Advances in Genetics and Breeding of Grain Shape in Rice
Abstract
1. Introduction
2. Genetic Regulation Mechanisms of Rice Grain Shape
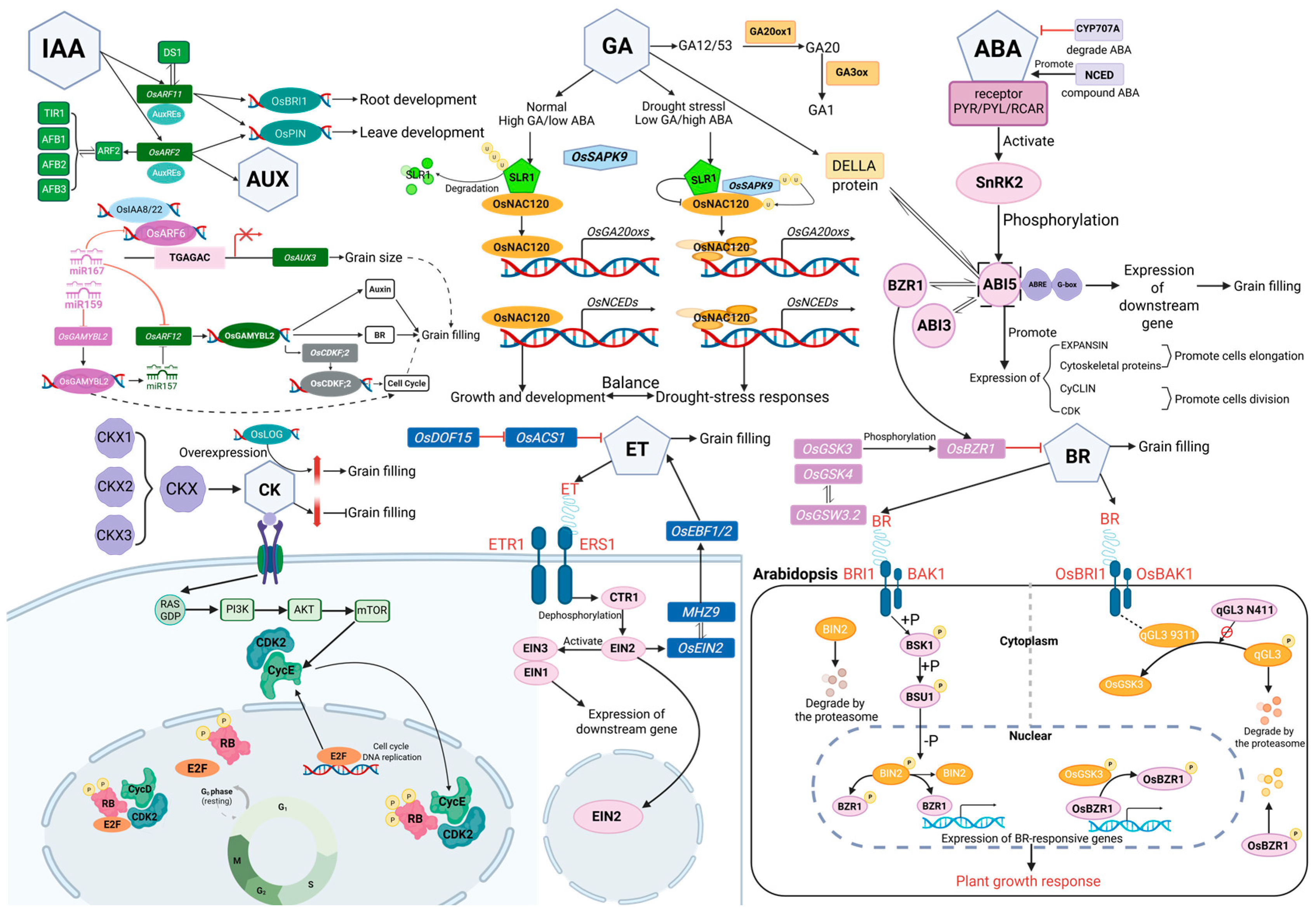
2.1. Hormone Signaling Pathways
2.1.1. Auxin
2.1.2. Cytokinin
2.1.3. Gibberellin
2.1.4. Abscisic Acid
2.1.5. Brassinosteroids
2.1.6. Ethylene
| Gene | Type | Function | Reference |
|---|---|---|---|
| OsARF2 | Transcription factor | An auxin response factor that positively regulates grain size | [29,30] |
| OsARF11 | Transcription factor | An auxin response factor that positively regulates grain size | [21,25] |
| OsIAA1 | Member of the rice Aux/IAA family | An auxin response factor that negatively regulates the auxin signaling pathway | [26] |
| OsPIN1 | Auxin transport protein | Determines rice grain size by “establishing and maintaining the auxin (IAA) concentration gradient between vascular bundles and endosperm” | [31] |
| OsCyclinD1 | Cell cycle regulatory protein | Positively regulates grain size | [20,32] |
| OsCDKA | Cell cycle regulatory protein | Positively regulates grain size | [20,32] |
| OsCKX1 | Member of the cytokinin oxidase/dehydrogenase gene family | Negatively regulates grain length and width by degrading active cytokinins | [33] |
| OsCKX2 | Member of the cytokinin oxidase/dehydrogenase gene family | Negatively regulates grain length and width by degrading active cytokinins | [33] |
| OsCKX3 | Member of the cytokinin oxidase/dehydrogenase gene family | Negatively regulates grain length and width by degrading active cytokinins | [33] |
| OsRR1 | Member of the type-A cytokinin response regulator family | Negatively regulates grain size by repressing cytokinin signaling | [30] |
| OsRR2 | Member of the type-A cytokinin response regulator family | Negatively regulates grain size by repressing cytokinin signaling | [36] |
| OsRR3 | Member of the type-A cytokinin response regulator family | Negatively regulates grain size by repressing cytokinin signaling | [36] |
| OsLOG | Cytokinin activation/biosynthesis gene | Promotes cytokinin biosynthesis, thereby positively regulating grain size | [37] |
| OsGA20ox1 | Key late-stage enzyme gene in gibberellin biosynthesis | Negatively regulates grain size by reducing gibberellin activity | [41,45,46] |
| OsGA3ox2 | Key late-stage enzyme gene in gibberellin biosynthesis | Positively regulates grain size by enhancing gibberellin activity | [42] |
| OsABI5 | bZIP transcription factor gene in the ABA signaling pathway | Negatively determines grain size by repressing the cell cycle, expansins, and sugar transport | [52,53,54] |
| OsNCED | Member of the 9-cis-epoxycarotenoid dioxygenase gene family | Positively regulates ABA biosynthesis, promoting cell differentiation and early grain filling | [55] |
| OsCYP707A | Member of the cytochrome P450 monooxygenase A-type family | Negatively regulates ABA biosynthesis, preventing premature termination of grain filling | [56] |
| OsBRI1 | Key receptor in the brassinosteroid (BR) signaling pathway | Perceives brassinosteroids and initiates downstream signaling, thereby positively regulating grain size | [63,64] |
| OsBZR1 | Core transcription factor in the BR signaling pathway | Functions with OsBRI1 to positively regulate grain size | [65,66] |
| OsGSK3 | Key kinase in the BR signaling pathway | Positively regulates grain size by controlling cell growth and division | [67] |
| OsACS1 | Key enzyme gene in ethylene biosynthesis | Positively regulates ethylene biosynthesis, thereby influencing grain size | [76,77] |
| OsETR1 | Member of the ethylene receptor gene family | Perceives ethylene and activates signaling cascades, inducing downstream regulatory factors to control grain size | [78] |
| OsERS1 | Member of the ethylene receptor gene family | Regulates cell division and expansion via ethylene signaling, thereby influencing grain size | [78] |
| OsEIN2 | Core transducer in the ethylene signaling pathway | Transmits signals to nuclear transcription factors, activating or repressing downstream genes, thus positively regulating grain size | [79,80] |
| OsCTR1 | Negative regulator RAF-like serine/threonine protein kinase gene in the ethylene signaling pathway | Suppresses abnormal responses triggered by excessive ethylene, thereby negatively regulating grain size | [81,82] |
2.2. Transcriptional Regulatory Pathways
2.3. Multigenic Interactions and Regulatory Networks
2.3.1. Multigenic Regulatory Networks
2.3.2. Conservation and Functional Divergence of Genes Across Species
2.4. Mechanisms of Protein Metabolism and Ubiquitination Regulation
2.4.1. Functions of the Ubiquitin–Proteasome System (UPS) in Seed Development
2.4.2. Molecular Mechanisms Linking Protein Degradation and Signal Transduction
2.5. Epigenetic and Non-Coding RNA Regulatory Mechanisms
2.5.1. Epigenetic Modifications in Seed Development
2.5.2. Regulatory Roles and Biological Significance of miRNAs and lncRNAs
2.5.3. Synergistic Effects of Non-Coding RNAs and Epigenetic Modifications
2.5.4. Role of Chromatin Remodeling in Seed Size Regulation
| Gene | Type | Function | Reference |
|---|---|---|---|
| OsDDM1b | Chromatin remodeling enzyme gene of the WI2/SNF2 family | Loss-of-function mutation significantly reduces overall DNA methylation, activating multiple genes that negatively regulate cell division, ultimately leading to smaller grain size | [148] |
| OsHDT701 | Histone deacetylase | Loss of activity causes premature expansion and division of endosperm cells, resulting in poor grain filling | [142] |
| H3K27me3 | Trimethylation of lysine 27 on histone H3 | Acts as an epigenetic “brake”; its level and distribution determine the balance between cell proliferation and grain filling, directly regulating grain size | [150,151] |
| H3K4me3 | Trimethylation of lysine 4 on histone H3 | Enhances transcriptional activity, promoting grain growth and filling | [152] |
| miRNA156 | MicroRNA (miRNA) | Regulates SPL expression to control plant responses to auxin, thereby influencing grain size | [87,88] |
| lncRNA | Long non-coding RNA | Interacts with chromatin remodeling proteins to precisely regulate the transcription of specific genes involved in cell division and proliferation during grain filling | [155] |
| SWI/SNF complex | Chromatin remodeling factor | Open chromatin structure promotes transcription of genes related to grain filling and cell division | [164] |
2.6. Environmental Influences on Seed Development and Adaptive Mechanisms
2.6.1. Impact of High-Temperature Stress, Light, and Nutrients on Seed Size
2.6.2. Effects of Local Environmental Conditions on Seed Development
2.6.3. Agricultural Implications of Regulating Seed Size
3. Application of Grain Shape Research in Rice Breeding
3.1. Molecular Regulatory Networks Lay the Foundation for Precision Breeding
3.2. Gene-Based Breeding Strategies and Germplasm Innovation
4. Perspectives
4.1. Multi-Omics Platforms Promote Intelligent Breeding of Grain Shape
