Automated Phenotypic Analysis of Mature Soybean Using Multi-View Stereo 3D Reconstruction and Point Cloud Segmentation
Abstract
1. Introduction
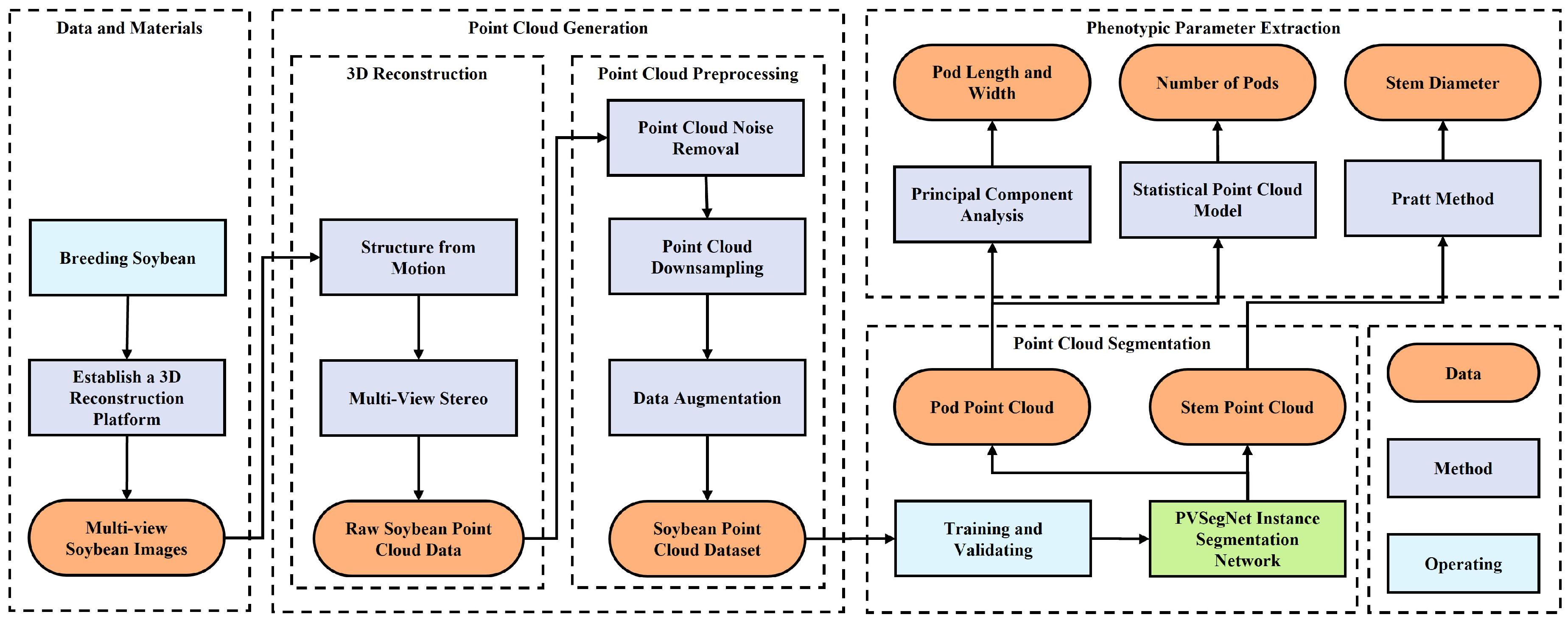
2. Materials and Methods
2.1. Soybean Experimental Samples
2.2. Point Cloud Data Generation
2.2.1. Image Acquisition
2.2.2. Three-Dimensional Reconstruction
2.2.3. Point Cloud Preprocessing
Point Cloud Denoising
Point Cloud Downsampling
Data Augmentation
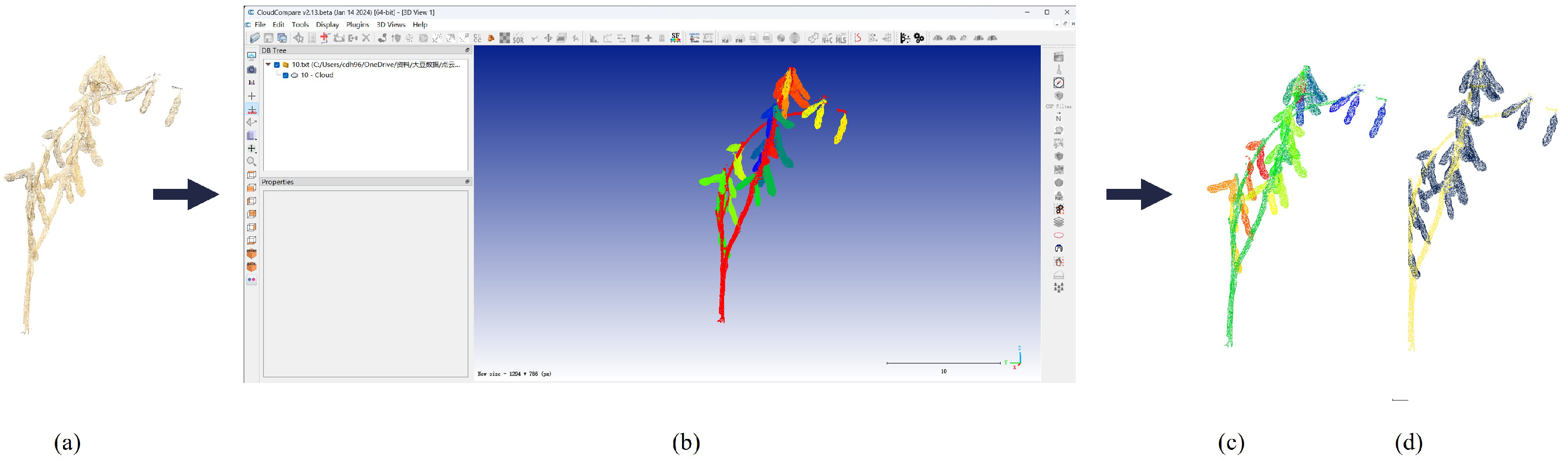
2.3. Point Cloud Segmentation
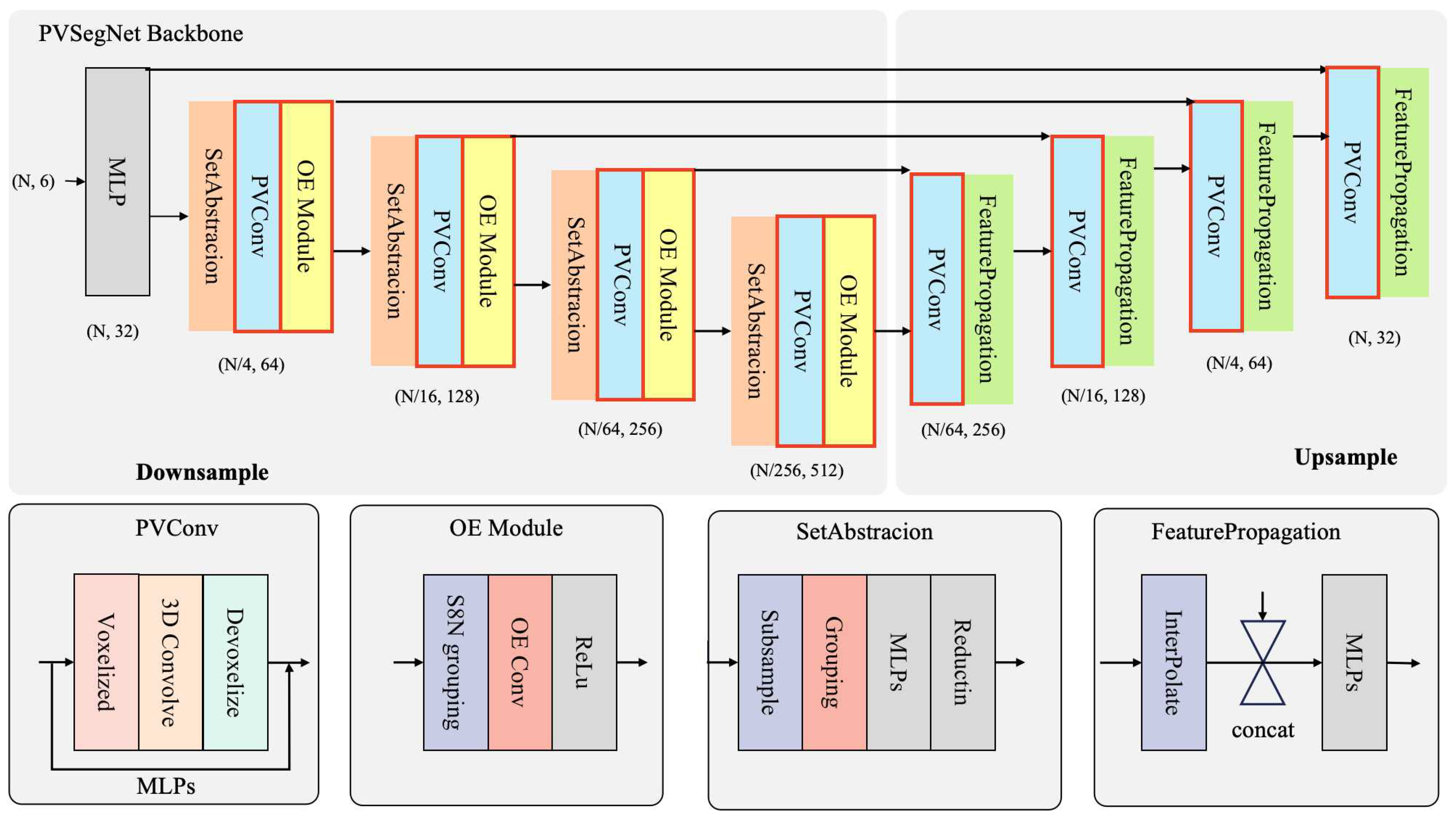
2.3.1. Network Architecture
2.3.2. Evaluation Metrics
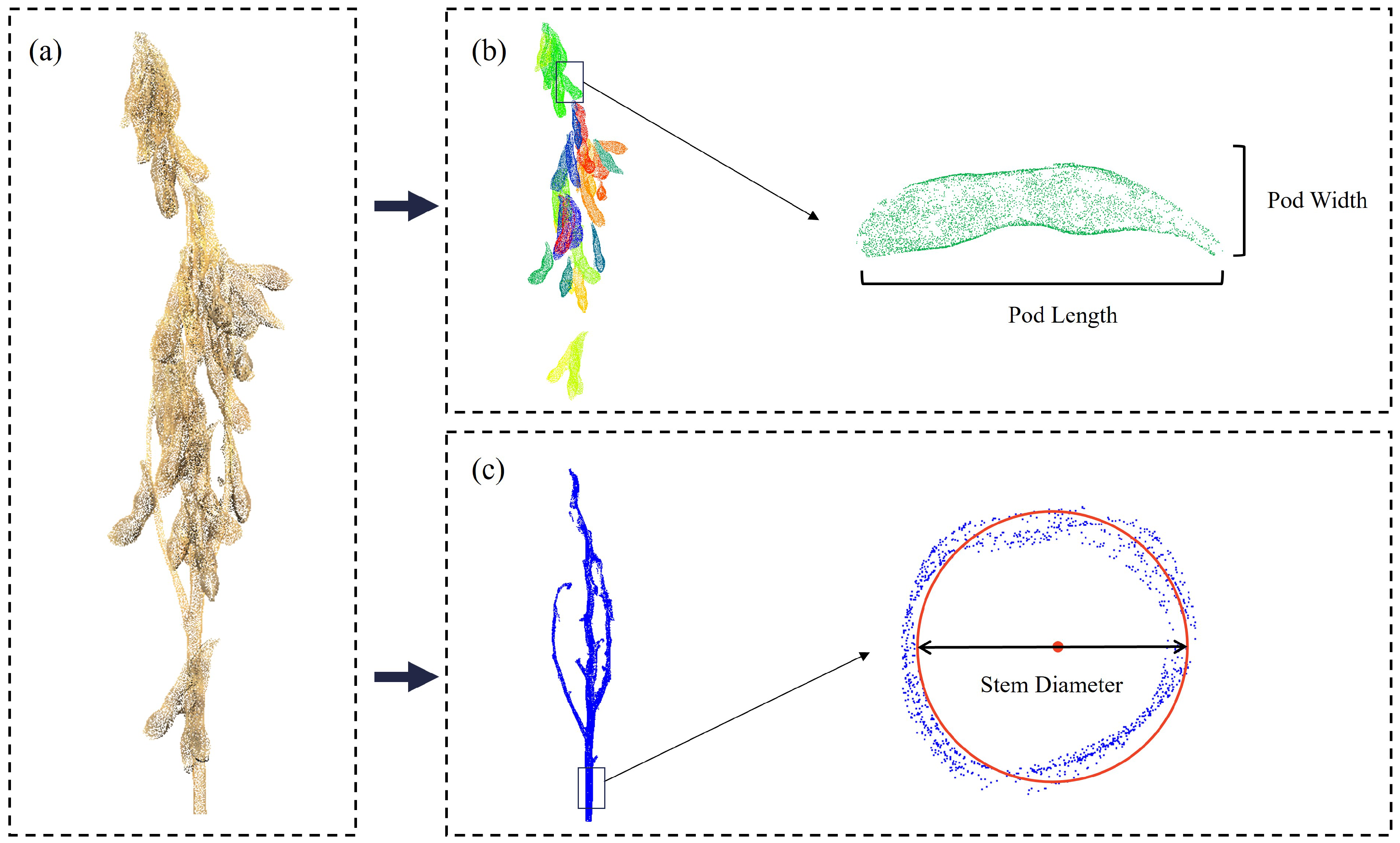
2.4. Morphological Parameter Extraction
3. Results
3.1. Experimental Setup
3.2. Ablation Study on the Effectiveness of the Method
3.3. Segmentation Results and Comparison
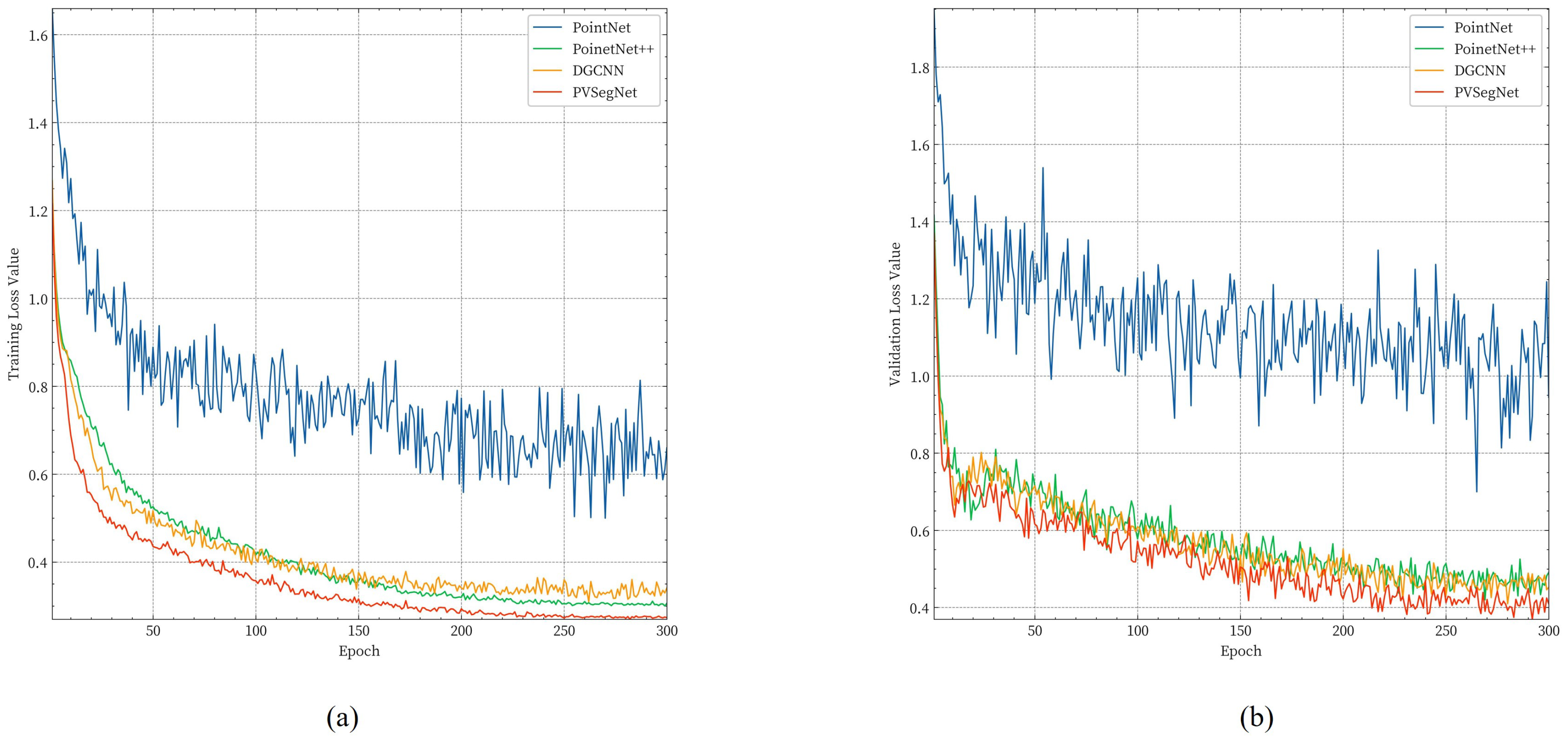
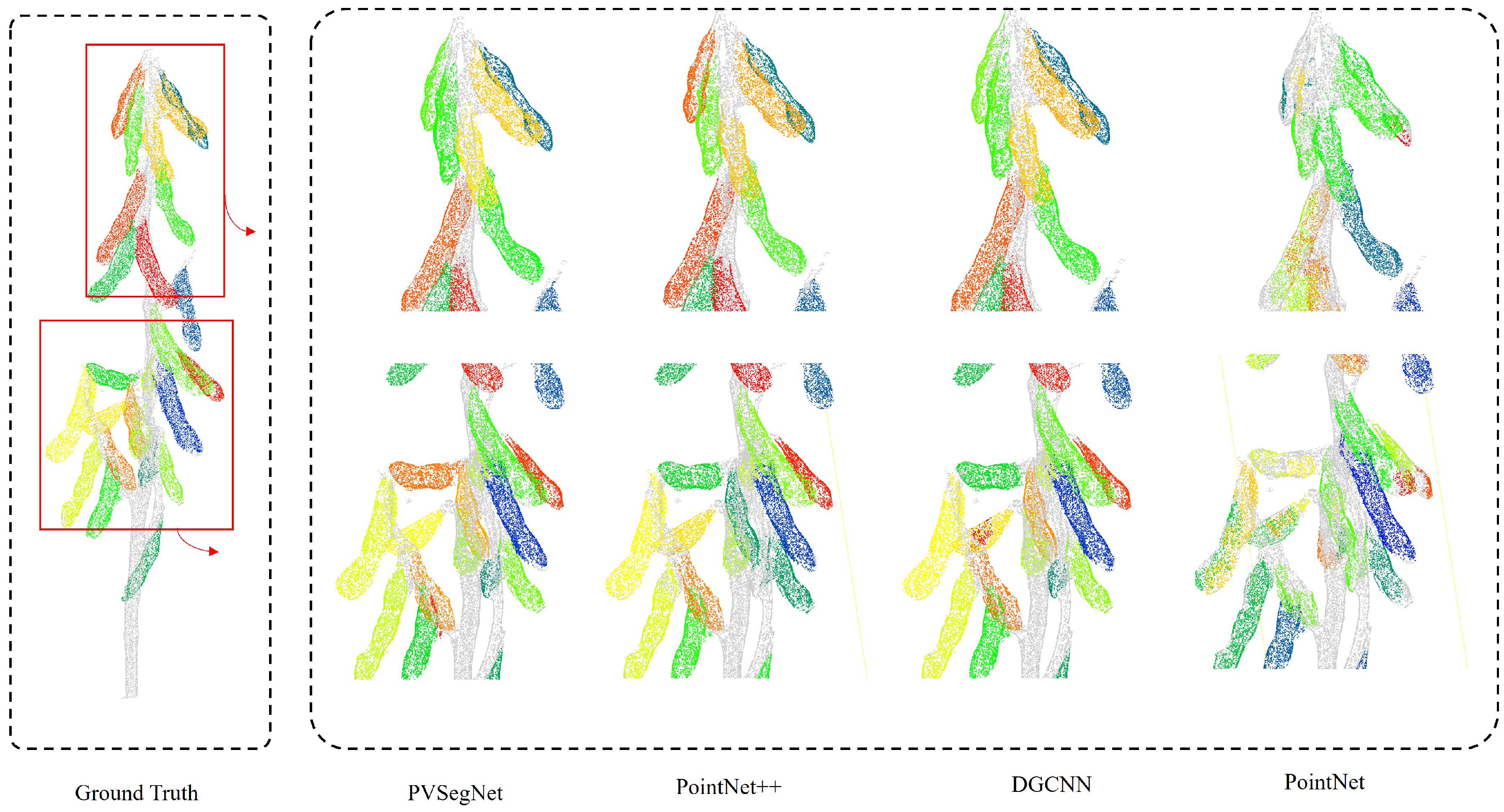
3.3.1. Comparison of Semantic Segmentation Methods
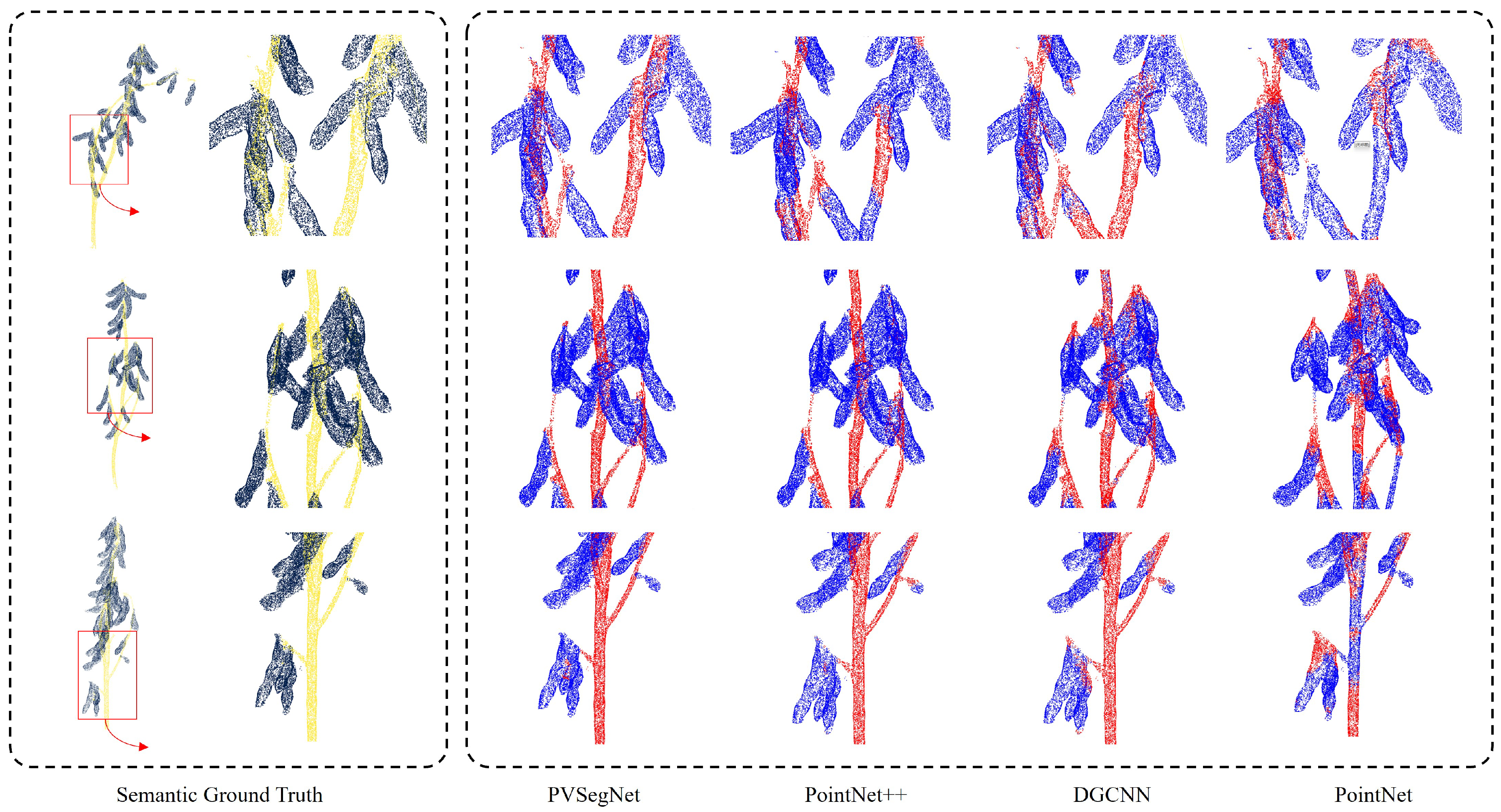
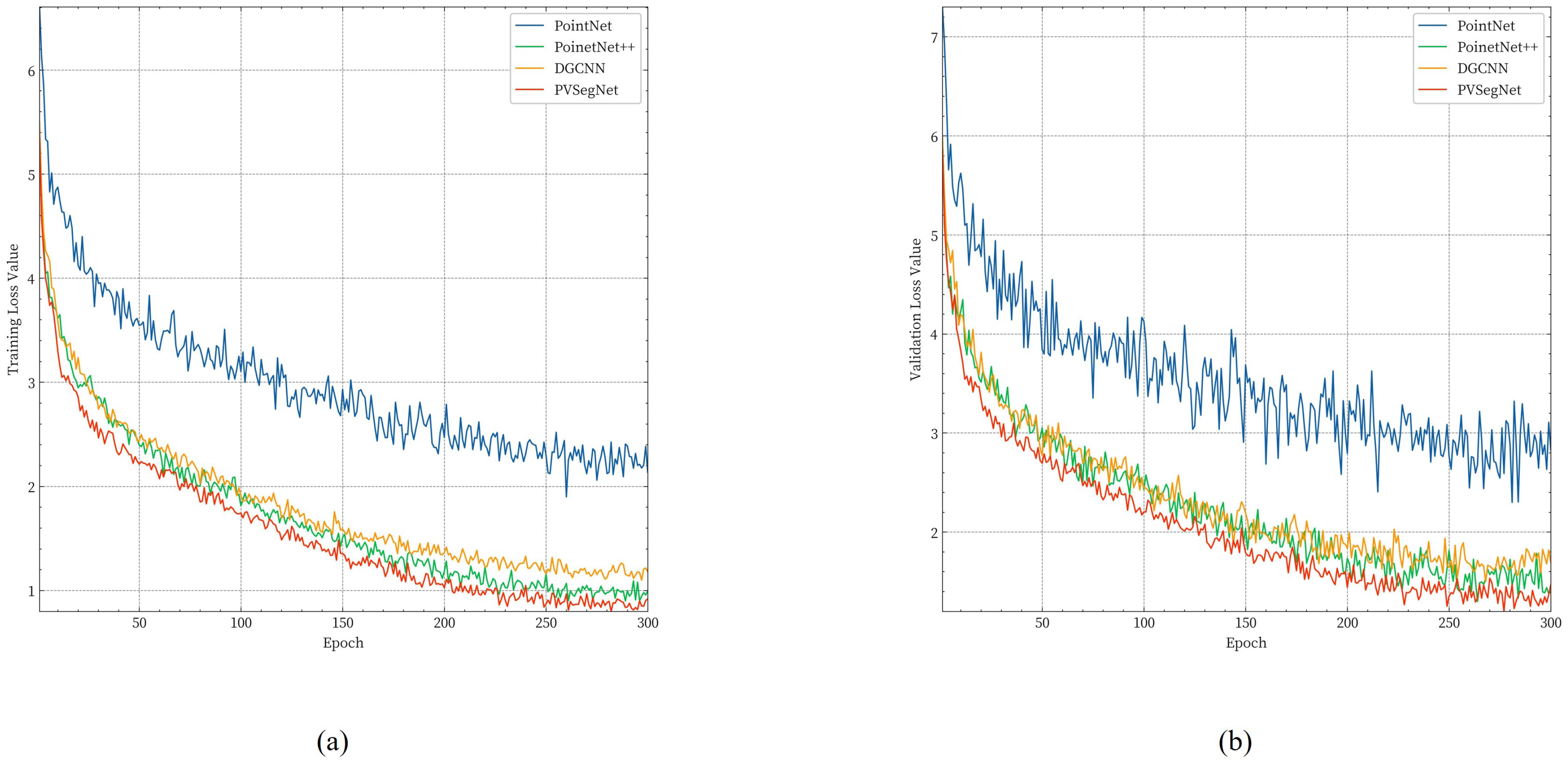
3.3.2. Comparison of Instance Segmentation Methods
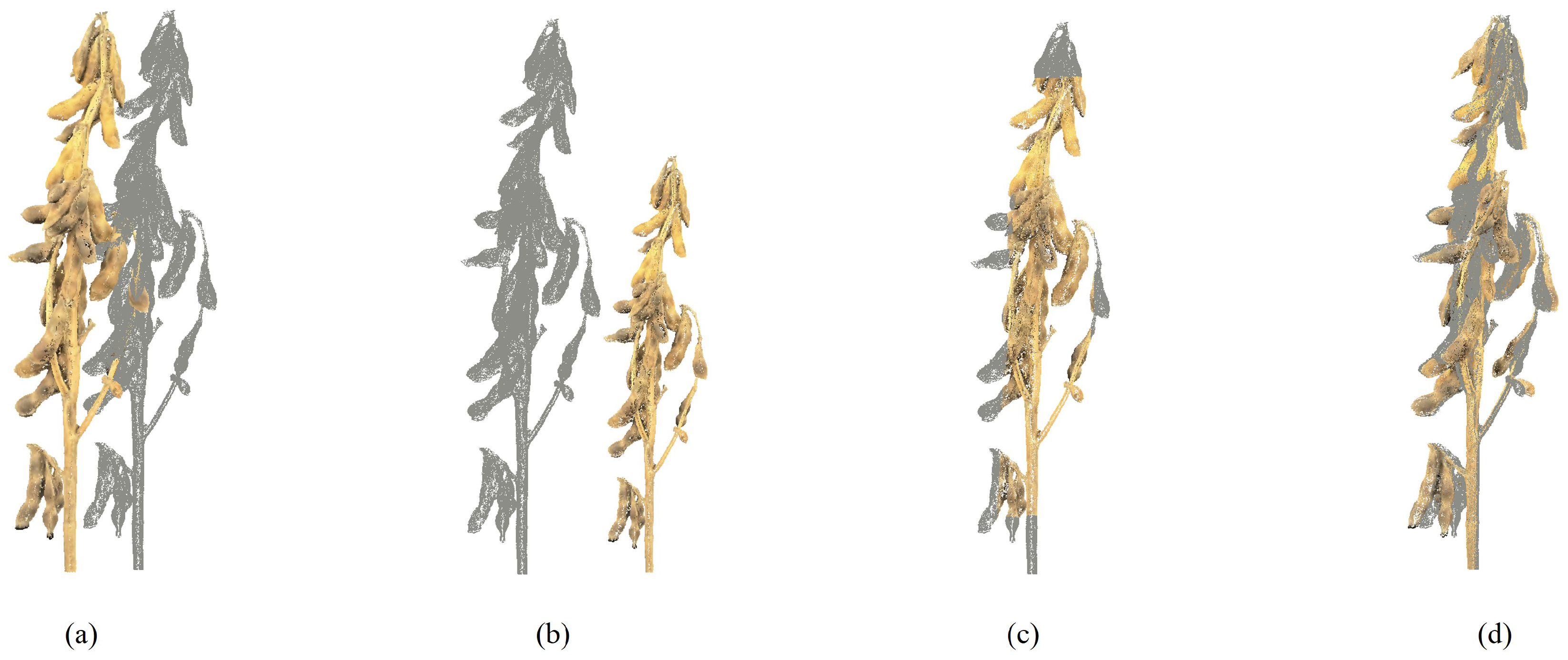
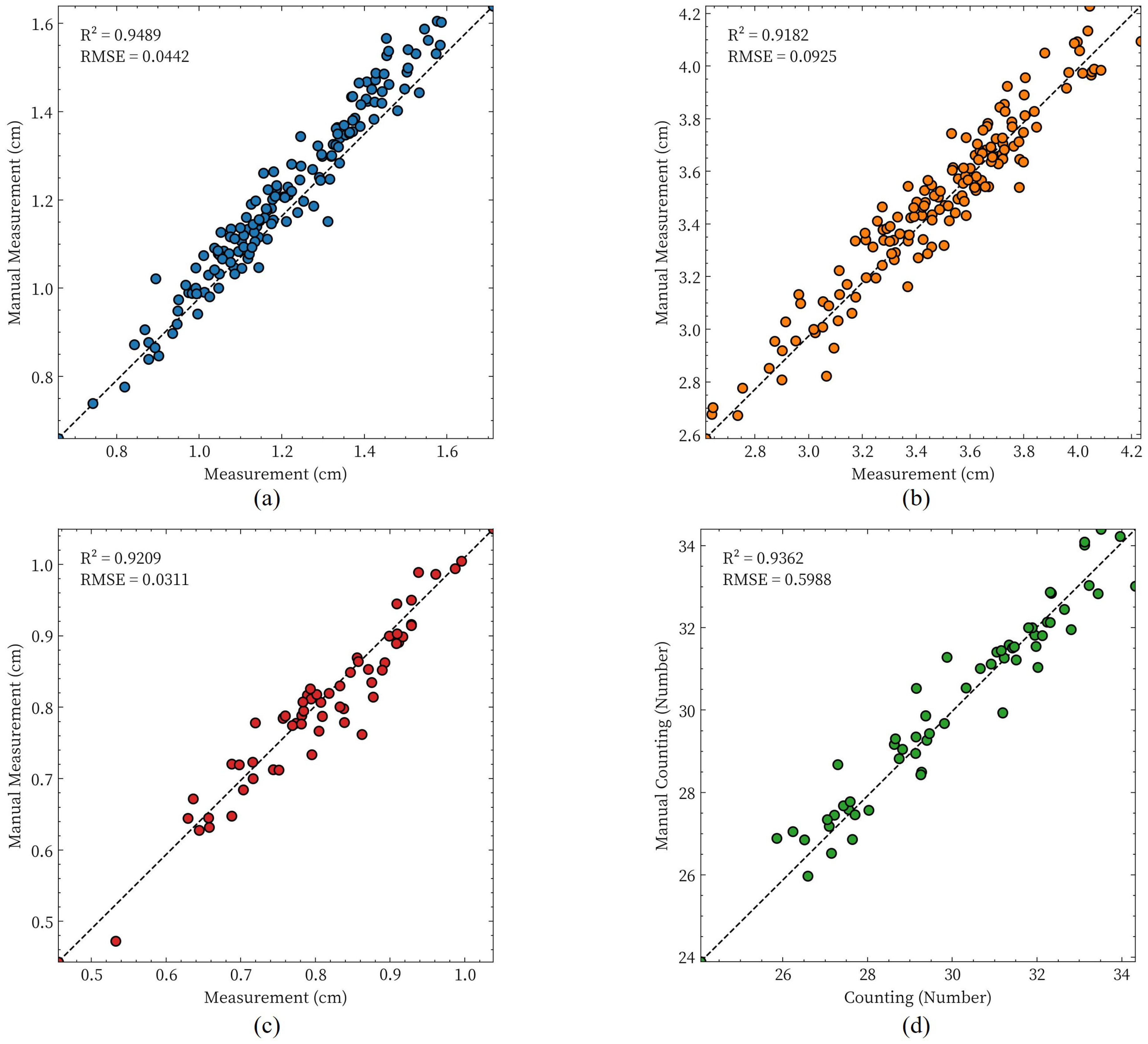
3.4. Results of Phenotypic Parameter Extraction
4. Discussion
4.1. Point Cloud Generation
4.2. Downsampling the Number of Point Clouds
4.3. Future Work
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Jiang, G.; Chen, P.; Zhang, J.; Florez-Palacios, L.; Zeng, A.; Wang, X.; Bowen, R.A.; Miller, A.M.; Berry, H. Genetic Analysis of Sugar Composition and Its Relationship with Protein, Oil, and Fiber in Soybean. Crop Sci. 2018, 58, 2413–2421. [Google Scholar] [CrossRef]
- Florou-Paneri, P.; Christaki, E.; Giannenas, I.; Bonos, E.; Skoufos, I.; Tsinas, A.; Tzora, A.; Peng, J. Alternative Protein Sources to Soybean Meal in Pig Diets. J. Food Agric. Environ. 2014, 12, 655–660. [Google Scholar]
- Zhang, M.; Liu, S.; Wang, Z.; Yuan, Y.; Zhang, Z.; Liang, Q.; Yang, X.; Duan, Z.; Liu, Y.; Kong, F.; et al. Progress in Soybean Functional Genomics over the Past Decade. Plant Biotechnol. J. 2021, 20, 256–282. [Google Scholar] [CrossRef] [PubMed]
- Bao, X.; Yao, X. Genetic Improvements in the Root Traits and Fertilizer Tolerance of Soybean Varieties Released during Different Decades. Agronomy 2023, 14, 2. [Google Scholar] [CrossRef]
- Chang, F.; Lv, W.; Lv, P.; Xiao, Y.; Yan, W.; Chen, S.; Zheng, L.; Xie, P.; Wang, L.; Karikari, B.; et al. Exploring Genetic Architecture for Pod-Related Traits in Soybean Using Image-Based Phenotyping. Mol. Breed. 2021, 41, 28. [Google Scholar] [CrossRef] [PubMed]
