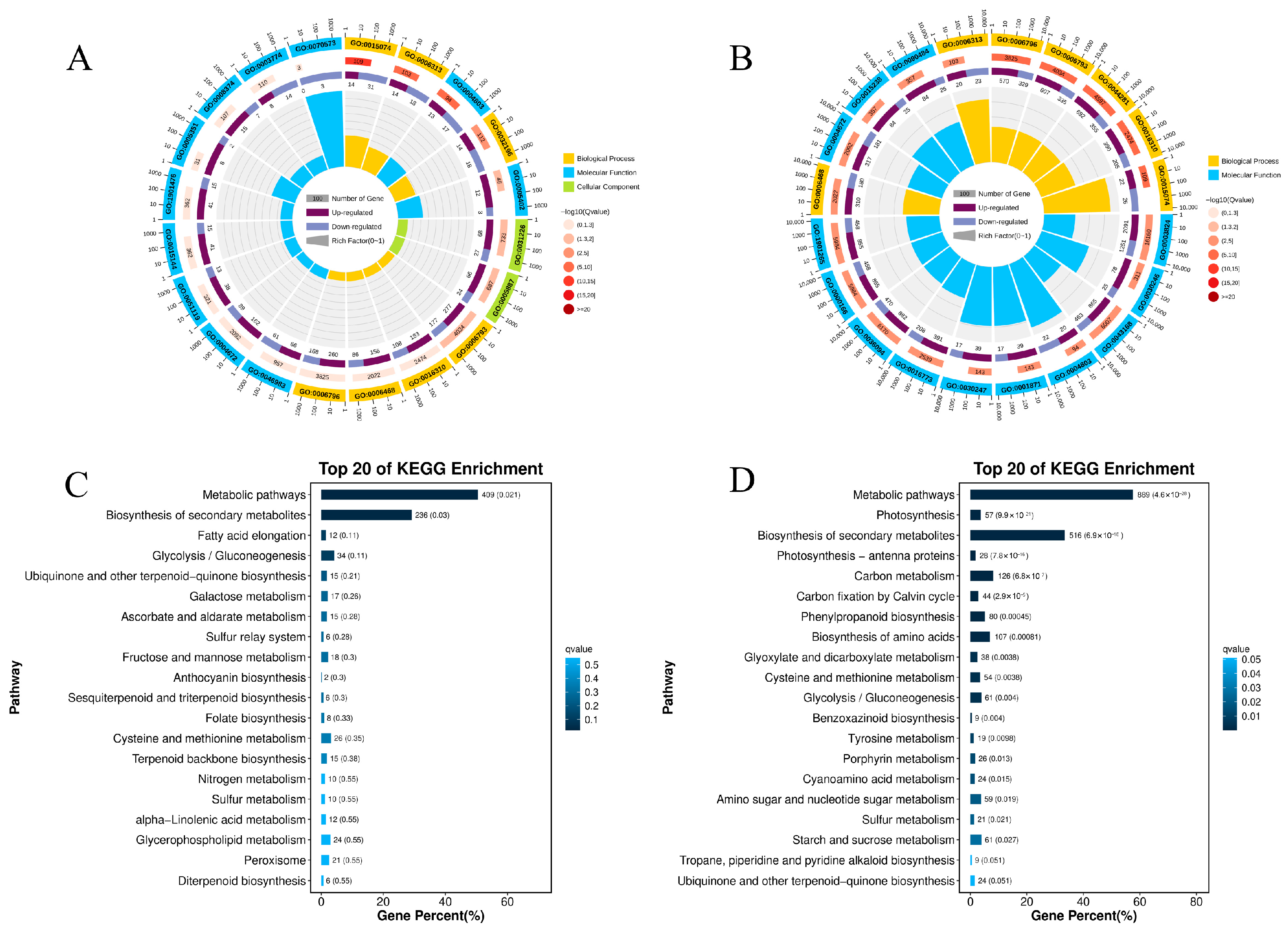
3.3. DEGs Enrichment Analysis
3.4. Metabolomic Analysis and Metabolite Identification
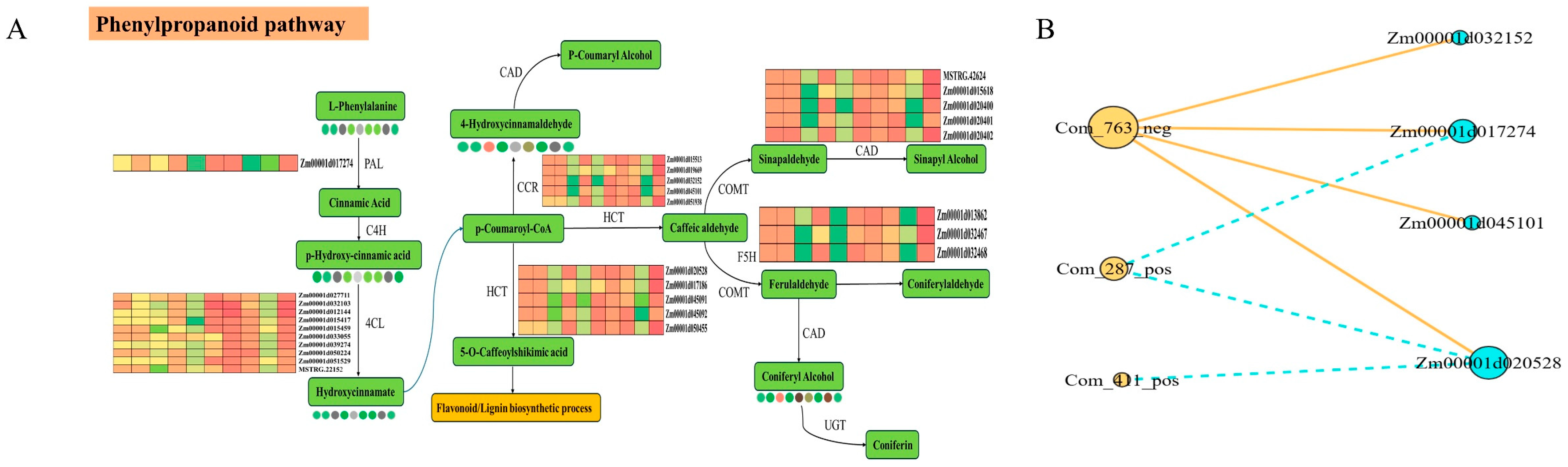
3.5. Integrated Transcriptomic and Metabolomic Analysis Reveals a Core Regulatory Module in the Phenylpropanoid Pathway
4. Discussion
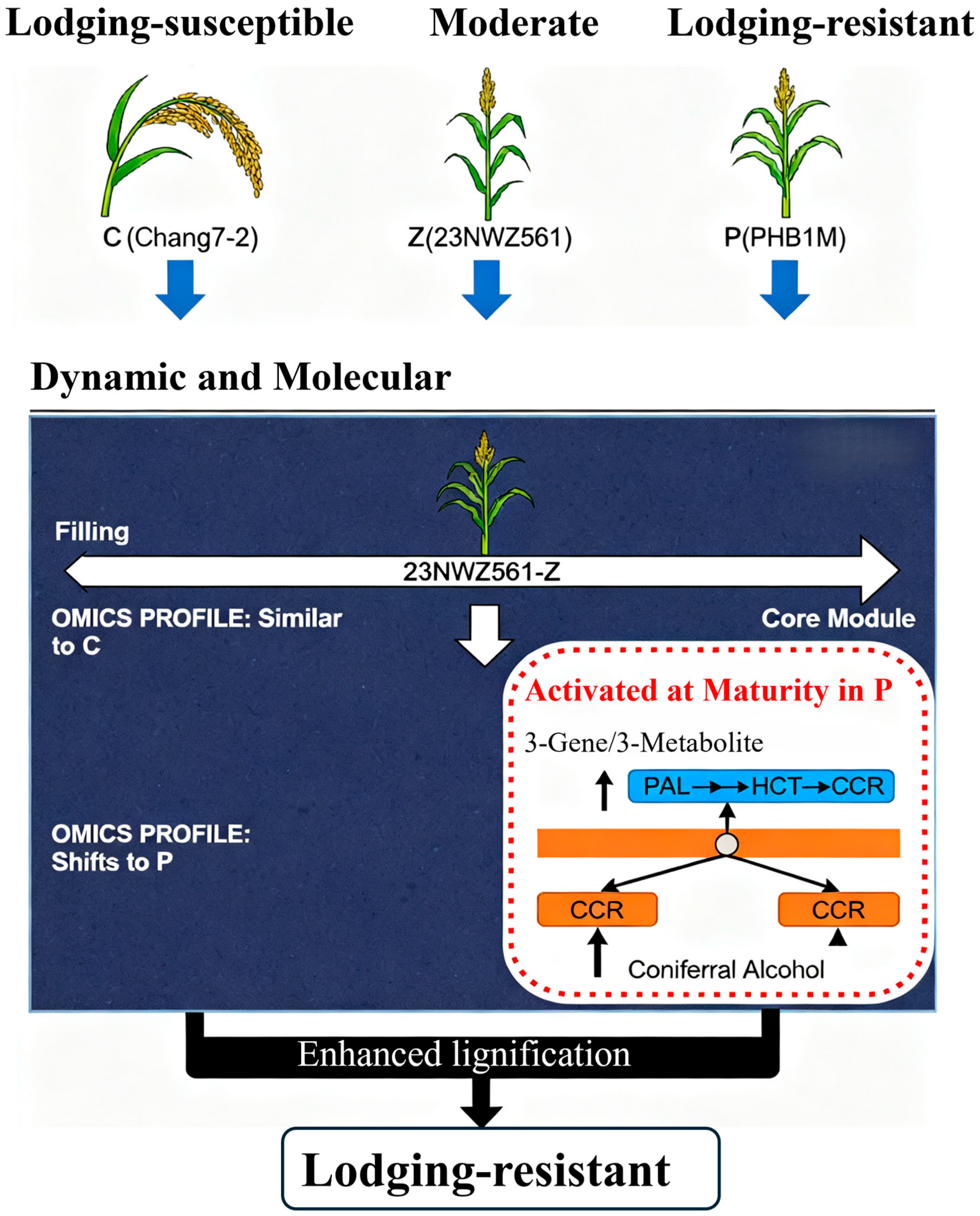
4.1. Lodging Resistance Is a Developmentally Regulated Trait
4.2. The Central Role of Phenylpropanoid Biosynthesis in Establishing Lodging Resistance at Maturity
4.3. Synergistic Functions of Kinase Signaling and Cell Wall Metabolism in Stalk Reinforcement
4.4. The Value of Multi-Omics Integration and Implications for Breeding
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wu, L.; Zheng, Y.; Jiao, F.; Wang, M.; Zhang, J.; Zhang, Z.; Huang, Y.; Jia, X.; Zhu, L.; Zhao, Y.; et al. Identification of quantitative trait loci for related traits of stalk lodging resistance using genome-wide association studies in maize (Zea mays L.). BMC Genom. Data 2022, 23, 76. [Google Scholar] [CrossRef]
- Liu, J.; Sun, C.; Guo, S.; Yin, X.; Yuan, Y.; Fan, B.; Lv, Q.; Cai, X.; Zhong, Y.; Xia, Y.; et al. Genomic and Transcriptomic Analyses Reveal Pathways and Genes Associated With Brittle Stalk Phenotype in Maize. Front. Plant Sci. 2022, 13, 849421. [Google Scholar] [CrossRef]
- Li, T.; Zhang, X.; Liu, Q.; Yan, P.; Liu, J.; Chen, Y.; Sui, P. Yield and yield stability of single cropping maize under different sowing dates and the corresponding changing trends of climatic variables. Field Crops Res. 2022, 285, 108589. [Google Scholar] [CrossRef]
- Zhai, J.; Zhang, Y.; Zhang, G.; Tian, M.; Xie, R.; Ming, B.; Hou, P.; Wang, K.; Xue, J.; Li, S. Effects of Nitrogen Fertilizer Management on Stalk Lodging Resistance Traits in Summer Maize. Agriculture 2022, 12, 162. [Google Scholar] [CrossRef]
- Tilman, D.; Balzer, C.; Hill, J.; Berort, B.L. Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. USA 2011, 108, 20260–20264. [Google Scholar] [CrossRef]