4.2. Challenges and Future Outlook
4.2.1. Challenges
4.2.2. Future Outlook
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fu, Y.; Zhao, H.; Xiao, M.; Zhang, G.; Wang, S. Research progress on elongation of cooked rice. J. South China Agric. Univ. 2023, 44, 670–678. [Google Scholar]
- Shoukat, R.; Cappai, M.; Pilia, L.; Pia, G. Rice Starch Chemistry, Functional Properties, and Industrial Applications: A Review. Polymers 2025, 17, 110. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wang, Z.; Liu, Q.; Zhao, D. Genetic Improvement of rice Grain size Using the CRISPR/Cas9 System. Rice 2025, 18, 3. [Google Scholar] [CrossRef]
- Jamil, M.; Ahmad, W.; Sanwal, M.; Maqsood, M.F. Gene editing and GWAS for digital imaging analysis of wheat grain weight, size and shape are inevitable to enhance the yield. Cereal Res. Commun. 2025, 53, 1199–1218. [Google Scholar] [CrossRef]
- Li, T.; Cao, H.; Xiong, L.; Wang, F.; Li, S.; Gu, H.; Luo, W.; He, G.; Liang, S. Research Progress on the Function of Rice Grain Type Genes. Guangdong Agric. Sci. 2023, 50, 12–28. [Google Scholar]
- Khush, G.S. Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 1997, 35, 25–34. [Google Scholar] [CrossRef]
- Juliano, B.O.; Villareal, C.P. Grain Quality Evaluation of World Rices; International Rice Research Institute: Los Baños, Philippines, 1993. [Google Scholar]
- Heinemann, R.J.B.; Fagundes, P.L.; Pinto, E.A.; Penteado, M.V.C.; Lanfer-Marquez, U.M. Comparative study of nutrient composition of commercial brown, parboiled and milled rice from Brazil. J. Food Compos. Anal. 2005, 18, 287–296. [Google Scholar] [CrossRef]
- Díaz-Benito, P.; Banakar, R.; Rodríguez-Menéndez, S.; Capell, T.; Pereiro, R.; Christou, P.; Abadía, J.; Fernández, B.; Álvarez-Fernández, A. Iron and Zinc in the Embryo and Endosperm of Rice (Oryza sativa L.) Seeds in Contrasting 2′-Deoxymugineic Acid/Nicotianamine Scenarios. Front. Plant Sci. 2018, 9, 1190. [Google Scholar] [CrossRef]
- Wang, X.; Sun, J.; Yi, Z.; Dong, S. Effects of seed size on soybean performance: Germination, growth, stress resistance, photosynthesis, and yield. BMC Plant Biol. 2025, 25, 219. [Google Scholar] [CrossRef] [PubMed]
- Li, W.W.; Wang, L.; Xue, H.; Zhang, M.M.; Song, H.; Qin, M.; Dong, Q.Z. Molecular and genetic basis of plant architecture in soybean. Front. Plant Sci. 2024, 15, 1477616. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Peng, J.Z.; Qiao, Y.C.; Wang, G.P. Natural Allelic Variations of Bch10G006400 Controlling Seed Size in Chieh-qua (Benincasa hispida Cogn. var. Chieh-qua How). Int. J. Mol. Sci. 2024, 25, 4236. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; He, Y.Q.; Huang, S.L.; Wang, Z.F. Advances in the Identification of Quantitative Trait Loci and Genes Involved in Seed Vigor in Rice. Front. Plant Sci. 2021, 12, 659307. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.W.; Kopittke, P.M.; Zhao, F.J.; Wang, P. Nutrient accumulation and transcriptome patterns during grain development in rice. J. Exp. Bot. 2023, 74, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Yang, L.; Fang, Z.; Zhang, Y.; Zhuang, M.; Lv, H.; Wang, Y. Plant SWEET Family of Sugar Transporters: Structure, Evolution and Biological Functions. Biomolecules 2022, 12, 205. [Google Scholar] [CrossRef]
- Barreda, L.; Boutet, S.; De Vos, D.; Boulard, C.; Grain, D.; Lepiniec, L.; Corso, M. Specialized metabolome and transcriptome atlas of developing Arabidopsis thaliana seed under warm temperatures. Sci. Data 2025, 12, 306. [Google Scholar] [CrossRef]
- Pankaj, R.; Lima, R.B.; Figueiredo, D.D. Hormonal regulation and crosstalk during early endosperm and seed coat development. Plant Reprod. 2024, 38, 5. [Google Scholar] [CrossRef]
- Zhang, Z.; Han, H.; Zhao, J.; Liu, Z.; Deng, L.; Wu, L.; Niu, J.; Guo, Y.; Wang, G.; Gou, X.; et al. Peptide hormones in plants. Mol. Hortic. 2025, 5, 7. [Google Scholar] [CrossRef]
- Lorenzo-Manzanarez, J.L.; Enríquez-Valencia, A.J.; Olivares-García, C.A.; Ibarra-Laclette, E.; Velázquez-López, O.; Ruiz-May, E.; Loyola-Vargas, V.M.; Kú-González, A.F.; Arteaga-Vázquez, M.A.; Mata-Rosas, M. Genome-wide analysis of ARF gene family and miR160 in avocado (Persea americana Mill.) and their roles in somatic embryogenesis from zygotic embryos. Planta 2025, 261, 61. [Google Scholar] [CrossRef]
- Choudhary, P.; Aggarwal, P.R.; Salvi, P.; Muthamilarasan, M. Molecular insight into auxin signaling and associated network modulating stress responses in rice. Plant Physiol. Biochem. 2025, 219, 109452. [Google Scholar] [CrossRef]
- Sims, K.; Abedi-Samakush, F.; Szulc, N.; Honti, M.G.M.; Mattsson, J. OsARF11 Promotes Growth, Meristem, Seed, and Vein Formation during Rice Plant Development. Int. J. Mol. Sci. 2021, 22, 4089. [Google Scholar] [CrossRef]
- Wang, F.; Lin, J.; Yang, F.; Chen, X.; Liu, Y.; Yan, L.; Chen, J.; Wang, Z.; Xie, H.; Zhang, J.; et al. The OsMAPK5-OsWRKY72 module negatively regulates grain length and grain weight in rice. J. Integr. Plant Biol. 2024, 66, 2648–2663. [Google Scholar] [CrossRef]
- Qiao, J.; Jiang, H.; Lin, Y.; Shang, L.; Wang, M.; Li, D.; Fu, X.; Geisler, M.; Qi, Y.; Gao, Z.; et al. A novel miR167a-OsARF6-OsAUX3 module regulates grain length and weight in rice. Mol. Plant 2021, 14, 1683–1698. [Google Scholar] [CrossRef]
- Huang, G.; Hu, H.; van de Meene, A.; Zhang, J.; Dong, L.; Zheng, S.; Zhang, F.; Betts, N.S.; Liang, W.; Bennett, M.J.; et al. AUXIN RESPONSE FACTORS 6 and 17 control the flag leaf angle in rice by regulating secondary cell wall biosynthesis of lamina joints. Plant Cell 2021, 33, 3120–3133. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yang, C.Y.; Miao, R.; Zhou, C.L.; Cao, P.H.; Lan, J.; Zhu, X.J.; Mou, C.L.; Huang, Y.S.; Liu, S.J.; et al. DS1/OsEMF1 interacts with OsARF11 to control rice architecture by regulation of brassinosteroid signaling. Rice 2018, 11, 46. [Google Scholar] [CrossRef]
- Parry, G.; Calderon-Villalobos, L.I.; Prigge, M.; Peret, B.; Dharmasiri, S.; Itoh, H.; Lechner, E.; Gray, W.M.; Bennett, M.; Estelle, M. Complex regulation of the TIR1/AFB family of auxin receptors. Proc. Natl. Acad. Sci. USA 2009, 106, 22540–22545. [Google Scholar] [CrossRef] [PubMed]
- Thakur, J.K.; Tyagi, A.K.; Khurana, J.P. OsIAA1, an Aux/IAA cDNA from rice, and changes in its expression as influenced by auxin and light. DNA Res. 2001, 8, 193–203. [Google Scholar] [CrossRef]
- Zhou, Y.; Ma, B.; Tao, J.J.; Yin, C.C.; Hu, Y.; Huang, Y.H.; Wei, W.; Xin, P.Y.; Chu, J.F.; Zhang, W.K.; et al. Rice EIL1 interacts with OsIAAs to regulate auxin biosynthesis mediated by the tryptophan aminotransferase MHZ10/OsTAR2 during root ethylene responses. Plant Cell 2022, 34, 4366–4387. [Google Scholar] [CrossRef] [PubMed]
- Hernández-García, J.; Carrillo-Carrasco, V.P.; Rienstra, J.; Tanaka, K.; de Roij, M.; Dipp-Álvarez, M.; Freire-Ríos, A.; Crespo, I.; Boer, R.; van den Berg, W.A.M.; et al. Evolutionary origins and functional diversification of Auxin Response Factors. Nat. Commun. 2024, 15, 10909. [Google Scholar] [CrossRef]