- Parmley, K.A.; Nagasubramanian, K.; Sarkar, S.; Ganapathysubramanian, B.; Singh, A.K. Development of Optimized Phenomic Predictors for Efficient Plant Breeding Decisions Using Phenomic-Assisted Selection in Soybean. Plant Phenomics 2019, 2019, 5809404. [Google Scholar] [CrossRef] [PubMed]
- Chouhan, S.S.; Singh, U.; Jain, S. Applications of Computer Vision in Plant Pathology: A Survey. Arch. Comput. Methods Eng. 2020, 27, 611–632. [Google Scholar] [CrossRef]
- Ngugi, L.C.; Abdelwahab, M.; Abo-Zahhad, M. Tomato Leaf Segmentation Algorithms for Mobile Phone Applications Using Deep Learning. Comput. Electron. Agric. 2020, 178, 105788. [Google Scholar] [CrossRef]
- Miao, C.; Guo, A.; Thompson, A.M.; Yang, J.; Ge, Y.; Schnable, J.C. Automation of Leaf Counting in Maize and Sorghum Using Deep Learning. Plant Phenome J. 2021, 4, e20022. [Google Scholar] [CrossRef]
- Zhang, Z.; Pope, M.; Shakoor, N.; Pless, R.; Mockler, T.; Stylianou, A. Comparing Deep Learning Approaches for Understanding Genotype × Phenotype Interactions in Biomass Sorghum. Front. Artif. Intell. 2022, 5, 872858. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Wang, D.; Zhu, B.; Liu, T.; Sun, C.; Zhang, Z. Estimation of Nitrogen Content in Wheat Using Indices Derived from RGB and Thermal Infrared Imaging. Field Crops Res. 2022, 289, 108735. [Google Scholar] [CrossRef]
- Wu, T.; Shen, P.; Dai, J.; Ma, Y.; Feng, Y. A Pathway to Assess Genetic Variation of Wheat Germplasm by Multidimensional Traits with Digital Images. Plant Phenomics 2023, 5, 119. [Google Scholar] [CrossRef] [PubMed]
- Feldmann, M.J.; Tabb, A. Cost-Effective, High-Throughput Phenotyping System for 3D Reconstruction of Fruit Form. bioRxiv 2021. [Google Scholar] [CrossRef]
- Wu, S.; Wen, W.; Gou, W.; Lu, X.; Zhang, W.; Zheng, C.; Xiang, Z.; Chen, L.; Guo, X. A Miniaturized Phenotyping Platform for Individual Plants Using Multi-View Stereo 3D Reconstruction. Front. Plant Sci. 2022, 13, 897746. [Google Scholar] [CrossRef] [PubMed]
- Ziamtsov, I.; Navlakha, S. Plant 3D (P3D): A Plant Phenotyping Toolkit for 3D Point Clouds. Bioinformatics 2020, 36, 3949–3950. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.; Sun, N.; Bai, H.; Wang, N.; Fan, Z.; Wang, Y.; Meng, Z.; Li, B.; Cong, Y. Field-Based High-Throughput Phenotyping for Maize Plant Using 3D LiDAR Point Cloud Generated With a “Phenomobile”. Front. Plant Sci. 2019, 10, 554. [Google Scholar] [CrossRef] [PubMed]
