Tomato Stem and Leaf Segmentation and Phenotype Parameter Extraction Based on Improved Red Billed Blue Magpie Optimization Algorithm
Abstract
1. Introduction
- deep learning models generally have the problem of large parameter sizes. although the demand for computational resources can be significantly reduced through model lightweighting techniques, this process often leads to a decline in model performance, i.e., loss of accuracy, which affects the accuracy of the final segmentation results.
- the segmentation accuracy of some deep learning models shows a certain degree of instability in complex environments, and this instability limits the applicability of the model in practical application scenarios, making it difficult to meet the demand for highly reliable segmentation results in agricultural production.
- the Red-billed Blue Magpie Optimization (RBMO) Algorithm is optimized using an elite strategy to improve the stability and search capability of the algorithm. The elite strategy improves the overall performance of the algorithm by retaining the best individuals in successive generations and guiding the evolution of the algorithm towards better solutions.
- the optimized red-billed blue magpie algorithm is fused with a deep learning network model to optimize the effect of stem and leaf segmentation of tomato plants. This fusion strategy aims to ensure the robustness of the model when dealing with complex point cloud data while reducing the dependence on a large number of parameters.
- the algorithm encapsulate customized convolutional layers by combining geometric and curvature features of the point cloud data. This approach makes the network model more attuned to the processing characteristics of 3D spatial data, thus improving the ability to recognize the details of plant structures while keeping the model lightweight.
2. Materials and Methods
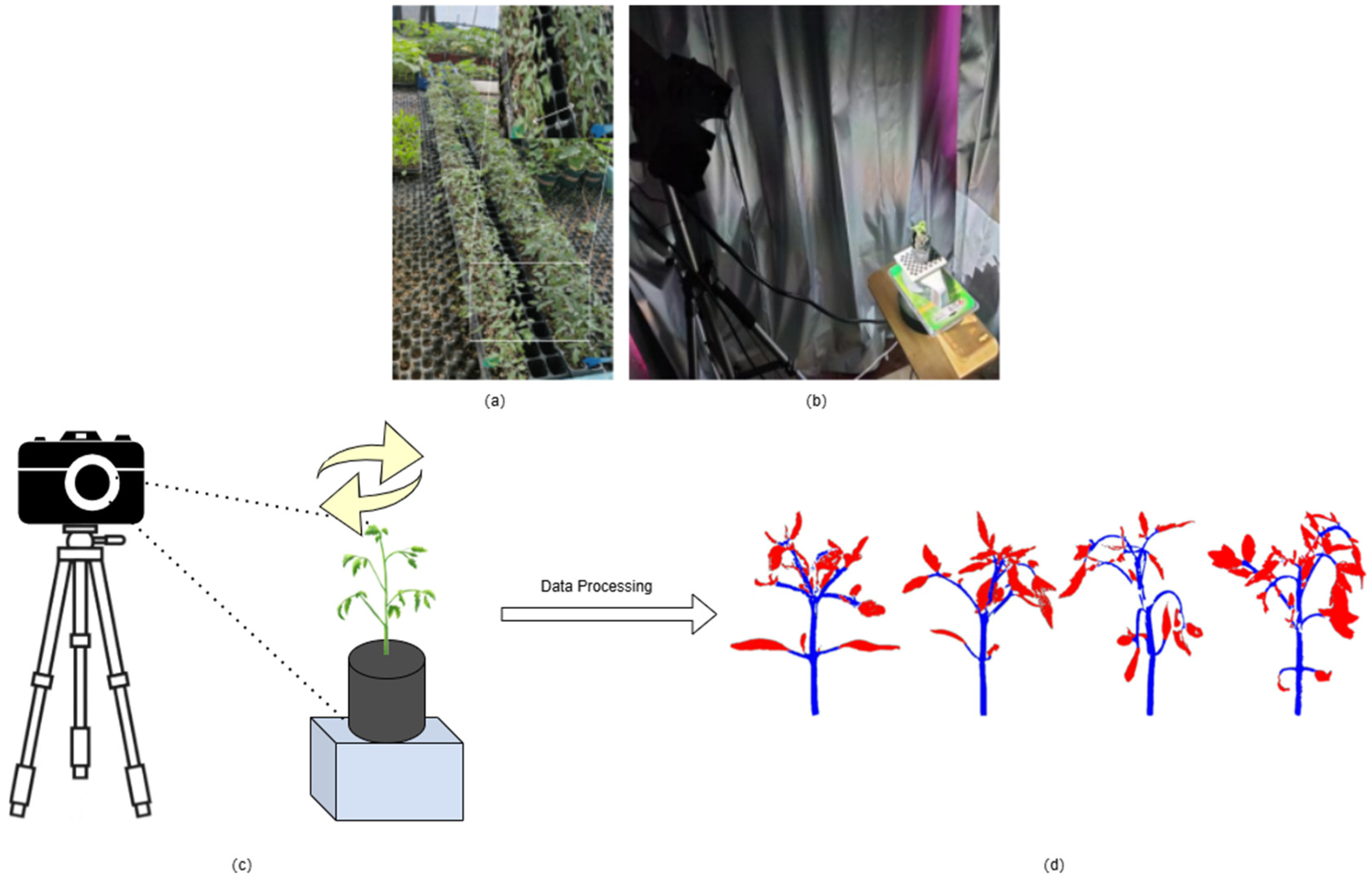
2.1. Data Sources and Collection
2.2. Methodologies
2.2.1. Algorithmic Principles of the Red-Billed Blue Magpie Algorithm for Elite Strategy Optimization
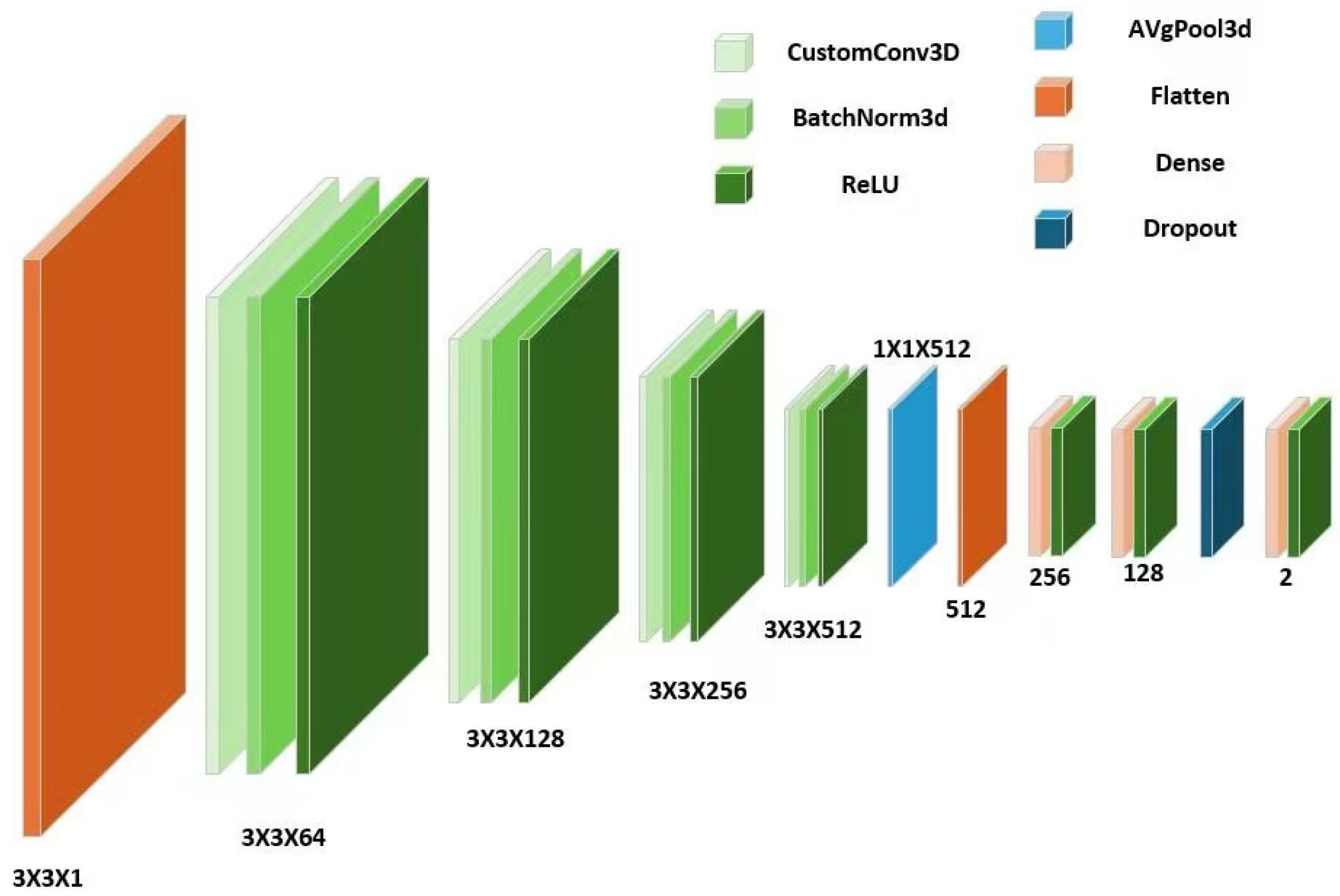
2.2.2. Algorithmic Principles of 3DCNN