- Huang, S.; Zhou, B.; Gao, Z.; Li, H.; Ding, Z. Increased expression of OsSAUR23 and OsRR9 regulates rice plant and organ size. Crop J. 2025, 13, 350–359. [Google Scholar] [CrossRef]
- Li, Y.; Ren, M.; Wu, Y.; Wang, L.; Zhao, K.; Gao, H.; Li, M.; Liu, Y.; Zhu, J.; Xu, J.; et al. A root system architecture regulator modulates OsPIN2 polar localization in rice. Nat. Commun. 2025, 16, 15. [Google Scholar] [CrossRef]
- Jiang, J.Z.; Kuo, C.H.; Chen, B.H.; Chen, M.K.; Lin, C.S.; Ho, S.L. Effects of OsCDPK1 on the Structure and Physicochemical Properties of Starch in Developing Rice Seeds. Int. J. Mol. Sci. 2018, 19, 3247. [Google Scholar] [CrossRef]
- Hou, M.; Zhang, Y.; Xu, X.; Ai, H. Advances in auxin synthesis, transport, and signaling in rice: Implications for stress resilience and crop improvement. Front. Plant Sci. 2025, 15, 1516884. [Google Scholar] [CrossRef]
- Saha, S.R.; Islam, S.M.S.; Itoh, K. Identification of abiotic stress-responsive genes: A genome-wide analysis of the cytokinin response regulator gene family in rice. Genes Genet. Syst. 2024, 99, 24-00068. [Google Scholar] [CrossRef]
- Liu, Y.; Peng, X.; Ma, A.; Liu, W.; Liu, B.; Yun, D.J.; Xu, Z.Y. Type-B response regulator OsRR22 forms a transcriptional activation complex with OsSLR1 to modulate OsHKT2;1 expression in rice. Sci. China Life Sci. 2023, 66, 2922–2934. [Google Scholar] [CrossRef]
- Pellarin, I.; Dall’Acqua, A.; Favero, A.; Segatto, I.; Rossi, V.; Crestan, N.; Karimbayli, J.; Belletti, B.; Baldassarre, G. Cyclin-dependent protein kinases and cell cycle regulation in biology and disease. Signal Transduct. Target. Ther. 2025, 10, 11. [Google Scholar] [CrossRef] [PubMed]
- Rathore, R.S.; Mishra, M.; Pareek, A.; Singla-Pareek, S.L. Concurrent improvement of rice grain yield and abiotic stress tolerance by overexpression of cytokinin activating enzyme LONELY GUY (OsLOG). Plant Physiol. Biochem. 2024, 211, 108635. [Google Scholar] [CrossRef]
- Sosnowski, J.; Truba, M.; Vasileva, V. The Impact of Auxin and Cytokinin on the Growth and Development of Selected Crops. Agriculture 2023, 13, 724. [Google Scholar] [CrossRef]
- Kotov, A.A.; Kotova, L.M. Auxin/cytokinin antagonism in shoot development: From moss to seed plants. J. Exp. Bot. 2023, 74, 6391–6395. [Google Scholar] [CrossRef] [PubMed]
- Amiri, S.; Mohammadi, R. The Effects of Culture Media and Plant Growth Regulators on Micropropagation of Cornelian Cherry (Cornus mas L.). Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2025, 95, 439–445. [Google Scholar] [CrossRef]
- Oikawa, T.; Koshioka, M.; Kojima, K.; Yoshida, H.; Kawata, M. A role of OsGA20ox1, encoding an isoform of gibberellin 20-oxidase, for regulation of plant stature in rice. Plant Mol. Biol. 2004, 55, 687–700. [Google Scholar] [CrossRef]
- Hao, X.H.; Hu, S.; Zhao, D.; Tian, L.F.; Xie, Z.J.; Wu, S.; Hu, W.L.; Lei, H.; Li, D.P. OsGA3ox genes regulate rice fertility and plant height by synthesizing diverse active GA. Yi Chuan 2023, 45, 845–855. [Google Scholar] [PubMed]
- Hu, J.; Su, H.; Cao, H.; Wei, H.; Fu, X.; Jiang, X.; Song, Q.; He, X.; Xu, C.; Luo, K. AUXIN RESPONSE FACTOR7 integrates gibberellin and auxin signaling via interactions between DELLA and AUX/IAA proteins to regulate cambial activity in poplar. Plant Cell 2022, 34, 2688–2707. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Okada, K.; Fukazawa, J.; Takahashi, Y. DELLA-dependent and -independent gibberellin signaling. Plant Signal Behav. 2018, 13, e1445933. [Google Scholar] [CrossRef]
- Xie, Z.; Jin, L.; Sun, Y.; Zhan, C.; Tang, S.; Qin, T.; Liu, N.; Huang, J. OsNAC120 balances plant growth and drought tolerance by integrating GA and ABA signaling in rice. Plant Commun. 2024, 5, 100782. [Google Scholar] [CrossRef]
- Abe, A.; Takagi, H.; Fujibe, T.; Aya, K.; Kojima, M.; Sakakibara, H.; Uemura, A.; Matsuoka, M.; Terauchi, R. OsGA20ox1, a candidate gene for a major QTL controlling seedling vigor in rice. Theor. Appl. Genet. 2012, 125, 647–657. [Google Scholar] [CrossRef]
- Wang, Y.; Deng, D. Molecular basis and evolutionary pattern of GA-GID1-DELLA regulatory module. Mol. Genet. Genom. 2014, 289, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.P. The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr. Biol. 2011, 21, R338–R345. [Google Scholar] [CrossRef]
- Liu, X.; Chen, Y.; Yuan, B.; Peng, M.; Zhao, Y.; Chen, T.; Lu, J.; Li, F.; Lu, X.; Yang, J. Identification of the citrus GRF gene family and its expression in fruit peel thickening mediated by gibberellin. BMC Plant Biol. 2025, 25, 216. [Google Scholar] [CrossRef]
- Wang, J.D.; Wang, J.; Huang, L.C.; Kan, L.J.; Wang, C.X.; Xiong, M.; Zhou, P.; Zhou, L.H.; Chen, C.; Zhao, D.S.; et al. ABA-mediated regulation of rice grain quality and seed dormancy via the NF-YB1-SLRL2-bHLH144 Module. Nat. Commun. 2024, 15, 4493. [Google Scholar] [CrossRef]
- Zhao, F.; Ma, Q.; Li, Y.; Jiang, M.; Zhou, Z.; Meng, S.; Peng, Y.; Zhang, J.; Ye, N.; Liu, B. OsNAC2 regulates seed dormancy and germination in rice by inhibiting ABA catabolism. Biochem. Biophys. Res. Commun. 2023, 682, 335–342. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, J.; Li, Z.; Qiao, J.; Quan, R.; Wang, J.; Huang, R.; Qin, H. SALT AND ABA RESPONSE ERF1 improves seed germination and salt tolerance by repressing ABA signaling in rice. Plant Physiol. 2022, 189, 1110–1127. [Google Scholar] [CrossRef]
- Jiang, M.; Song, Y.; Yang, R.; Zheng, C.; Zheng, Y.; Zhang, H.; Li, S.; Tan, Y.; Huang, J.; Shu, Q.; et al. Melatonin activates the OsbZIP79-OsABI5 module that orchestrates nitrogen and ROS homeostasis to alleviate nitrogen-limitation stress in rice. Plant Commun. 2023, 4, 100674. [Google Scholar] [CrossRef]
- Zou, M.; Guan, Y.; Ren, H.; Zhang, F.; Chen, F. A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol. Biol. 2008, 66, 675–683. [Google Scholar] [CrossRef]
- Chen, Y.; Xiang, Z.; Liu, M.; Wang, S.; Zhang, L.; Cai, D.; Huang, Y.; Mao, D.; Fu, J.; Chen, L. ABA biosynthesis gene OsNCED3 contributes to preharvest sprouting resistance and grain development in rice. Plant Cell Environ. 2023, 46, 1384–1401. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Cheng, J.; Wang, J.; Cheng, Y.; He, Y.; Zhang, H.; Wang, Z. Physiological characteristics of cold stratification on seed dormancy release in rice. Plant Growth Regul. 2019, 89, 131–141. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, J.; Wang, L.; Zhang, C.; Xu, P.; Li, Y.; Yu, S.; Li, Y. BR signalling haplotypes contribute to indica-japonica differentiation for grain yield and quality in rice. Plant Biotechnol. J. 2025, 23, 1618–1636. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Shi, J.; Persson, S.; Huang, G.; Zhang, D. RMD and Its Suppressor MAPK6 Control Root Circumnutation and Obstacle Avoidance via BR Signaling. Int. J. Mol. Sci. 2024, 25, 10543. [Google Scholar] [CrossRef]