- Rivera, G.; Porras, R.; Florencia, R.; Sanchez-Solis, J.P. LiDAR Applications in Precision Agriculture for Cultivating Crops: A Review of Recent Advances. Comput. Electron. Agric. 2023, 207, 107737. [Google Scholar] [CrossRef]
- Jin, S.; Sun, X.; Wu, F.; Su, Y.; Li, Y.; Song, S.; Xu, K.; Ma, Q.; Baret, F.; Jiang, D.; et al. Lidar Sheds New Light on Plant Phenomics for Plant Breeding and Management: Recent Advances and Future Prospects. ISPRS-J. Photogramm. Remote Sens. 2021, 171, 202–223. [Google Scholar] [CrossRef]
- Song, P.; Li, Z.; Yang, M.; Shao, Y.; Pu, Z.; Yang, W.; Zhai, R. Dynamic Detection of Three-Dimensional Crop Phenotypes Based on a Consumer-Grade RGB-D Camera. Front. Plant Sci. 2023, 14, 1097725. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Duan, Y.; Shi, Y.; Kato, Y.; Ninomiya, S.; Guo, W. EasyIDP: A Python Package for Intermediate Data Processing in UAV-Based Plant Phenotyping. Remote Sens. 2021, 13, 2622. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, Z.; Sun, K.; Li, S.; Yu, J.; Miao, L.; Zhang, Z.; Li, Y.; Zhao, H.; Hu, Z.; et al. Soybean-MVS: Annotated Three-Dimensional Model Dataset of Whole Growth Period Soybeans for 3D Plant Organ Segmentation. Agriculture 2023, 13, 1321. [Google Scholar] [CrossRef]
- Luo, L.; Jiang, X.; Yang, Y.; Samy, E.R.A.; Lefsrud, M.; Hoyos-Villegas, V.; Sun, S. Eff-3DPSeg: 3D Organ-Level Plant Shoot Segmentation Using Annotation-Efficient Deep Learning. Plant Phenomics 2023, 5, 80. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Ruifang, Z.; Pujuan, S.; Pengfei, W. Segmentation of Crop Organs through Region Growing in 3D Space. In Proceedings of the 2016 Fifth International Conference on Agro-Geoinformatics (Agro-Geoinformatics), Tianjin, China, 18–20 July 2016; pp. 1–6. [Google Scholar] [CrossRef]
- Cuevas-Velasquez, H.; Gallego, A.J.; Fisher, R.B. Segmentation and 3D Reconstruction of Rose Plants from Stereoscopic Images. Comput. Electron. Agric. 2020, 171, 105296. [Google Scholar] [CrossRef]
- Vo, A.; Truong-Hong, L.; Laefer, D.; Bertolotto, M. Octree-Based Region Growing for Point Cloud Segmentation. ISPRS J. Photogramm. Remote Sens. 2015, 104, 88–100. [Google Scholar] [CrossRef]
- Han, H.; Han, X.; Gao, T. 3D Mesh Model Segmentation Based on Skeleton Extraction. Imaging Sci. J. 2021, 69, 153–163. [Google Scholar] [CrossRef]