3. Results
3.1. Comparison Experiment
3.2. Ablation Experiment
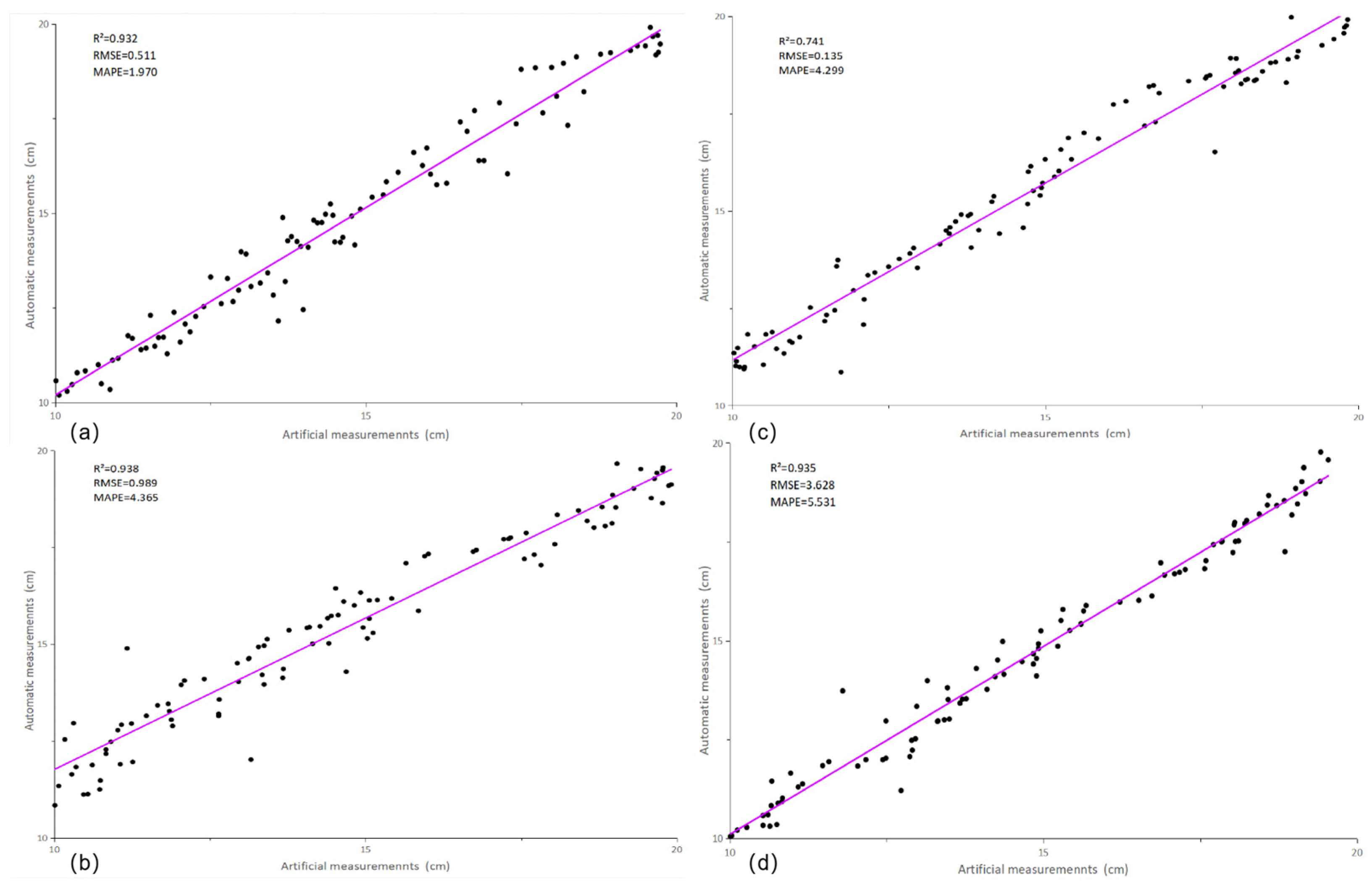
3.3. Phenotypic Parameter Measurement Results and Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Boogaard, F.; Henten, E.; Kootstra, G. The added value of 3D point clouds for digital plant phenotyping—A case study on internode length measurements in cucumber. Biosyst. Eng. 2023, 234, 1–12. [Google Scholar] [CrossRef]
- Alighaleh, P.; Mesri Gundoshmian, T.; Alighaleh, S.; Rohani, A. Feasibility and reliability of agricultural crop height measurement using the laser sensor array. Inf. Process. Agric. 2024, 11, 228–236. [Google Scholar] [CrossRef]
- Anshori, M.F.; Dirpan, A.; Sitaresmi, T.; Rossi, R.; Farid, M.; Hairmansis, A.; Purwoko, B.; Suwarno, W.B.; Nugraha, Y. An overview of image-based phenotyping as an adaptive 4. 0 technology for studying plant abiotic stress: A bibliometric and literature review. Heliyon 2023, 9, e21650. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Sun, L.; Han, T.; Cai, M.; Gao, C. PestLite: A Novel YOLO-Based Deep Learning Technique for Crop Pest Detection. Agriculture 2024, 14, 228. [Google Scholar] [CrossRef]
- Li, R.; Li, Y.; Qin, W.; Abbas, A.; Li, S.; Ji, R.; Wu, Y.; He, Y.; Yang, J. Lightweight Network for Corn Leaf Disease Identification Based on Improved YOLO v8s. Agriculture 2024, 14, 220. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, M.; Shen, Y. Identifying the Growth Status of Hydroponic Lettuce Based on YOLO-EfficientNet. Plants 2024, 13, 372. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhao, Y.; Liu, T.; Deng, H. A Weakly Supervised Semantic Segmentation Model of Maize Seedlings and Weed Images Based on Scrawl Labels. Sensors 2023, 23, 9846. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Guo, X.; Yang, H. Win-Former: Window-Based Transformer for Maize Plant Point Cloud Semantic Segmentation. Agronomy 2023, 13, 2723. [Google Scholar] [CrossRef]
- Luo, L.; Jiang, X.; Yang, Y.; Samy, E.R.; Lefsrud, M.; Hoyos-Villegas, V.; Sun, S. Eff-3DPSeg: 3D organ-level plant shoot segmentation using annotation-efficient point clouds. arXiv 2022, arXiv:2212.10263. [Google Scholar]