- Su, X.; Liu, X.; Li, C.; Zhang, Y. 24-epibrassinolide as a multidimensional regulator of rice (Oryza sativa) physiological and molecular responses under isoproturon stress. Ecotoxicol. Environ. Saf. 2024, 281, 116575. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Meng, W.; Liu, D.; Pan, D.; Yang, Y.; Chen, Z.; Ma, X.; Yin, W.; Niu, M.; Dong, N.; et al. Enhancing rice panicle branching and grain yield through tissue-specific brassinosteroid inhibition. Science 2024, 383, eadk8838. [Google Scholar] [CrossRef]
- Pan, Y.H.; Gao, L.J.; Liang, Y.T.; Zhao, Y.; Liang, H.F.; Chen, W.W.; Yang, X.H.; Qing, D.J.; Gao, J.; Wu, H.; et al. OrMKK3 Influences Morphology and Grain Size in Rice. J. Plant Biol. 2023, 66, 269–282. [Google Scholar] [CrossRef]
- Huang, J.; Chen, Z.; Lin, J.; Chen, J.; Wei, M.; Liu, L.; Yu, F.; Zhang, Z.; Chen, F.; Jiang, L.; et al. Natural variation of the BRD2 allele affects plant height and grain size in rice. Planta 2022, 256, 27. [Google Scholar] [CrossRef]
- Blanco-Touriñán, N.; Rana, S.; Nolan, T.M.; Li, K.; Vukašinović, N.; Hsu, C.W.; Russinova, E.; Hardtke, C.S. The brassinosteroid receptor gene BRI1 safeguards cell-autonomous brassinosteroid signaling across tissues. Sci. Adv. 2024, 10, eadq3352. [Google Scholar] [CrossRef]
- Luo, Y.; Takagi, J.; Claus, L.A.N.; Zhang, C.; Yasuda, S.; Hasegawa, Y.; Yamaguchi, J.; Shan, L.; Russinova, E.; Sato, T. Deubiquitinating enzymes UBP12 and UBP13 stabilize the brassinosteroid receptor BRI1. EMBO Rep. 2022, 23, e53354. [Google Scholar] [CrossRef] [PubMed]
- Bai, M.Y.; Zhang, L.Y.; Gampala, S.S.; Zhu, S.W.; Song, W.Y.; Chong, K.; Wang, Z.Y. Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA 2007, 104, 13839–13844. [Google Scholar] [CrossRef]
- Cao, X.H.; Wei, Y.N.; Shen, B.D.; Liu, L.C.; Mao, J. Interaction of the Transcription Factors BES1/BZR1 in Plant Growth and Stress Response. Int. J. Mol. Sci. 2024, 25, 6836. [Google Scholar] [CrossRef] [PubMed]
- Pal, A.K.; Gandhivel, V.H.-S.; Nambiar, A.B.; Shivaprasad, P.V. Upstream regulator of genomic imprinting in rice endosperm is a small RNA-associated chromatin remodeler. Nat. Commun. 2024, 15, 7807. [Google Scholar] [CrossRef]
- Gao, X.; Zhang, J.Q.; Zhang, X.; Zhou, J.; Jiang, Z.; Huang, P.; Tang, Z.; Bao, Y.; Cheng, J.; Tang, H.; et al. Rice qGL3/OsPPKL1 Functions with the GSK3/SHAGGY-Like Kinase OsGSK3 to Modulate Brassinosteroid Signaling. Plant Cell 2019, 31, 1077–1093. [Google Scholar] [CrossRef]
- Yan, L.; Jiao, B.; Duan, P.; Guo, G.; Zhang, B.; Jiao, W.; Zhang, H.; Wu, H.; Zhang, L.; Liang, H.; et al. Control of grain size and weight by the RNA-binding protein EOG1 in rice and wheat. Cell Rep. 2024, 43, 114856. [Google Scholar] [CrossRef] [PubMed]
- Tian, P.; Liu, J.; Yan, B.; Li, S.; Lei, B.; Shen, R.; Lei, C.; Xu, M. OsBSK3 Positively Regulates Grain Length and Weight by Inhibiting the Phosphatase Activity of OsPPKL1. Plants 2022, 11, 1586. [Google Scholar] [CrossRef]
- Liang, X.; Liu, Y.; Tian, M.; Jiang, W.; Zheng, Y.; Chen, Z.; Liu, X.; Wang, L. The natural variation allele OsGSW3.2 in Oryza rufipogon is involved in brassinosteroid signaling and influences grain size and weight. Plant J. 2025, 121, e70110. [Google Scholar] [CrossRef]
- Liu, C.; Ma, T.; Yuan, D.; Zhou, Y.; Long, Y.; Li, Z.; Dong, Z.; Duan, M.; Yu, D.; Jing, Y.; et al. The OsEIL1-OsERF115-target gene regulatory module controls grain size and weight in rice. Plant Biotechnol. J. 2022, 20, 1470–1486. [Google Scholar] [CrossRef]
- Qin, H.; Xiao, M.; Li, Y.; Huang, R. Ethylene Modulates Rice Root Plasticity under Abiotic Stresses. Plants 2024, 13, 432. [Google Scholar] [CrossRef]
- Li, Z.; Chen, H.; Yuan, P.; Jiang, X.; Li, Z.M.; Wang, S.T.; Zhou, T.G.; Zhu, H.Y.; Bian, Q.; Zhu, X.F.; et al. IDD10-NAC079 transcription factor complex regulates sheath blight resistance by inhibiting ethylene signaling in rice. J. Adv. Res. 2024, 71, 93–106. [Google Scholar] [CrossRef]
- Xie, W.; Ding, C.; Hu, H.; Dong, G.; Zhang, G.; Qian, Q.; Ren, D. Molecular Events of Rice AP2/ERF Transcription Factors. Int. J. Mol. Sci. 2022, 23, 12013. [Google Scholar] [CrossRef]
- Qin, H.; Wang, J.; Chen, X.; Wang, F.; Peng, P.; Zhou, Y.; Miao, Y.; Zhang, Y.; Gao, Y.; Qi, Y.; et al. Rice OsDOF15 contributes to ethylene-inhibited primary root elongation under salt stress. New Phytol. 2019, 223, 798–813. [Google Scholar] [CrossRef]
- Lee, H.Y.; Yoon, G.M. Regulation of Ethylene Biosynthesis by Phytohormones in Etiolated Rice (Oryza sativa L.) Seedlings. Mol. Cells 2018, 41, 311–319. [Google Scholar]
- Rzewuski, G.; Sauter, M. Ethylene biosynthesis and signaling in rice. Plant Sci. 2008, 175, 32–42. [Google Scholar] [CrossRef]
- Huang, Y.H.; Han, J.Q.; Ma, B.; Cao, W.Q.; Li, X.K.; Xiong, Q.; Zhao, H.; Zhao, R.; Zhang, X.; Zhou, Y.; et al. A translational regulator MHZ9 modulates ethylene signaling in rice. Nat. Commun. 2023, 14, 4674. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; He, S.J.; Duan, K.X.; Yin, C.C.; Chen, H.; Yang, C.; Xiong, Q.; Song, Q.X.; Lu, X.; Chen, H.W.; et al. Identification of rice ethylene-response mutants and characterization of MHZ7/OsEIN2 in distinct ethylene response and yield trait regulation. Mol. Plant 2013, 6, 1830–1848. [Google Scholar] [CrossRef]
- Li, X.-K.; Huang, Y.-H.; Zhao, R.; Cao, W.-Q.; Lu, L.; Han, J.-Q.; Zhou, Y.; Zhang, X.; Wu, W.-A.; Tao, J.-J.; et al. Membrane protein MHZ3 regulates the on-off switch of ethylene signaling in rice. Nat. Commun. 2024, 15, 5987. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Wang, H.; Liu, S.; Lu, S.; Hua, J.; Zou, B. Ethylene antagonizes ABA and inhibits stomatal closure and chilling tolerance in rice. J. Exp. Bot. 2025. [Google Scholar] [CrossRef]
- Yan, Y.; Wei, M.X.; Li, Y.; Tao, H.; Wu, H.Y.; Chen, Z.F.; Li, C.; Xu, J.H. MiR529a controls plant height, tiller number, panicle architecture and grain size by regulating SPL target genes in rice (Oryza sativa L.). Plant Sci. 2021, 302, 110728. [Google Scholar] [CrossRef]
- Zhang, X.F.; Yang, C.Y.; Lin, H.X.; Wang, J.W.; Xue, H.W. Rice SPL12 coevolved with GW5 to determine grain shape. Sci Bull. 2021, 66, 2353–2357. [Google Scholar] [CrossRef]
- Si, L.; Chen, J.; Huang, X.; Gong, H.; Luo, J.; Hou, Q.; Zhou, T.; Lu, T.; Zhu, J.; Shangguan, Y.; et al. OsSPL13 controls grain size in cultivated rice. Nat. Genet. 2016, 48, 447–456. [Google Scholar] [CrossRef]
- Guo, X.; Chen, Y.; Hu, Y.; Feng, F.; Zhu, X.; Sun, H.; Li, J.; Zhao, Q.; Sun, H. OsMADS5 interacts with OsSPL14/17 to inhibit rice root elongation by restricting cell proliferation of root meristem under ammonium supply. Plant J. 2023, 116, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; He, Y.; Liu, Z.; Qin, T.; Wang, L.; Chen, Z.; Zhang, B.; Zhang, H.; Li, H.; Liu, L.; et al. OsSPL14 acts upstream of OsPIN1b and PILS6b to modulate axillary bud outgrowth by fine-tuning auxin transport in rice. Plant J. 2022, 111, 1167–1182. [Google Scholar] [CrossRef]
- Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X.; et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef]
- Wang, S.; Li, S.; Liu, Q.; Wu, K.; Zhang, J.; Wang, S.; Wang, Y.; Chen, X.; Zhang, Y.; Gao, C.; et al. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 2015, 47, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, K.; Yuan, Q.; Liu, X.; Liu, Z.; Lin, X.; Zeng, R.; Zhu, H.; Dong, G.; Qian, Q.; et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 2012, 44, 950–954. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Qin, P.; Hu, L.; Zhan, S.; Wang, S.; Gao, P.; Li, J.; Jin, M.; Xu, Z.; Gao, Q.; et al. OsSPL18 controls grain weight and grain number in rice. J. Genet. Genom. 2019, 46, 41–51. [Google Scholar] [CrossRef]
- Tang, J.; Mei, E.; He, M.; Bu, Q.; Tian, X. Functions of OsWRKY24, OsWRKY70 and OsWRKY53 in regulating grain size in rice. Planta 2022, 255, 92. [Google Scholar] [CrossRef]
- Tian, X.; He, M.; Mei, E.; Zhang, B.; Tang, J.; Xu, M.; Liu, J.; Li, X.; Wang, Z.; Tang, W.; et al. WRKY53 integrates classic brassinosteroid signaling and the mitogen-activated protein kinase pathway to regulate rice architecture and seed size. Plant Cell 2021, 33, 2753–2775. [Google Scholar] [CrossRef]
- Zhang, C.Q.; Xu, Y.; Lu, Y.; Yu, H.X.; Gu, M.H.; Liu, Q.Q. The WRKY transcription factor OsWRKY78 regulates stem elongation and seed development in rice. Planta 2011, 234, 541–554. [Google Scholar] [CrossRef]
- Mathew, I.E.; Das, S.; Mahto, A.; Agarwal, P. Three Rice NAC Transcription Factors Heteromerize and Are Associated with Seed Size. Front. Plant Sci. 2016, 7, 1638. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Liu, Y.C.; Hao, C.Y.; Li, T.; Majeed, U.; Liu, H.X.; Li, H.F.; Hou, J.; Zhang, X.Y. Wheat NAC-A18 regulates grain starch and storage proteins synthesis and affects grain weight. Theor. Appl. Genet. 2023, 136, 123. [Google Scholar] [CrossRef]
- Li, Z.; Wei, X.; Tong, X.; Zhao, J.; Liu, X.; Wang, H.; Tang, L.; Shu, Y.; Li, G.; Wang, Y.; et al. The OsNAC23-Tre6P-SnRK1a feed-forward loop regulates sugar homeostasis and grain yield in rice. Mol. Plant 2022, 15, 706–722. [Google Scholar] [CrossRef]
- Dwivedi, N.; Maji, S.; Waseem, M.; Thakur, P.; Kumar, V.; Parida, S.K.; Thakur, J.K. The Mediator subunit OsMED15a is a transcriptional co-regulator of seed size/weight-modulating genes in rice. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 194432. [Google Scholar] [CrossRef]
- Mathew, I.E.; Priyadarshini, R.; Mahto, A.; Jaiswal, P.; Parida, S.K.; Agarwal, P. SUPER STARCHY1/ONAC025 participates in rice grain filling. Plant Direct 2020, 4, e00249. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.K.; Zhang, M.Q.; Leng, Y.J.; Xu, L.N.; Jia, S.W.; Wang, S.L.; Song, T.; Wang, R.A.; Yang, Q.Q.; Tao, T.; et al. OsNAC129 Regulates Seed Development and Plant Growth and Participates in the Brassinosteroid Signaling Pathway. Front. Plant Sci. 2022, 13, 905148. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Huang, K.; Zhang, L.; Zhang, B.; Duan, P.; Zhang, G.; Huang, X.; Zhou, C.; Han, N.; Zheng, L.; et al. A molecular framework for the GS2-SUG1 module-mediated control of grain size and weight in rice. Nat. Commun. 2025, 16, 3944. [Google Scholar] [CrossRef]
- Wang, Y.; Lv, Y.; Wen, Y.; Wang, J.; Hu, P.; Wu, K.; Chai, B.; Gan, S.; Liu, J.; Wu, Y.; et al. GS2 cooperates with IPA1 to control panicle architecture. New Phytol. 2025, 245, 2726–2743. [Google Scholar] [CrossRef]
- Chen, W.; Hu, X.; Hu, L.; Hou, X.; Xu, Z.; Yang, F.; Yuan, M.; Chen, F.; Wang, Y.; Tu, B.; et al. Wide Grain 3, a GRAS Protein, Interacts with DLT to Regulate Grain Size and Brassinosteroid Signaling in Rice. Rice 2022, 15, 55. [Google Scholar] [CrossRef]
- Xiao, Y.; Liu, D.; Zhang, G.; Tong, H.; Chu, C. Brassinosteroids Regulate OFP1, a DLT Interacting Protein, to Modulate Plant Architecture and Grain Morphology in Rice. Front. Plant Sci. 2017, 8, 1698. [Google Scholar] [CrossRef]
- Huang, Y.; Dong, H.; Mou, C.; Wang, P.; Hao, Q.; Zhang, M.; Wu, H.; Zhang, F.; Ma, T.; Miao, R.; et al. Ribonuclease H-like gene SMALL GRAIN2 regulates grain size in rice through brassinosteroid signaling pathway. J. Integr. Plant Biol. 2022, 64, 1883–1900. [Google Scholar] [CrossRef] [PubMed]
- Usman, B.; Nawaz, G.; Zhao, N.; Liao, S.; Qin, B.; Liu, F.; Liu, Y.; Li, R. Programmed Editing of Rice (Oryza sativa L.) OsSPL16 Gene Using CRISPR/Cas9 Improves Grain Yield by Modulating the Expression of Pyruvate Enzymes and Cell Cycle Proteins. Int. J. Mol. Sci. 2020, 22, 249. [Google Scholar] [CrossRef]
- Jangam, A.P.; Pathak, R.R.; Raghuram, N. Microarray Analysis of Rice d1 (RGA1) Mutant Reveals the Potential Role of G-Protein Alpha Subunit in Regulating Multiple Abiotic Stresses Such as Drought, Salinity, Heat, and Cold. Front. Plant Sci. 2016, 7, 11. [Google Scholar] [CrossRef]
- Yang, X.; Lu, J.; Shi, W.J.; Chen, Y.H.; Yu, J.W.; Chen, S.H.; Zhao, D.S.; Huang, L.C.; Fan, X.L.; Zhang, C.Q.; et al. RGA1 regulates grain size, rice quality and seed germination in the small and round grain mutant srg5. BMC Plant Biol. 2024, 24, 167. [Google Scholar] [CrossRef]
- Hao, J.; Wang, D.; Wu, Y.; Huang, K.; Duan, P.; Li, N.; Xu, R.; Zeng, D.; Dong, G.; Zhang, B.; et al. The GW2-WG1-OsbZIP47 pathway controls grain size and weight in rice. Mol. Plant 2021, 14, 1266–1280. [Google Scholar] [CrossRef]
- Liu, S.; Wu, J.; Mawia, A.M.; Wei, X.; Cao, R.; Jiao, G.; Wu, Y.; Zhang, J.; Xie, L.; Sheng, Z.; et al. A novel transcription factor OsMYB73 affects grain size and chalkiness by regulating endosperm storage substances’ accumulation-mediated auxin biosynthesis signalling pathway in rice. Plant Biotechnol. J. 2025, 23, 1021–1038. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Zheng, C.; Shi, C.; Lu, X.; She, Z.; Jiang, S.; Tian, D.; Qin, Y. The OsZHD1 and OsZHD2, Two Zinc Finger Homeobox Transcription Factor, Redundantly Control Grain Size by Influencing Cell Proliferation in Rice. Rice 2025, 18, 20. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.; Wang, H.; Zhang, Q.; Zhou, K.; Li, M.; Li, R.; Xiang, S.; Zhang, T.; Ling, Y.; Yang, Z.; et al. Identification and Pyramiding of QTLs for Rice Grain Size Based on Short-Wide Grain CSSL-Z563 and Fine-Mapping of qGL3-2. Rice 2021, 14, 35. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Jia, J.; Liu, R.; Wei, R.; Guo, Z.; Cai, Z.; Chen, B.; Liang, F.; Xia, Q.; Nian, H.; et al. Identification of major QTLs for soybean seed size and seed weight traits using a RIL population in different environments. Front. Plant Sci. 2022, 13, 1094112. [Google Scholar] [CrossRef] [PubMed]
- Arif, M.A.R.; Komyshev, E.G.; Genaev, M.A.; Koval, V.S.; Shmakov, N.A.; Börner, A.; Afonnikov, D.A. QTL Analysis for Bread Wheat Seed Size, Shape and Color Characteristics Estimated by Digital Image Processing. Plants 2022, 11, 2105. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Shao, D.; Qiu, X.J.; Sun, L.; Yan, W.H.; Zhou, X.C.; Yang, L.; He, Y.Q.; Yu, S.B.; Xing, Y.Z. Natural variation and artificial selection in four genes determine grain shape in rice. New Phytol. 2013, 200, 1269–1280. [Google Scholar] [CrossRef]