- Dutagaci, H.; Rasti, P.; Galopin, G.; Rousseau, D. ROSE-X: An Annotated Data Set for Evaluation of 3D Plant Organ Segmentation Methods. Plant Methods 2020, 16, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Chaudhury, A.; Ward, C.D.W.; Talasaz, A.; Ivanov, A.; Brophy, M.; Grodzinski, B.; Hüner, N.; Patel, R.V.; Barron, J. Machine Vision System for 3D Plant Phenotyping. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017, 16, 2009–2022. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, S.; Mondal, D.; Panjvani, K.; Kochian, L.; Stavness, I. Explainable Deep Learning in Plant Phenotyping. Front. Artif. Intell. 2023, 6, 1203546. [Google Scholar] [CrossRef]
- Charles, R.Q.; Su, H.; Kaichun, M.; Guibas, L.J. PointNet: Deep Learning on Point Sets for 3D Classification and Segmentation. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 77–85. [Google Scholar] [CrossRef]
- Li, Y.; Wen, W.; Miao, T.; Wu, S.; Yu, Z.; Wang, X.; Guo, X.; Zhao, C. Automatic Organ-Level Point Cloud Segmentation of Maize Shoots by Integrating High-Throughput Data Acquisition and Deep Learning. Comput. Electron. Agric. 2022, 193, 106702. [Google Scholar] [CrossRef]
- Li, D.; Shi, G.; Li, J.; Chen, Y.; Zhang, S.; Xiang, S.; Jin, S. PlantNet: A Dual-Function Point Cloud Segmentation Network for Multiple Plant Species. ISPRS J. Photogramm. Remote Sens. 2022, 184, 243–263. [Google Scholar] [CrossRef]
- Zhou, Y.; Tuzel, O. VoxelNet: End-to-End Learning for Point Cloud Based 3D Object Detection. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 4490–4499. [Google Scholar] [CrossRef]
- Jin, S.; Su, Y.; Gao, S.; Wu, F.; Ma, Q.; Xu, K.; Ma, Q.; Hu, T.; Liu, J.; Pang, S.; et al. Separating the Structural Components of Maize for Field Phenotyping Using Terrestrial LiDAR Data and Deep Convolutional Neural Networks. IEEE Trans. Geosci. Remote Sens. 2020, 58, 2644–2658. [Google Scholar] [CrossRef]
- Li, D.; Li, J.; Xiang, S.; Pan, A. PSegNet: Simultaneous Semantic and Instance Segmentation for Point Clouds of Plants. Plant Phenomics 2022, 2022. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Lu, D.; Raghavan, B.; Breitkopf, P.; Dutta, S.; Xiao, M.; Zhang, W. Accelerating Large-scale Topology Optimization: State-of-the-Art and Challenges. Arch. Comput. Methods Eng. 2021, 28, 4549–4571. [Google Scholar] [CrossRef]
- Howe, M.; Repasky, B.; Payne, T. Effective Utilisation of Multiple Open-Source Datasets to Improve Generalisation Performance of Point Cloud Segmentation Models. In Proceedings of the 2022 International Conference on Digital Image Computing: Techniques and Applications (DICTA), Sydney, Australia, 30 November–2 December 2022; pp. 1–7. [Google Scholar] [CrossRef]