- Wang, Y.; Liu, Q.; Yang, J.; Ren, G.; Wang, W.; Zhang, W.; Li, F. A Method for Tomato Plant Stem and Leaf Segmentation and Phenotypic Extraction Based on Skeleton Extraction and Supervoxel Clustering. Agronomy 2024, 14, 198. [Google Scholar] [CrossRef]
- Morteza, G.; Kevin, W.; Corke, F.M.; Tiddeman, B.; Liu, Y.; Doonan, J.H. Deep Segmentation of Point Clouds of Wheat. Front. Plant Sci. 2021, 12, 608732. [Google Scholar]
- Yonatan, L.; Ofri, R.; Merav, A. Dissecting the roles of supervised and unsupervised learning in perceptual discrimination judgments. J. Neurosci. 2020, 41, 757–765. [Google Scholar]
- Yao, D.; Chuanchuan, Y.; Hao, C.; Yan, W.; Li, H. Low-complexity point cloud denoising for LiDAR by PCA-based dimension reduction. Opt. Commun. 2021, 482, 126567. [Google Scholar]
- Alkadri, F.M.; Yuliana, Y.; Agung, C.R.M.; Rahman, M.A.; Hein, C. Enhancing preservation: Addressing humidity challenges in Indonesian heritage buildings through advanced detection methods point cloud data. Results Eng. 2024, 24, 103292. [Google Scholar] [CrossRef]
- Jing, R.; Shao, Y.; Zeng, Q.; Liu, Y.; Wei, W.; Gan, B.; Duan, X. Multimodal feature integration network for lithology identification from point cloud data. Comput. Geosci. 2025, 194, 105775. [Google Scholar] [CrossRef]
- Chen, T.; Ying, X. FPSMix: Data augmentation strategy for point cloud classification. Front. Comput. Sci. 2024, 19, 192701. [Google Scholar] [CrossRef]
- Miao, T.; Zhu, C.; Xu, T.; Yang, T.; Li, N.; Zhou, Y.; Deng, H. Automatic stem-leaf segmentation of maize shoots using three-dimensional point cloud. Comput. Electron. Agric. 2021, 187, 106310. [Google Scholar] [CrossRef]
- Shen, F.; Lu, Z.-M.; Lu, Z.; Wang, Z. Dual semantic-guided model for weakly-supervised zero-shot semantic segmentation. Multimed. Tools Appl. 2021, 81, 5443–5458. [Google Scholar] [CrossRef]
- Wu, J.; Sun, M.; Xu, H.; Jiang, C.; Ma, W.; Zhang, Q. Class agnostic and specific consistency learning for weakly-supervised point cloud semantic segmentation. Pattern Recognit. 2025, 158, 111067. [Google Scholar] [CrossRef]