- Bodirsky, B.L.; Dietrich, J.P.; Martinelli, E.; Stenstad, A.; Pradhan, P.; Gabrysch, S.; Mishra, A.; Weindl, I.; Le Mouël, C.; Rolinski, S.; et al. The ongoing nutrition transition thwarts long-term targets for food security, public health and environmental protection. Sci. Rep. 2020, 10, 19778. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Zhang, C.; Zhang, Z.; Wen, M.; Qiu, H. QTL mapping of maize (Zea mays L.) kernel traits under low-phosphorus stress. Physiol. Mol. Biol. Plants 2023, 29, 435–445. [Google Scholar] [CrossRef]
- Yang, J.; Li, M.; Yin, Y.; Liu, Y.; Gan, X.; Mu, X.; Li, H.; Li, J.; Li, H.; Zheng, J.; et al. Spatial accumulation of lignin monomers and cellulose underlying stalk strength in maize. Plant Physiol. Biochem. 2024, 214, 108918. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Xie, R.-Z.; Zhang, W.-F.; Wang, K.-R.; Hou, P.; Ming, B.; Gou, L.; Li, S. Research progress on reduced lodging of high-yield and -density maize. J. Integr. Agric. 2017, 16, 2717–2725. [Google Scholar] [CrossRef]
- Liu, L.; Liu, S.; Lu, H.; Tian, Z.; Zhao, H.; Wei, D.; Wang, S.; Huang, Z. Integration of transcriptome and metabolome analyses reveals key lodging-resistance-related genes and metabolic pathways in maize. Front. Genet. 2022, 13, 1001195. [Google Scholar] [CrossRef]
- Shah, A.N.; Tanveer, M.; Rehman, A.U.; Anjum, S.A.; Iqbal, J.; Ahmad, R. Lodging stress in cereal-effects and management: An overview. Environ. Sci. Pollut. Res. Int. 2017, 24, 5222–5237. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Zhang, Y.; Guo, X.; Ma, L.; Shao, M.; Pan, X.; Zhao, C. Micron-scale phenotyping quantification and three-dimensional microstructure reconstruction of vascular bundles within maize stalks based on micro-CT scanning. Funct. Plant Biol. 2016, 44, 10–22. [Google Scholar] [CrossRef]