- Schunck, D.; Magistri, F.; Rosu, R.; Cornelissen, A.; Chebrolu, N.; Paulus, S.; Léon, J.; Behnke, S.; Stachniss, C.; Kuhlmann, H.; et al. Pheno4D: A Spatio-Temporal Dataset of Maize and Tomato Plant Point Clouds for Phenotyping and Advanced Plant Analysis. PLoS ONE 2021, 16, e0256340. [Google Scholar] [CrossRef]
- Boogaard, F.P.; van Henten, E.J.; Kootstra, G. Improved Point-Cloud Segmentation for Plant Phenotyping Through Class-Dependent Sampling of Training Data to Battle Class Imbalance. Front. Plant Sci. 2022, 13, 838190. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Liu, F.; Che, Y.; Hui, F.; Ma, Y. Three-Dimensional Quantification of Intercropping Crops in Field by Ground and Aerial Photography. In Proceedings of the 2018 6th International Symposium on Plant Growth Modeling, Simulation, Visualization and Applications (PMA), Hefei, China, 4–8 November 2018; pp. 1–5. [Google Scholar] [CrossRef]
- Chakraborty, S.; Banerjee, A.; Gupta, S.; Christensen, P. Region of Interest Aware Compressive Sensing of THEMIS Images and Its Reconstruction Quality. In Proceedings of the 2018 IEEE Aerospace Conference, Big Sky, MT, USA, 3–10 March 2018; pp. 1–11. [Google Scholar] [CrossRef]
- López-Torres, C.V.; Salazar-Colores, S.; Kells, K.; Ortega, J.; Arreguín, J.M.R. Improving 3D Reconstruction Accuracy in Wavelet Transform Profilometry by Reducing Shadow Effects. IET Image Process. 2020, 14, 310–317. [Google Scholar] [CrossRef]
- Liu, Y.; Dai, Q.; Xu, W. A Point-Cloud-Based Multiview Stereo Algorithm for Free-Viewpoint Video. IEEE Trans. Vis. Comput. Graph. 2010, 16, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Condorelli, F.; Higuchi, R.; Nasu, S.; Rinaudo, F.; Sugawara, H. Improving Performance Of Feature Extraction In Sfm Algorithms For 3d Sparse Point Cloud. ISPRS Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2019, 42, 101–106. [Google Scholar] [CrossRef]
- Dascăl, A.; Popa, M. Possibilities of 3D Reconstruction of the Vehicle Collision Scene in the Photogrammetric Environment Agisoft Metashape 1.6.2. J. Physics: Conf. Ser. 2021, 1781, 012053. [Google Scholar] [CrossRef]
- Kim, J.; Ko, Y.; Seo, J. Construction of Machine-Labeled Data for Improving Named Entity Recognition by Transfer Learning. IEEE Access 2020, 8, 59684–59693. [Google Scholar] [CrossRef]
- Armeni, I.; Sener, O.; Zamir, A.R.; Jiang, H.; Brilakis, I.; Fischer, M.; Savarese, S. 3D Semantic Parsing of Large-Scale Indoor Spaces. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 1534–1543. [Google Scholar]