- Samoaa, P.; Aronsson, L.; Longa, A.; Leitner, P.; Chehreghani, M.H. A unified active learning framework for annotating graph data for regression task. Eng. Appl. Artif. Intell. 2024, 138, 109383. [Google Scholar] [CrossRef]
- Bicheng, S.; Peng, Z.; Liang, D.; Li, X. Active deep image clustering. Knowl.-Based Syst. 2022, 252, 109346. [Google Scholar]
- Sun, R.; Guo, S.; Guo, J.; Li, W.; Zhang, X.; Guo, X.; Pan, Z. GraphMoCo: A graph momentum contrast model for large-scale binary function representation learning. Neurocomputing 2024, 575, 127273. [Google Scholar] [CrossRef]
- Shengwei, F.; Ke, L.; Haisong, H.; Ma, C.; Fan, Q.; Zhu, Y. Red-billed blue magpie optimizer: A novel metaheuristic algorithm for 2D/3D UAV path planning and engineering design problems. Artif. Intell. Rev. 2024, 57, 134. [Google Scholar]
- Hassen, L.; Ali, L.; Slim, B.; Kariri, E. Joint filter and channel pruning of convolutional neural networks as a bi-level optimization problem. Memetic Comput. 2024, 16, 71–90. [Google Scholar]
- Baljon, M. A Framework for Agriculture Plant Disease Prediction using Deep Learning Classifier. Int. J. Adv. Comput. Sci. Appl. IJACSA 2023, 14, 1098–1111. [Google Scholar] [CrossRef]
- Xiaodan, L.; Zijian, Z. A Whale Optimization Algorithm with Convergence and Exploitability Enhancement and Its Application. Math. Probl. Eng. 2022, 2022, 2904625. [Google Scholar]
- Anderson, J.P.; Stephens, D.W.; Dunbar, S.R. Saltatory search: A theoretical analysis. Behav. Ecol. 1997, 8, 307–317. [Google Scholar] [CrossRef]
- Fergany EA, A.; Agwa, M.A. Red-Billed Blue Magpie Optimizer for Electrical Characterization of Fuel Cells with Prioritizing Estimated Parameters. Technologies 2024, 12, 156. [Google Scholar] [CrossRef]
- Wang, P.; Liu, Z.; Wang, Z.; Zhao, Z.; Yang, D.; Yan, W. Graph generative adversarial networks with evolutionary algorithm. Appl. Soft Comput. 2024, 164, 111981. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, H.; Cui, Z.; Chen, J. Hybrid multi-objective cuckoo search with dynamical local search. Memetic Comput. 2018, 10, 199–208. [Google Scholar] [CrossRef]
- Li, C.; Priemer, R.; Cheng, K.H. Optimization by random search with jumps. Int. J. Numer. Methods Eng. 2004, 60, 1301–1315. [Google Scholar] [CrossRef]