- Yamaguchi, K.; Yamamoto, T.; Segami, S.; Horikawa, M.; Chaya, G.; Kitano, H.; Iwasaki, Y.; Miura, K. gw2 mutation increases grain width and culm thickness in rice (Oryza sativa L.). Breed. Sci. 2020, 70, 456–461. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Zhu, A.K.; Xue, P.; Wen, X.X.; Cao, Y.R.; Wang, B.F.; Zhang, Y.; Shah, L.; Cheng, S.H.; Cao, L.Y.; et al. Effects of GS3 and GL3.1 for Grain Size Editing by CRISPR/Cas9 in Rice. Rice Sci. 2020, 27, 405–413. [Google Scholar] [CrossRef]
- Takano-Kai, N.; Doi, K.; Yoshimura, A. GS3 participates in stigma exsertion as well as seed length in rice. Breed. Sci. 2011, 61, 244–250. [Google Scholar] [CrossRef]
- Liu, J.F.; Chen, J.; Zheng, X.M.; Wu, F.Q.; Lin, Q.B.; Heng, Y.Q.; Tian, P.; Cheng, Z.J.; Yu, X.W.; Zhou, K.N.; et al. GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants 2017, 3, 17043. [Google Scholar] [CrossRef]
- Rasheed, H.; Fiaz, S.; Khan, M.A.; Mehmood, S.; Ullah, F.; Saeed, S.; Khan, S.U.; Yaseen, T.; Hussain, R.M.; Qayyum, A. Characterization of functional genes GS3 and GW2 and their effect on the grain size of various landraces of rice (Oryza sativa). Mol. Biol. Rep. 2022, 49, 5397–5403. [Google Scholar] [CrossRef]
- Shomura, A.; Izawa, T.; Ebana, K.; Ebitani, T.; Kanegae, H.; Konishi, S.; Yano, M. Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 2008, 40, 1023–1028. [Google Scholar] [CrossRef]
- Gao, X.Y.; Zhang, X.J.; Lan, H.X.; Huang, J.; Wang, J.F.; Zhang, H.S. The additive effects of GS3 and qGL3 on rice grain length regulation revealed by genetic and transcriptome comparisons. BMC Plant Biol. 2015, 15, 156. [Google Scholar] [CrossRef]
- Liu, R.; Zhao, D.; Li, P.; Xia, D.; Feng, Q.; Wang, L.; Wang, Y.; Shi, H.; Zhou, Y.; Chen, F.; et al. Natural variation in OsMADS1 transcript splicing affects rice grain thickness and quality by influencing monosaccharide loading to the endosperm. Plant Commun. 2025, 6, 101178. [Google Scholar] [CrossRef] [PubMed]
- Abbas, W.; Shalmani, A.; Zhang, J.; Sun, Q.; Zhang, C.; Li, W.; Cui, Y.; Xiong, M.; Li, Y. The GW5-WRKY53-SGW5 module regulates grain size variation in rice. New Phytol. 2024, 242, 2011–2025. [Google Scholar] [CrossRef]
- Ruan, B.; Shang, L.; Zhang, B.; Hu, J.; Wang, Y.; Lin, H.; Zhang, A.; Liu, C.; Peng, Y.; Zhu, L.; et al. Natural variation in the promoter of TGW2 determines grain width and weight in rice. New Phytol. 2020, 227, 629–640. [Google Scholar] [CrossRef] [PubMed]
- He, J.B.; Meng, S.; Zhao, T.J.; Xing, G.N.; Yang, S.P.; Li, Y.; Guan, R.Z.; Lu, J.J.; Wang, Y.F.; Xia, Q.J.; et al. An innovative procedure of genome-wide association analysis fits studies on germplasm population and plant breeding. Theor. Appl. Genet. 2017, 130, 2327–2343. [Google Scholar] [CrossRef]
- Zhu, L.; Shen, Y.; Dai, Z.; Miao, X.; Shi, Z. Gγ-protein GS3 Function in Tight Genetic Relation with OsmiR396/GS2 to Regulate Grain Size in Rice. Rice 2024, 17, 59. [Google Scholar] [CrossRef]
- Ma, S.; Sun, Y.; Chen, X.; Guo, J.; Wu, S.; Wu, G.; Huang, G.; Kabore, M.A.F.; Woldegiorgis, S.T.; Ai, Y.; et al. Gα Solicits OsNYC4 and GW2-WG1-OsbZIP47 Modules to Regulate Grain Size in Rice (Oryza sativa L.). Agronomy 2024, 14, 1514. [Google Scholar] [CrossRef]
- Liu, H.; Li, H.; Hao, C.; Wang, K.; Wang, Y.; Qin, L.; An, D.; Li, T.; Zhang, X. TaDA1, a conserved negative regulator of kernel size, has an additive effect with TaGW2 in common wheat (Triticum aestivum L.). Plant Biotechnol. J. 2020, 18, 1330–1342. [Google Scholar] [CrossRef]
- Li, L.; Shi, F.; Wang, Y.Q.; Yu, X.F.; Zhi, J.J.; Guan, Y.B.; Zhao, H.Y.; Chang, J.L.; Chen, M.J.; Yang, G.X.; et al. TaSPL13 regulates inflorescence architecture and development in transgenic wheat (Triticum aestivum L.). Plant Sci. 2020, 296, 110516. [Google Scholar] [CrossRef]
- Jin, T.; Zhang, K.; Hao, X.; Zhang, X.; Wang, S.; Long, W. Genome-Wide Analysis of Soybean GS3-Family Genes and Functional Characterization of GmGS3-1 Responses to Saline and Drought Stresses. Agriculture 2025, 15, 443. [Google Scholar] [CrossRef]
- Peng, Y.-B.; Du, C.-Y.; He, Y.-N.; Zheng, C.-K.; Sun, W.; Zhou, J.-J.; Xie, L.-X.; Jiang, C.-H.; Xu, J.-D.; Wang, F.; et al. Natural variation of Grain size 3 allele differentially functions in regulating grain length in xian/indica and geng/japonica rice. Euphytica 2024, 220, 44. [Google Scholar] [CrossRef]
- Wang, J.L.; Tang, M.Q.; Chen, S.; Zheng, X.F.; Mo, H.X.; Li, S.J.; Wang, Z.; Zhu, K.M.; Ding, L.N.; Liu, S.Y.; et al. Down-regulation of BnDA1, whose gene locus is associated with the seeds weight, improves the seeds weight and organ size in Brassica napus. Plant Biotechnol. J. 2017, 15, 1024–1033. [Google Scholar] [CrossRef]
- Wei, B.; Jiao, Y. Grain size control in wheat: Toward a molecular understanding. Seed Biol. 2024, 3, e007. [Google Scholar] [CrossRef]
- Liang, S.; Duan, Z.; He, X.; Yang, X.; Yuan, Y.; Liang, Q.; Pan, Y.; Zhou, G.; Zhang, M.; Liu, S.; et al. Natural variation in GmSW17 controls seed size in soybean. Nat. Commun. 2024, 15, 7417. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Liu, H.J.; Zhou, C.; Wang, J.X.; Wang, J.Q.; Han, Y.H.; Zheng, N.; Zhang, M.; Li, X.M. The ubiquitin-proteasome system in the plant response to abiotic stress: Potential role in crop resilience improvement. Plant Sci. 2024, 342, 112035. [Google Scholar] [CrossRef]
- Verma, A.; Prakash, G.; Ranjan, R.; Tyagi, A.K.; Agarwal, P. Silencing of an Ubiquitin Ligase Increases Grain Width and Weight in indica Rice. Front. Genet. 2020, 11, 600378. [Google Scholar] [CrossRef]
- Wen, Y.; Hu, P.; Fang, Y.; Tan, Y.; Wang, Y.; Wu, H.; Wang, J.; Wu, K.; Chai, B.; Zhu, L.; et al. GW9 determines grain size and floral organ identity in rice. Plant Biotechnol. J. 2024, 22, 915–928. [Google Scholar] [CrossRef]
- Xie, Y.H.; Fan, Z.P.; Liang, X.Y.; Teng, K.C.; Huang, Z.J.; Huang, M.Y.; Zhao, H.; Xu, K.Z.; Li, J.X. OsPUB9 modulates leaf angle and grain size through the brassinosteroid signaling pathway in rice. Plant J. 2025, 121, e17230. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Ren, Y.; Liu, L.; Wang, F.; Zhang, H.; Tian, P.; Pan, T.; Wang, Y.; Jing, R.; Liu, T.; et al. Ubiquitin Specific Protease 15 Has an Important Role in Regulating Grain Width and Size in Rice. Plant Physiol. 2019, 180, 381–391. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, Z.; Lv, M.; Luan, Z.; Li, T.; Hu, J. Novel histone modifications and liver cancer: Emerging frontiers in epigenetic regulation. Clin. Epigenet. 2025, 17, 30. [Google Scholar] [CrossRef]