- Zhang, H.; Zhu, L.; Cai, X.; Dong, L. Noise Removal Algorithm Based on Point Cloud Classification. In Proceedings of the 2022 International Seminar on Computer Science and Engineering Technology (SCSET), Indianapolis, IN, USA, 8–9 January 2022; pp. 93–96. [Google Scholar] [CrossRef]
- Wu, Z.; Zeng, Y.; Li, D.; Liu, J.; Feng, L. High-Volume Point Cloud Data Simplification Based on Decomposed Graph Filtering. Autom. Constr. 2021, 129, 103815. [Google Scholar] [CrossRef]
- Chen, S.; Wang, J.; Pan, W.; Gao, S.; Wang, M.; Lu, X. Towards Uniform Point Distribution in Feature-Preserving Point Cloud Filtering. Comput. Vis. Media 2022, 9, 249–263. [Google Scholar] [CrossRef]
- Zhou, J.; Fu, X.; Zhou, S.; Zhou, J.; Ye, H.; Nguyen, H. Automated Segmentation of Soybean Plants from 3D Point Cloud Using Machine Learning. Comput. Electron. Agric. 2019, 162, 143–153. [Google Scholar] [CrossRef]
- Cortés-Ciriano, I.; Bender, A. Improved Chemical Structure-Activity Modeling Through Data Augmentation. J. Chem. Inf. Model. 2015, 55 12, 2682–2692. [Google Scholar] [CrossRef]
- Wang, M.; Ju, M.; Fan, Y.; Guo, S.; Liao, M.; Yang, H.j.; He, D.; Komura, T. 3D Incomplete Point Cloud Surfaces Reconstruction With Segmentation and Feature-Enhancement. IEEE Access 2019, 7, 15272–15281. [Google Scholar] [CrossRef]
- Xin, B.; Sun, J.; Bartholomeus, H.; Kootstra, G. 3D Data-Augmentation Methods for Semantic Segmentation of Tomato Plant Parts. Front. Plant Sci. 2023, 14, 1045545. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Tang, H.; Lin, Y.; Han, S. Point-Voxel CNN for Efficient 3D Deep Learning. arXiv 2019, arXiv:cs/1907.03739. [Google Scholar]
- Jiang, M.; Wu, Y.; Zhao, T.; Zhao, Z.; Lu, C. PointSIFT: A SIFT-like Network Module for 3D Point Cloud Semantic Segmentation. arXiv 2018, arXiv:cs/1807.00652. [Google Scholar]
- Magistri, F.; Chebrolu, N.; Stachniss, C. Segmentation-Based 4D Registration of Plants Point Clouds for Phenotyping. In Proceedings of the 2020 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS), Las Vegas, NV, USA, 25–29 October 2020; pp. 2433–2439. [Google Scholar] [CrossRef]
- Wahabzada, M.; Paulus, S.; Kersting, K.; Mahlein, A.K. Automated Interpretation of 3D Laserscanned Point Clouds for Plant Organ Segmentation. BMC Bioinform. 2015, 16, 1–11. [Google Scholar] [CrossRef]
- Jiang, L.; Zhao, H.; Shi, S.; Liu, S.; Fu, C.W.; Jia, J. PointGroup: Dual-Set Point Grouping for 3D Instance Segmentation. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; pp. 4866–4875. [Google Scholar] [CrossRef]
- Saeed, F.; Sun, S.; Rodriguez-Sanchez, J.; Snider, J.; Liu, T.; Li, C. Cotton plant part 3D segmentation and architectural trait extraction using point voxel convolutional neural networks. Plant Methods 2023, 19, 33. [Google Scholar] [CrossRef]