- Shan, D.; Zhang, X.; Shi, W.; Li, L. Neural Architecture Search for a Highly Efficient Network with Random Skip Connections. Appl. Sci. 2020, 10, 3712. [Google Scholar] [CrossRef]
- Gan, W.; Li, H.; Hao, P. Many-objective optimization algorithm based on the similarity principle and multi-mechanism collaborative search. J. Supercomput. 2024, 81, 124. [Google Scholar] [CrossRef]
- Fang, G.; Weibin, Z.; Guofu, L.; Zhang, X.; Luo, L.; Wu, Y.; Guo, P. A point cloud registration method based on multiple-local-feature matching. Optik 2023, 295, 171511. [Google Scholar]
- Jingtao, W.; Changcai, Y.; Lifang, W.; Chen, R. CSCE-Net: Channel-Spatial Contextual Enhancement Network for Robust Point Cloud Registration. Remote Sens. 2022, 14, 5751. [Google Scholar] [CrossRef]
- Wang, L. High-precision point cloud registration method based on volume image correlation. Meas. Sci. Technol. 2024, 35, 035024. [Google Scholar] [CrossRef]
- Chuang, T.Y.; Jaw, J.J. Multi-feature registration of point clouds. Remote Sens. 2017, 9, 281. [Google Scholar] [CrossRef]
- Yu, F.; Chen, Z.; Cao, J.; Jiang, M. Redundant same sequence point cloud registration. Vis. Comput. 2023, 40, 7719–7730. [Google Scholar] [CrossRef]
- Xu, S. An Introduction to Scientific Computing with Matlab and Python Tutorials; CRC Press: Boca Raton, FL, USA, 2021. [Google Scholar]
- Garcia, H.A.; Zhu, W. Building an Accessible and Flexible Multi-User Robotic Simulation Framework with Unity-MATLAB Bridge. Computers 2024, 13, 282. [Google Scholar] [CrossRef]
- Gasmi, K.; Hasnaoui, S. Dataflow-based automatic parallelization of MATLAB/Simulink models for fitting modern multicore architectures. Clust. Comput. 2024, 27, 6579–6590. [Google Scholar] [CrossRef]
- Yang, D. An improved particle swarm optimization algorithm for parameter optimization. Comput. Informatiz. Mech. Syst. 2022, 5, 35–38. [Google Scholar]
- Mohan, B.G.; Kumar, P.R.; Elakkiya, R. Enhancing pre-trained models for text summarization: A multi-objective genetic algorithm optimization approach. Multimed. Tools Appl. 2024, 1–17. [Google Scholar] [CrossRef]