- Sarki, Y.N.; Singh, H.B.; Keot, A.K.; Marwein, R.; Singha, D.L.; Dehury, B.; Chikkaputtaiah, C. Investigating the Role of OsHDT701 and Other Blast-Associated Negative Regulatory Genes in Indica Rice Cultivar Ranjit Using Combined Wet Lab and Computational Approaches. Mol. Biotechnol. 2024. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Lu, Y.; Li, Q.; Luo, S.; Shen, S.; Li, N.; Chen, X. TIR1/AFB proteins: Active players in abiotic and biotic stress signaling. Front. Plant Sci. 2022, 13, 1083409. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.; Sun, J.; Shabbir, S.; Khattak, W.A.; Ren, G.; Nie, X.; Bo, Y.; Javed, Q.; Du, D.; Sonne, C. A review of plants strategies to resist biotic and abiotic environmental stressors. Sci. Total Environ. 2023, 900, 165832. [Google Scholar] [CrossRef]
- Wang, X.; Liu, X.; Su, Y.; Shen, H. Rice Responses to Abiotic Stress: Key Proteins and Molecular Mechanisms. Int. J. Mol. Sci. 2025, 26, 896. [Google Scholar] [CrossRef]
- Li, Y.; Yin, M.; Wang, J.; Zhao, X.; Xu, J.; Wang, W.; Fu, B. Epitranscriptome profiles reveal participation of the RNA methyltransferase gene OsMTA1 in rice seed germination and salt stress response. BMC Plant Biol. 2025, 25, 115. [Google Scholar] [CrossRef]
- Nau, T.; Cutts, S.; Naidoo, N. DNA methylation and its influence on the pathogenesis of osteoarthritis: A systematic literature review. EFORT Open Rev. 2025, 10, 66–74. [Google Scholar] [CrossRef]
- Guo, M.; Zhang, W.; Mohammadi, M.A.; He, Z.; She, Z.; Yan, M.; Shi, C.; Lin, L.; Wang, A.; Liu, J.; et al. OsDDM1b Controls Grain Size by Influencing Cell Cycling and Regulating Homeostasis and Signaling of Brassinosteroid in Rice. Front. Plant Sci. 2022, 13, 873993. [Google Scholar] [CrossRef]
- Wang, M.; He, Y.; Zhong, Z.; Papikian, A.; Wang, S.; Gardiner, J.; Ghoshal, B.; Feng, S.; Jami-Alahmadi, Y.; Wohlschlegel, J.A.; et al. Histone H3 lysine 4 methylation recruits DNA demethylases to enforce gene expression in Arabidopsis. Nat. Plants 2025, 11, 206–217. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.Q.; Jiang, Z.J.; Chen, J.Y.; Xie, M.Y.; Huang, W.D.; Li, J.; Zhuang, C.X.; Liu, Z.L.; Zheng, S.Y. SET DOMAIN GROUP 711-mediated H3K27me3 methylation of cytokinin metabolism genes regulates organ size in rice. Plant Physiol. 2024, 194, 2069–2085. [Google Scholar] [CrossRef]
- Wang, H.; Tong, X.; Tang, L.; Wang, Y.; Zhao, J.; Li, Z.; Liu, X.; Shu, Y.; Yin, M.; Adegoke, T.V.; et al. RLB (RICE LATERAL BRANCH) recruits PRC2-mediated H3K27 tri-methylation on OsCKX4 to regulate lateral branching. Plant Physiol. 2022, 188, 460–476. [Google Scholar] [CrossRef]
- Huang, Y.; Bai, X.; Cheng, N.; Xiao, J.; Li, X.; Xing, Y. Wide Grain 7 increases grain width by enhancing H3K4me3 enrichment in the OsMADS1 promoter in rice (Oryza sativa L.). Plant J. 2020, 102, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Huang, X.; Wen, M.; Yin, W.; Chen, Y.; Liu, Y.; Liu, X. Cytological observation and RNA-seq analysis reveal novel miRNAs high expression associated with the pollen fertility of neo-tetraploid rice. BMC Plant Biol. 2023, 23, 434. [Google Scholar] [CrossRef]
- Jiang, C.; Wang, Y.; He, Y.; Peng, Y.; Xie, L.; Li, Y.; Sun, W.; Zhou, J.; Zheng, C.; Xie, X. Identification and Characterization of miRNAs and lncRNAs Associated with Salinity Stress in Rice Panicles. Int. J. Mol. Sci. 2024, 25, 8247. [Google Scholar] [CrossRef]
- Xue, Y.; Muhammad, S.; Yang, J.; Wang, X.; Zhao, N.; Qin, B.; Qiu, Y.; Du, Z.; Ulhassan, Z.; Zhou, W.; et al. Comparative transcriptome-wide identification and differential expression of genes and lncRNAs in rice near-isogenic line (KW-Bph36-NIL) in response to BPH feeding. Front. Plant Sci. 2022, 13, 1095602. [Google Scholar] [CrossRef]
- Nalbant, E.; Akkaya-Ulum, Y.Z. Exploring regulatory mechanisms on miRNAs and their implications in inflammation-related diseases. Clin. Exp. Med. 2024, 24, 142. [Google Scholar] [CrossRef]
- Arshad, M.; Feyissa, B.A.; Amyot, L.; Aung, B.; Hannoufa, A. MicroRNA156 improves drought stress tolerance in alfalfa (Medicago sativa) by silencing SPL13. Plant Sci. 2017, 258, 122–136. [Google Scholar] [CrossRef]
- Much, C.; Lasda, E.L.; Pereira, I.T.; Vallery, T.K.; Ramirez, D.; Lewandowski, J.P.; Dowell, R.D.; Smallegan, M.J.; Rinn, J.L. The temporal dynamics of lncRNA Firre-mediated epigenetic and transcriptional regulation. Nat. Commun. 2024, 15, 6821. [Google Scholar] [CrossRef]
- Lone, J.K.; Bhardwaj, R. Posttranscriptional Gene Regulation: The Role of Noncoding RNAs. In Non-Coding RNAs for Crop Improvement: Concepts and Applications; Goswami, K., Gelaw, T.A., Sanan-Mishra, N., Eds.; Springer Nature: Singapore, 2025; pp. 41–56. [Google Scholar]
- Ferrer, J.; Dimitrova, N. Transcription regulation by long non-coding RNAs: Mechanisms and disease relevance. Nat. Rev. Mol. Cell Biol. 2024, 25, 396–415. [Google Scholar] [CrossRef]
- Pawłasek, N.; Sokołowska, A.; Koter, M.; Oracz, K. The interaction between miR165/166 and miR160 regulates Arabidopsis thaliana seed size, weight, and number in a ROS-dependent manner. Planta 2024, 260, 72. [Google Scholar] [CrossRef] [PubMed]
- Liao, D.; Zhang, Y.; Li, S.; Tang, H.; Bai, X. miRNAs in neurodegenerative diseases: From target screening to precision therapy. Neurol. Sci. 2025, 46, 2393–2399. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.K. Epigenetic regulation of angiogenesis and its therapeutics. Genom. Inform. 2025, 23, 4. [Google Scholar] [CrossRef]
- Lu, Y.; Zeng, J.; Liu, Q. The Rice miR396-GRF-GIF-SWI/SNF Module: A Player in GA Signaling. Front. Plant Sci. 2021, 12, 786641. [Google Scholar] [CrossRef]
- Huang, F.; He, Y. Epigenetic control of gene expression by cellular metabolisms in plants. Curr. Opin. Plant Biol. 2024, 81, 102572. [Google Scholar] [CrossRef]
- Tremblay, B.J.; Qüesta, J.I. Non-coding and epigenetic mechanisms in the regulation of seed germination in Arabidopsis thaliana. J. Exp. Bot. 2025, 76, 2455–2467. [Google Scholar] [CrossRef]
- Luo, L.; Yang, M.; Zhou, Y. BMI1s interact with condensin complexes to regulate chromatin 3D structure and gene expression in Arabidopsis. aBIOTECH 2025. [Google Scholar] [CrossRef]
- Yu, J.; Du, T.; Zhang, P.; Ma, Z.; Chen, X.; Cao, J.; Li, H.; Li, T.; Zhu, Y.; Xu, F.; et al. Impacts of High Temperatures on the Growth and Development of Rice and Measures for Heat Tolerance Regulation: A Review. Agronomy 2024, 14, 2811. [Google Scholar] [CrossRef]
- Xing, Y.-H.; Lu, H.; Zhu, X.; Deng, Y.; Xie, Y.; Luo, Q.; Yu, J. How Rice Responds to Temperature Changes and Defeats Heat Stress. Rice 2024, 17, 73. [Google Scholar] [CrossRef] [PubMed]
- Pant, K.K.; Naik, J.; Barthakur, S.; Chandra, V. High-temperature stress in wheat (Triticum aestivum L.): Unfolding the impacts, tolerance and methods to mitigate the detrimental effects. Cereal Res. Commun. 2025, 53, 1171–1197. [Google Scholar] [CrossRef]
- Wang, H.; Charagh, S.; Dong, N.; Lu, F.; Wang, Y.; Cao, R.; Ma, L.; Wang, S.; Jiao, G.; Xie, L.; et al. Genome-Wide Analysis of Heat Shock Protein Family and Identification of Their Functions in Rice Quality and Yield. Int. J. Mol. Sci. 2024, 25, 11931. [Google Scholar] [CrossRef]