- Kotake, T.; Aohara, T.; Hirano, K.; Sato, A.; Kaneko, Y.; Tsumuraya, Y.; Tsumuraya, Y.; Takatsuji, H.; Kawasaki, S. Rice Brittle culm 6 encodes a dominant-negative form of CesA protein that perturbs cellulose synthesis in secondary cell walls. J. Exp. Bot. 2011, 62, 2053–2062. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, R.; Shi, Z.; Zhang, Y.; Sun, X.; Ji, Y.; Zhao, Y.; Wang, J.; Zhang, Y.; Xing, J.; et al. Multi-omics analysis of the development and fracture resistance for maize internode. Sci. Rep. 2019, 9, 8183. [Google Scholar] [CrossRef]
- Guo, Y.; Hu, Y.; Chen, H.; Yan, P.; Du, Q.; Wang, Y.; Wang, H.; Wang, Z.; Kang, D.; Li, W.-X. Identification of traits and genes associated with lodging resistance in maize. Crop J. 2021, 9, 1408–1417. [Google Scholar] [CrossRef]
- Jiao, S.; Hazebroek, J.P.; Chamberlin, M.A.; Perkins, M.; Sandhu, A.S.; Gupta, R.; Simcox, K.D.; Yinghong, L.; Prall, A.; Heetland, L.; et al. Chitinase-like1 Plays a Role in Stalk Tensile Strength in Maize. Plant Physiol. 2019, 181, 1127–1147. [Google Scholar] [CrossRef]
- Sindhu, A.; Langewisch, T.; Olek, A.; Multani, D.S.; McCann, M.C.; Vermerris, W.; Carpita, N.C.; Johal, G. Maize Brittle stalk2 encodes a COBRA-like protein expressed in early organ development but required for tissue flexibility at maturity. Plant Physiol. 2007, 145, 1444–1459. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Qian, Q.; Zhou, Y.; Yan, M.; Sun, L.; Zhang, M.; Fu, Z.; Wang, Y.; Han, B.; Pang, X.; et al. BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell 2003, 15, 2020–2031. [Google Scholar] [CrossRef]
- Manga-robles, A.; Santiago, R.; Malvar, R.A.; Moreno-González, V.; Fornalé, S.; López, I.; Centeno, M.L.; Acebes, J.L.; Álvarez, J.M.; Caparros-Ruiz, D.; et al. Elucidating compositional factors of maize cell walls contributing to stalk strength and lodging resistance. Plant Sci. 2021, 307, 110882. [Google Scholar] [CrossRef]
- Sato, K.; Ito, S.; Fujii, T.; Suzuki, R.; Takenouchi, S.; Nakaba, S.; Funada, R.; Sano, Y.; Kajita, S.; Hidemi, K.; et al. The carbohydrate-binding module (CBM)-like sequence is crucial for rice CWA1/BC1 function in proper assembly of secondary cell wall materials. Plant Signal Behav. 2010, 5, 1433–1436. [Google Scholar] [CrossRef]