- Chaudhury, A. Multilevel Optimization for Registration of Deformable Point Clouds. IEEE Trans. Image Process. 2020, 29, 8735–8746. [Google Scholar] [CrossRef]
- Tianyuan, L.; Jiacheng, W.; Xiaodi, H.; Lu, Y.; Bao, J. 3DSMDA-Net: An improved 3DCNN with separable structure and multi-dimensional attention for welding status recognition. J. Manuf. Syst. 2021, 62, 811–822. [Google Scholar]
- Peyman, A.; Keyhan, G.; Atieh, A.; Deb, P.; Moradkhani, H. Bayesian Multi-modeling of Deep Neural Nets for Probabilistic Crop Yield Prediction. Agric. For. Meteorol. 2022, 314, 108773. [Google Scholar]
- Zhu, G.; Zhang, L.; Shen, P.; Song, J.; Shah, S.A.; Bennamoun, M. Continuous Gesture Segmentation and Recognition Using 3DCNN and Convolutional LSTM. IEEE Trans. Multimed. 2019, 21, 1011–1021. [Google Scholar] [CrossRef]
- Zhihua, D.; Peiliang, G.; Baohua, Z.; Zhang, D.; Yan, J.; He, Z.; Zhao, S.; Zhao, C. Maize crop row recognition algorithm based on improved UNet network. Comput. Electron. Agric. 2023, 210, 107940. [Google Scholar]
- Yi, X.; Wang, J.; Wu, P.; Wang, G.; Mo, L.; Lou, X.; Liang, H.; Huang, H.; Lin, E.; Maponde, B.T. AC-UNet: An improved UNet-based method for stem and leaf segmentation in Betula luminifera. Front. Plant Sci. 2023, 14, 1268098. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Chen, S.; Huang, H.; Tian, X. Tree Species Classification of Backpack Laser Scanning Data Using the PointNet++ Point Cloud Deep Learning Method. Remote Sens. 2022, 14, 3809. [Google Scholar] [CrossRef]
- Xiang, R. Image segmentation for whole tomato plant recognition at night. Comput. Electron. Agric. 2018, 154, 434–442. [Google Scholar] [CrossRef]
- Zeng, W.; He, M. Rice disease segmentation method based on CBAM-CARAFE-DeepLabv3+. Crop Prot. 2024, 180, 106665. [Google Scholar] [CrossRef]



| P | R | F1 | IoU | ACC | |
|---|---|---|---|---|---|
| AC-UNet | 0.916 | 0.903 | 0.915 | 0.892 | 0.886 |
| UNet | 0.912 | 0.901 | 0.912 | 0.894 | 0.864 |
| PointNet++ | 0.927 | 0.911 | 0.896 | 0.881 | 0.873 |
| PCNN | 0.906 | 0.890 | 0.904 | 0.909 | 0.898 |
| DeepLabV3 | 0.877 | 0.904 | 0.885 | 0.821 | 0.853 |
| ES-RMBO | 0.965 | 0.965 | 0.965 | 0.965 | 0.933 |