- Shen, Q.; Xiao, J.; Wang, L.; Feng, J.; Chen, Y.; Zhu, B.; Li, H.; Lambers, H. Maize shows intraspecific facilitation under phosphorus deficiency but competition under nitrogen deficiency when grown under increased plant densities in alkaline soil. Plant Soil 2025. [Google Scholar] [CrossRef]
- Lin, G.; Wang, L.; Li, Y.; Li, J.; Qian, C.; Zhang, X.; Zuo, Q. Optimal Planting Density Increases the Seed Yield by Improving Biomass Accumulation and Regulating the Canopy Structure in Rapeseed. Plants 2024, 13, 1986. [Google Scholar] [CrossRef]
- Mathan, J.; Dwivedi, A.; Ranjan, A. Revisiting development and physiology of wild rice relatives for crop improvement and climate resilience. Plant Cell Rep. 2025, 44, 55. [Google Scholar] [CrossRef]
- Gann, P.J.I.; Nandy, S.; Botelho, F.B.S.; Vinzant, K.; Khodakovskaya, M.; Srivastava, V. A vacuolar proton pump controls the post-germinative growth of rice (Oryza sativa ssp. japonica). Plant Growth Regul. 2025, 105, 675–686. [Google Scholar] [CrossRef]
- Wu, C.; Acuña, A.; Florez-Palacios, L.; Harrison, D.; Rogers, D.; Mozzoni, L.; Mian, R.; Vieira, C.C. Across-environment seed protein stability and genetic architecture of seed components in soybean. Sci. Rep. 2024, 14, 16452. [Google Scholar] [CrossRef] [PubMed]
- Zarea, M.J. The Regulatory Roles of Phytohormones in the Wheat Grain-Filling Process. J. Plant Growth Regul. 2025, 44, 2609–2626. [Google Scholar] [CrossRef]
- Kannababu, N.; Nanjundappa, S.; Narayanan, N.; Vetriventhan, M.; Venkateswarlu, R.; Das, I.K.; Srikanth, A.; Viswanath, A.; Singh, S.; Malipatil, R.; et al. Role of functional genes for seed vigor related traits through genome-wide association mapping in finger millet (Eleusine coracana L. Gaertn.). Sci. Rep. 2025, 15, 5569. [Google Scholar]
- Whisnant, E.D.; Keith, C.; Smieska, L.; Chia, J.-C.; Bekele-Alemu, A.; Vatamaniuk, O.K.; VanBuren, R.; Ligaba-Osena, A. Biggest of tinies: Natural variation in seed size and mineral distribution in the ancient crop tef [Eragrostis tef (Zucc.) Trotter]. Front. Plant Sci. 2024, 15, 1485819. [Google Scholar] [CrossRef]
- Weng, Y.; Wang, Y.; Wang, K.; Wu, F.; Wei, Y.; Jiang, J.; Zhu, Y.; Wang, F.; Xie, H.; Xiao, Y.; et al. OsLOX1 positively regulates seed vigor and drought tolerance in rice. Plant Mol. Biol. 2025, 115, 16. [Google Scholar] [CrossRef] [PubMed]
- Miao, R.; Lin, Q.; Cao, P.; Zhou, C.; Feng, M.; Lan, J.; Luo, S.; Zhang, F.; Wu, H.; Hao, Q.; et al. SMALL AND ROUND GRAIN is involved in the brassinosteroid signaling pathway which regulates grain size in rice. J. Integr. Plant Biol. 2025, 67, 1290–1306. [Google Scholar] [CrossRef] [PubMed]
- Pell, C.J.; King, S.L.; Hawkins, T.; Symmank, M. Determining the effects of reduced water availability on seed germination of five bottomland hardwood tree species. For. Ecol. Manag. 2025, 577, 122410. [Google Scholar] [CrossRef]
- Upretee, P.; Bandara, M.S.; Tanino, K.K. The Role of Seed Characteristics on Water Uptake Preceding Germination. Seeds 2024, 3, 559–574. [Google Scholar] [CrossRef]
- Awasthi, R.; Devi, P.; Jha, U.C.; Sharma, K.D.; Roorkiwal, M.; Kumar, S.; Pareek, A.; Siddique, K.H.M.; Prasad, P.V.V.; Parida, S.K.; et al. Exploring the synergistic effects of drought and heat stress on chickpea seed development: Insights into nutritional quality and seed yield. Plant Stress 2024, 14, 100635. [Google Scholar] [CrossRef]
- Zhuang, T.; Ata-Ui-Karim, S.T.; Zhao, B.; Liu, X.; Tian, Y.; Zhu, Y.; Cao, W.; Cao, Q. Investigating the impacts of different degrees of deficit irrigation and nitrogen interactions on assimilate translocation, yield, and resource use efficiencies in winter wheat. Agric. Water Manag. 2024, 304, 109089. [Google Scholar] [CrossRef]
- Zhai, M.; Wei, X.; Pan, Z.; Xu, Q.; Qin, D.; Li, J.; Zhang, J.; Wang, L.; Wang, K.; Duan, X.; et al. Optimizing plant density and canopy structure to improve light use efficiency and cotton productivity: Two years of field evidence from two locations. Ind. Crops Prod. 2024, 222, 119946. [Google Scholar] [CrossRef]
- Tao, Y.; Li, Z.; Shah, F.; Wu, W. Optimizing biomass allocation for optimum balance of seed yield and lodging resistance in rapeseed. Field Crops Res. 2024, 316, 109493. [Google Scholar] [CrossRef]
- Gong, H.; Duan, D.; Suo, Y.; Jia, N.; Hu, K.; Chen, J.; Hu, N.; Zhao, G.; Wang, Z. Conservative allocation strategy of nitrogen and phosphorus among leaves, stems and roots of Artemisia species. BMC Plant Biol. 2025, 25, 182. [Google Scholar] [CrossRef]
- Li, R.; Xu, C.; Wu, Z.; Xu, Y.; Sun, S.; Song, W.; Wu, C. Optimizing canopy-spacing configuration increases soybean yield under high planting density. Crop J. 2024, 13, 233–245. [Google Scholar] [CrossRef]
- Zhao, J.; Xin, Y.; Cui, W.; Li, P.; Su, J.; Zhao, L.; Wang, Q. The ankyrin repeat-containing protein OsANK3 affects grain size and quality in rice. Planta 2025, 262, 21. [Google Scholar] [CrossRef]
- Zhou, C.; Lin, Q.; Ren, Y.; Lan, J.; Miao, R.; Feng, M.; Wang, X.; Liu, X.; Zhang, S.; Pan, T.; et al. A CYP78As-small grain4-coat protein complex II pathway promotes grain size in rice. Plant Cell 2023, 35, 4325–4346. [Google Scholar] [CrossRef]
- Ma, X.; Wang, H.; Yan, S.; Zhou, C.; Zhou, K.; Zhang, Q.; Li, M.; Yang, Y.; Li, D.; Song, P.; et al. Large-scale genomic and phenomic analyses of modern cultivars empower future rice breeding design. Mol. Plant 2025, 18, 651–668. [Google Scholar] [CrossRef]
- Bai, C.; Wang, G.J.; Feng, X.H.; Gao, Q.; Wang, W.Q.; Xu, R.; Guo, S.J.; Shen, S.Y.; Ma, M.; Lin, W.H.; et al. OsMAPK6 phosphorylation and CLG1 ubiquitylation of GW6a non-additively enhance rice grain size through stabilization of the substrate. Nat. Commun. 2024, 15, 4300. [Google Scholar] [CrossRef] [PubMed]
- Bai, F.; Ma, H.; Cai, Y.; Shahid, M.Q.; Zheng, Y.; Lang, C.; Chen, Z.; Wu, J.; Liu, X.; Wang, L. Natural allelic variation in GRAIN SIZE AND WEIGHT 3 of wild rice regulates the grain size and weight. Plant Physiol. 2023, 193, 502–518. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Zhang, N.; Wang, W.Q.; Shen, S.Y.; Bai, C.; Song, X.J. The ubiquitin-interacting motif-type ubiquitin receptor HDR3 interacts with and stabilizes the histone acetyltransferase GW6a to control the grain size in rice. Plant Cell 2021, 33, 3331–3347. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Q.; Zhu, Y.; Ruan, B.; Yu, Y. Advances in Genetics and Breeding of Grain Shape in Rice. Agriculture 2025, 15, 1944. https://doi.org/10.3390/agriculture15181944
Chen Q, Zhu Y, Ruan B, Yu Y. Advances in Genetics and Breeding of Grain Shape in Rice. Agriculture. 2025; 15(18):1944. https://doi.org/10.3390/agriculture15181944
Chicago/Turabian StyleChen, Qian, Yuheng Zhu, Banpu Ruan, and Yanchun Yu. 2025. "Advances in Genetics and Breeding of Grain Shape in Rice" Agriculture 15, no. 18: 1944. https://doi.org/10.3390/agriculture15181944
APA StyleChen, Q., Zhu, Y., Ruan, B., & Yu, Y. (2025). Advances in Genetics and Breeding of Grain Shape in Rice. Agriculture, 15(18), 1944. https://doi.org/10.3390/agriculture15181944