- Li, P.; Liu, Y.; Tan, W.; Chen, J.; Zhu, M.; Lv, Y.; Liu, Y.; Yu, S.; Zhang, W.; Cai, H. Brittle Culm 1 Encodes a COBRA-Like Protein Involved in Secondary Cell Wall Cellulose Biosynthesis in Sorghum. Plant Cell Physiol. 2019, 60, 788–801. [Google Scholar] [CrossRef] [PubMed]
- Flint-garcia, S.A.; Mcmullen, M.D.; Darrah, L.L. Genetic Relationship of Stalk Strength and Ear Height in Maize. Crop Sci. 2003, 43, 23–31. [Google Scholar] [CrossRef]
- Aohara, T.; Kotake, T.; Kaneko, Y.; Takatsuji, H.; Tsumuraya, Y.; Kawasaki, S. Rice BRITTLE CULM 5 (BRITTLE NODE) is involved in secondary cell wall formation in the sclerenchyma tissue of nodes. Plant Cell Physiol. 2009, 50, 1886–1897. [Google Scholar] [CrossRef]
- Ren, Z.; Wang, X.; Tao, Q.; Guo, Q.; Zhou, Y.; Yi, F.; Huang, G.; Li, Y.; Zhang, M.; Li, Z.; et al. Transcriptome dynamic landscape underlying the improvement of maize lodging resistance under coronatine treatment. BMC Plant Biol. 2021, 21, 202. [Google Scholar] [CrossRef]
- Xie, L.; Wen, D.; Wu, C.; Zhang, C. Transcriptome analysis reveals the mechanism of internode development affecting maize stalk strength. BMC Plant Biol. 2022, 22, 49. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zhao, Y.; Liu, Q.; Diao, Y.; Wang, Q.; Yu, J.; Jiang, E.; Zhang, Y.; Liu, B. Identification of ZmBK2 Gene Variation Involved in Regulating Maize Brittleness. Genes 2023, 14, 1126. [Google Scholar] [CrossRef]
- Le, L.; Guo, W.; Du, D.; Zhang, X.; Wang, W.; Yu, J.; Wang, H.; Qiao, H.; Zhang, C.; Pu, L. A spatiotemporal transcriptomic network dynamically modulates stalk development in maize. Plant Biotechnol. J. 2022, 20, 2313–2331. [Google Scholar] [CrossRef]
- He, Y.; Deng, Z.; He, S.; Qi, Z.; Chang, H.; Liu, P.; Chen, Z.; Zou, C.; Shen, Y.; Ma, L. Transcriptome and co-expression network analysis reveal the genetic basis of cell wall components in maize stalks. BMC Genom. 2025, 26, 620. [Google Scholar] [CrossRef]