- Miao, T.; Zhu, C.; Xu, T.; Yang, T.; Li, N.; Zhou, Y.; Deng, H. Automatic Stem-Leaf Segmentation of Maize Shoots Using Three-Dimensional Point Cloud. Comput. Electron. Agric. 2021, 187, 106310. [Google Scholar] [CrossRef]
- Qi, C.R.; Yi, L.; Su, H.; Guibas, L.J. Pointnet++: Deep hierarchical feature learning on point sets in a metric space. Adv. Neural Inf. Process. Syst. 2017, 30. [Google Scholar]
- Wang, Y.; Sun, Y.; Liu, Z.; Sarma, S.E.; Bronstein, M.M.; Solomon, J.M. Dynamic Graph CNN for Learning on Point Clouds. ACM Trans. Graph. 2019, 38, 146:1–146:12. [Google Scholar] [CrossRef]
- Rose, J.; Paulus, S.; Kuhlmann, H. Accuracy Analysis of a Multi-View Stereo Approach for Phenotyping of Tomato Plants at the Organ Level. Sensors 2015, 15, 9651–9665. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Wang, M.; Zhang, C.; Li, J.; Yan, K.; Liang, Z.; Wei, R. A Dynamic Multi-Branch Neural Network Module for 3D Point Cloud Classification and Segmentation Using Structural Re-parametertization. In Proceedings of the 2023 11th International Conference on Agro-Geoinformatics (Agro-Geoinformatics), Wuhan, China, 25–28 July 2023; pp. 1–6. [Google Scholar] [CrossRef]
- Zhai, R.; Li, X.; Wang, Z.; Guo, S.; Hou, S.; Hou, Y.; Gao, F.; Song, J. Point Cloud Classification Model Based on a Dual-Input Deep Network Framework. IEEE Access 2020, 8, 55991–55999. [Google Scholar] [CrossRef]


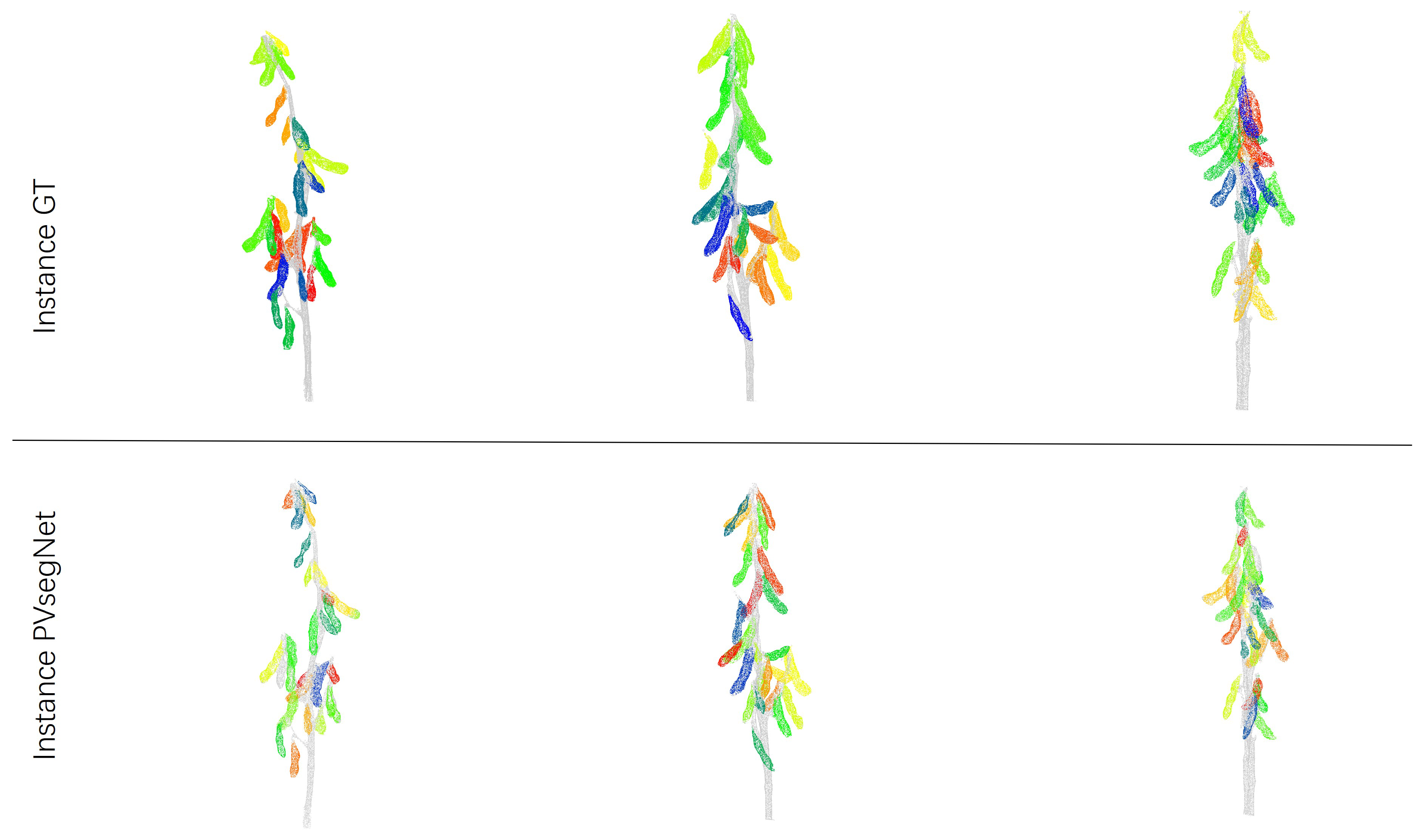
| Description | Value |
|---|---|
| Number of samples | 60 |
| Number of points | 61.12 × − 68.58 × |
| Plant coverage ([length, width, height]) | min: [−2.56, −2.66, 12.00], max: [1.73, 2.19, 31.39] |
| Average pods proportion (%) | 43.21 |
| Parameter | Value |
|---|---|
| Batch Size | 4 |
| Epochs | 300 |
| Learning Rate | 0.01 |
| Optimizer | Adam |
| Momentum | 0.9 |
| Method | IoU(%) | Prec (%) | Rec (%) | F1-Score (%) | |
|---|---|---|---|---|---|
| Baseline | Stem | 87.80 | 93.86 | 93.15 | 93.50 |
| Pod | 93.65 | 96.54 | 96.91 | 96.72 | |
| Mean | 90.73 | 95.20 | 95.03 | 95.11 | |
| +PVConv | Stem | 88.26 | 95.06 | 95.51 | 95.28 |
| Pod | 93.98 | 96.25 | 94.46 | 95.35 | |
| Mean | 91.12 | 95.65 | 94.98 | 95.31 | |
| +OE | Stem | 91.91 | 95.22 | 96.17 | 95.69 |
| Pod | 90.13 | 94.96 | 94.42 | 94.69 | |
| Mean | 91.02 | 95.09 | 95.30 | 95.19 | |
| +PVConv + OE | Stem | 89.51 | 96.47 | 92.54 | 94.46 |
| Pod | 94.70 | 96.29 | 98.28 | 97.28 | |