| Training Time (Hours) | Parameter Size (Millions) | Video Memory Usage (GB) | Computational Efficiency (E) | |
|---|---|---|---|---|
| AC-UNet | 14.0 | 22.8 | 6.7 | 9.84 |
| UNet | 14.2 | 23.4 | 6.5 | 9.56 |
| PointNet++ | 15.5 | 4.5 | 8.2 | 10.29 |
| PCNN | 15.0 | 15.2 | 7.8 | 11.96 |
| DeepLabV3 | 23.8 | 40.6 | 10.4 | 5.20 |
| ES-RMBO | 13.1 | 5.8 | 5.9 | 17.04 |
| P | R | F1 | IoU | ACC | |
|---|---|---|---|---|---|
| 3DCNN | 0.876 | 0.877 | 0.868 | 0.871 | 0.793 |
| RMBO | 0.913 | 0.915 | 0.924 | 0.928 | 0.818 |
| ES-RMBO | 0.965 | 0.965 | 0.965 | 0.965 | 0.933 |
| Training Time (Hours) | Parameter Size (Millions) | Video Memory Usage (GB) | Computational Efficiency (E) | |
|---|---|---|---|---|
| 3DCNN | 13.0 | 10.8 | 6.7 | 9.37 |
| RMBO | 10.2 | 11.4 | 6.5 | 11.36 |
| ES-RMBO | 13.1 | 5.8 | 5.9 | 17.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Huang, Z.; Yang, Z.; Yang, B.; Yu, S.; Zhao, S.; Zhang, X.; Li, X.; Yang, H.; Lin, Y.; et al. Tomato Stem and Leaf Segmentation and Phenotype Parameter Extraction Based on Improved Red Billed Blue Magpie Optimization Algorithm. Agriculture 2025, 15, 180. https://doi.org/10.3390/agriculture15020180
Zhang L, Huang Z, Yang Z, Yang B, Yu S, Zhao S, Zhang X, Li X, Yang H, Lin Y, et al. Tomato Stem and Leaf Segmentation and Phenotype Parameter Extraction Based on Improved Red Billed Blue Magpie Optimization Algorithm. Agriculture. 2025; 15(2):180. https://doi.org/10.3390/agriculture15020180
Chicago/Turabian StyleZhang, Lina, Ziyi Huang, Zhiyin Yang, Bo Yang, Shengpeng Yu, Shuai Zhao, Xingrui Zhang, Xinying Li, Han Yang, Yixing Lin, and et al. 2025. "Tomato Stem and Leaf Segmentation and Phenotype Parameter Extraction Based on Improved Red Billed Blue Magpie Optimization Algorithm" Agriculture 15, no. 2: 180. https://doi.org/10.3390/agriculture15020180
APA StyleZhang, L., Huang, Z., Yang, Z., Yang, B., Yu, S., Zhao, S., Zhang, X., Li, X., Yang, H., Lin, Y., & Yu, H. (2025). Tomato Stem and Leaf Segmentation and Phenotype Parameter Extraction Based on Improved Red Billed Blue Magpie Optimization Algorithm. Agriculture, 15(2), 180. https://doi.org/10.3390/agriculture15020180