- Xiong, W.; Wu, Z.; Liu, Y.; Li, Y.; Su, K.; Bai, Z.; Guo, S.; Hu, Z.; Zhang, Z.; Bao, Y.; et al. Mutation of 4-coumarate: Coenzyme A ligase 1 gene affects lignin biosynthesis and increases the cell wall digestibility in maize brown midrib5 mutants. Biotechnol. Biofuels 2019, 12, 82. [Google Scholar] [CrossRef]
- Wang, S.; Wang, X.; Yue, L.; Li, H.; Zhu, L.; Dong, Z.; Long, Y. Genome-Wide Identification and Characterization of Lignin Synthesis Genes in Maize. Int. J. Mol. Sci. 2024, 25, 6710. [Google Scholar] [CrossRef]
- Gomez-cano, L.; Gomez-cano, F.; Dillon, F.M.; Alers-Velazquez, R.; Doseff, A.I.; Grotewold, E.; Gray, J. Discovery of modules involved in the biosynthesis and regulation of maize phenolic compounds. Plant Sci. 2020, 291, 110364. [Google Scholar] [CrossRef]
- Andersen, J.R.; Zein, I.; Wenzel, G.; Darnhofer, B.; Eder, J.; Ouzunova, M.; Lübberstedt, T. Characterization of phenylpropanoid pathway genes within European maize (Zea mays L.) inbreds. BMC Plant Biol. 2008, 8, 2. [Google Scholar] [CrossRef]
- Wang, X.; Shi, Z.; Zhang, R.; Sun, X.; Wang, J.; Wang, S.; Zhang, Y.; Zhao, Y.; Su, A.; Li, C.; et al. Stalk architecture, cell wall composition, and QTL underlying high stalk flexibility for improved lodging resistance in maize. BMC Plant Biol. 2020, 20, 515. [Google Scholar] [CrossRef]
- Ishfaq, S.; Ding, Y.; Liang, X.; Guo, W. Advancing lodging resistance in maize: Integrating genetic, hormonal, and agronomic insights for sustainable crop productivity. Plant Stress 2025, 15, 100777. [Google Scholar] [CrossRef]
- Van Soest, P.J.; Robertson, J.B.; Lewis, B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef]
- Langmead, B.; Wilks, C.; Antonescu, V.; Charles, R. Scaling read aligners to hundreds of threads on general-purpose processors. Bioinformatics 2018, 35, 421–432. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innov. 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Fu, C.; Liang, C.; Ni, X.; Zhao, X.; Chen, M.; Ou, L. Crop Lodging and The Roles of Lignin, Cellulose, and Hemicellulose in Lodging Resistance. Agronomy 2022, 12, 1795. [Google Scholar] [CrossRef]