| Mean | 92.10 | 96.38 | 95.41 | 95.87 |
| Method | IoU (%) | Prec (%) | Rec (%) | F1-Score (%) | |
|---|---|---|---|---|---|
| PointNet | Stem | 48.38 | 70.40 | 62.31 | 62.64 |
| Pod | 78.51 | 85.62 | 90.44 | 87.96 | |
| Mean | 58.96 | 78.81 | 70.07 | 74.18 | |
| PointNet++ | Stem | 87.80 | 93.86 | 93.15 | 93.50 |
| Pod | 93.65 | 96.54 | 96.91 | 96.72 | |
| Mean | 90.73 | 95.20 | 95.03 | 95.11 | |
| DGCNN | Stem | 92.13 | 93.90 | 98.00 | 95.90 |
| Pod | 84.13 | 95.68 | 87.45 | 91.38 | |
| Mean | 88.13 | 94.79 | 92.72 | 93.64 | |
| PVSegNet (Ours) | Stem | 89.51 | 96.47 | 92.54 | 94.46 |
| Pod | 94.70 | 96.29 | 98.28 | 97.28 | |
| Mean | 92.10 | 96.38 | 95.41 | 95.87 |
| Method | AP@50 (%) | AP@25 (%) | AR@50 (%) | AR@25 (%) | |
|---|---|---|---|---|---|
| PointNet | Stem | 41.23 | 61.83 | 42.09 | 55.45 |
| Pod | 40.07 | 52.48 | 63.89 | 88.89 | |
| Mean | 40.65 | 57.15 | 52.99 | 72.17 | |
| DGCNN | Stem | 83.00 | 91.25 | 83.33 | 91.67 |
| Pod | 69.73 | 75.51 | 70.20 | 75.58 | |
| Mean | 76.37 | 83.38 | 76.77 | 83.62 | |
| PointNet++ | Stem | 74.97 | 98.02 | 86.79 | 89.86 |
| Pod | 86.75 | 89.54 | 86.11 | 97.22 | |
| Mean | 80.86 | 93.78 | 86.45 | 93.54 | |
| PVSegNet (Ours) | Stem | 78.97 | 97.96 | 86.12 | 90.17 |
| Pod | 87.97 | 90.15 | 88.02 | 97.23 | |
| Mean | 83.47 | 94.06 | 87.07 | 93.70 |
| Number of Points | mIoU (%) | Throughput (Items/s) |
|---|---|---|
| 6144 | 88.97 | 117.11 |
| 8192 | 91.02 | 84.92 |
| 10,240 | 92.10 | 65.27 |
| 12,288 | 92.23 | 52.50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, D.; Liu, P.; Liu, Y.; Zhao, Z.; Feng, J. Automated Phenotypic Analysis of Mature Soybean Using Multi-View Stereo 3D Reconstruction and Point Cloud Segmentation. Agriculture 2025, 15, 175. https://doi.org/10.3390/agriculture15020175
Cui D, Liu P, Liu Y, Zhao Z, Feng J. Automated Phenotypic Analysis of Mature Soybean Using Multi-View Stereo 3D Reconstruction and Point Cloud Segmentation. Agriculture. 2025; 15(2):175. https://doi.org/10.3390/agriculture15020175
Chicago/Turabian StyleCui, Daohan, Pengfei Liu, Yunong Liu, Zhenqing Zhao, and Jiang Feng. 2025. "Automated Phenotypic Analysis of Mature Soybean Using Multi-View Stereo 3D Reconstruction and Point Cloud Segmentation" Agriculture 15, no. 2: 175. https://doi.org/10.3390/agriculture15020175
APA StyleCui, D., Liu, P., Liu, Y., Zhao, Z., & Feng, J. (2025). Automated Phenotypic Analysis of Mature Soybean Using Multi-View Stereo 3D Reconstruction and Point Cloud Segmentation. Agriculture, 15(2), 175. https://doi.org/10.3390/agriculture15020175