| Genotype | Abbreviation | Developmental Stage | Stage Code | Sample Name | Biological Replicates (n) for RNA-seq | Biological Replicates (n) for Metabolomics |
|---|---|---|---|---|---|---|
| Chang 7-2 | C | Grain-filling | 7 | C7(M01-03) | 3 | 3 |
| C | Maturity | 9 | C9(M10-12) | 3 | 3 | |
| PHB1M | P | Grain-filling | 7 | P7(M04-06) | 3 | 3 |
| P | Maturity | 9 | P9(M13-15) | 3 | 3 | |
| 23NWZ561 | Z | Grain-filling | 7 | Z7(M07-09) | 3 | 3 |
| Z | Maturity | 9 | Z9(M16-18) | 3 | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, C.; Wu, H.; Zhang, X.; Sun, F.; Zhang, S.; Yu, Z.; Dong, Q.; Liu, Y.; Zhang, H.; Ma, Q.; et al. Integrated Transcriptomic and Metabolomic Analyses Implicate Key Genes and Metabolic Pathways in Maize Lodging Resistance. Agriculture 2025, 15, 2416. https://doi.org/10.3390/agriculture15232416
Xue C, Wu H, Zhang X, Sun F, Zhang S, Yu Z, Dong Q, Liu Y, Zhang H, Ma Q, et al. Integrated Transcriptomic and Metabolomic Analyses Implicate Key Genes and Metabolic Pathways in Maize Lodging Resistance. Agriculture. 2025; 15(23):2416. https://doi.org/10.3390/agriculture15232416
Chicago/Turabian StyleXue, Chunlei, Haiyan Wu, Xuting Zhang, Fengcheng Sun, Sainan Zhang, Zhonghao Yu, Qi Dong, Yanan Liu, Hailong Zhang, Qing Ma, and et al. 2025. "Integrated Transcriptomic and Metabolomic Analyses Implicate Key Genes and Metabolic Pathways in Maize Lodging Resistance" Agriculture 15, no. 23: 2416. https://doi.org/10.3390/agriculture15232416
APA StyleXue, C., Wu, H., Zhang, X., Sun, F., Zhang, S., Yu, Z., Dong, Q., Liu, Y., Zhang, H., Ma, Q., & Wang, L. (2025). Integrated Transcriptomic and Metabolomic Analyses Implicate Key Genes and Metabolic Pathways in Maize Lodging Resistance. Agriculture, 15(23), 2416. https://doi.org/10.3390/agriculture15232416






