Enhancement of Antioxidant Enzymatic Defenses in Salt-Adapted Rice Seedlings
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
Salt Treatments
2.2. Quantitation of Thiobarbituric Acid Reactive Substances
2.3. Immunological Analyses
2.4. Determination of GSH and GSSG Levels
2.5. Statistical Analysis
3. Results
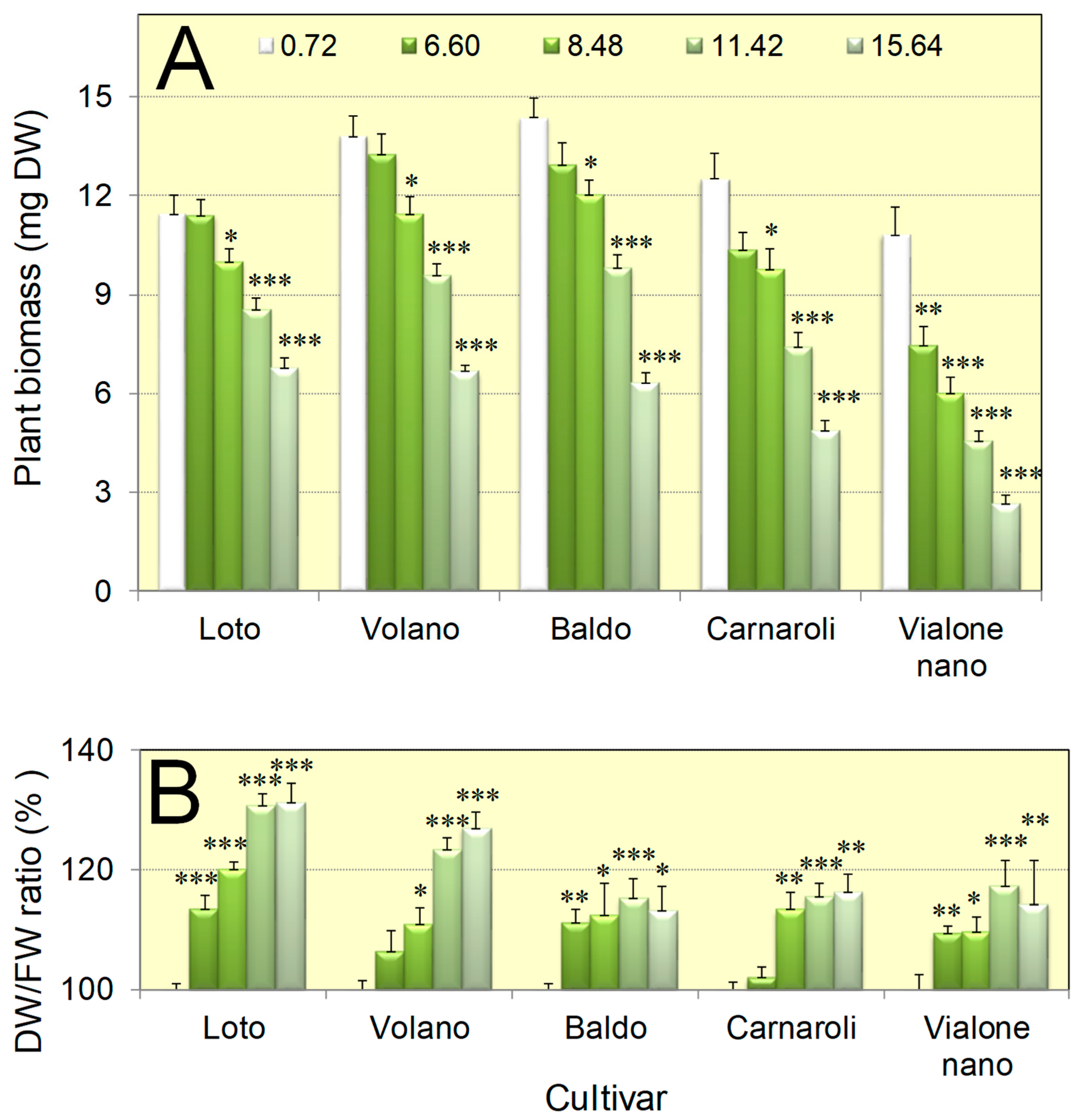
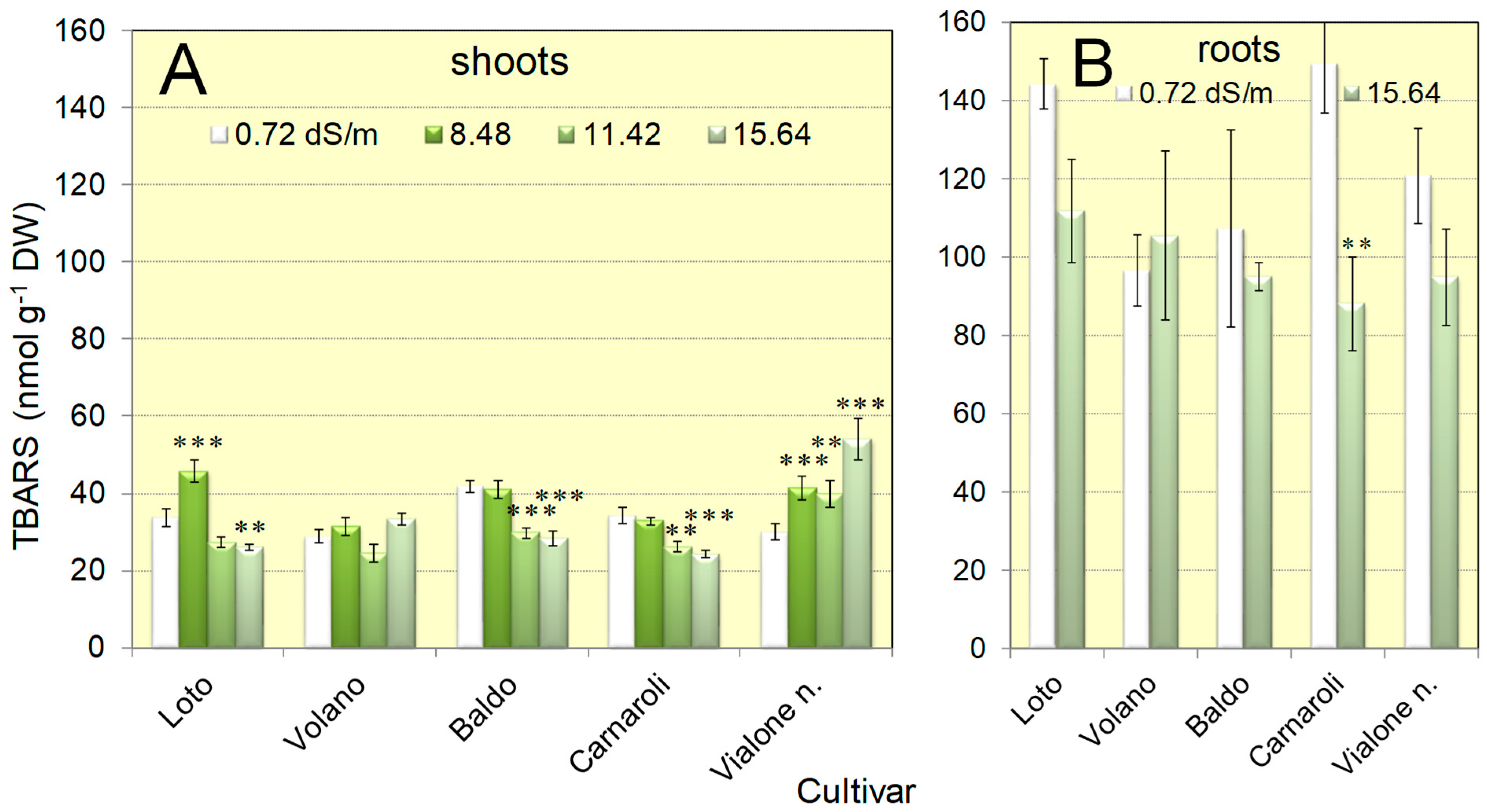
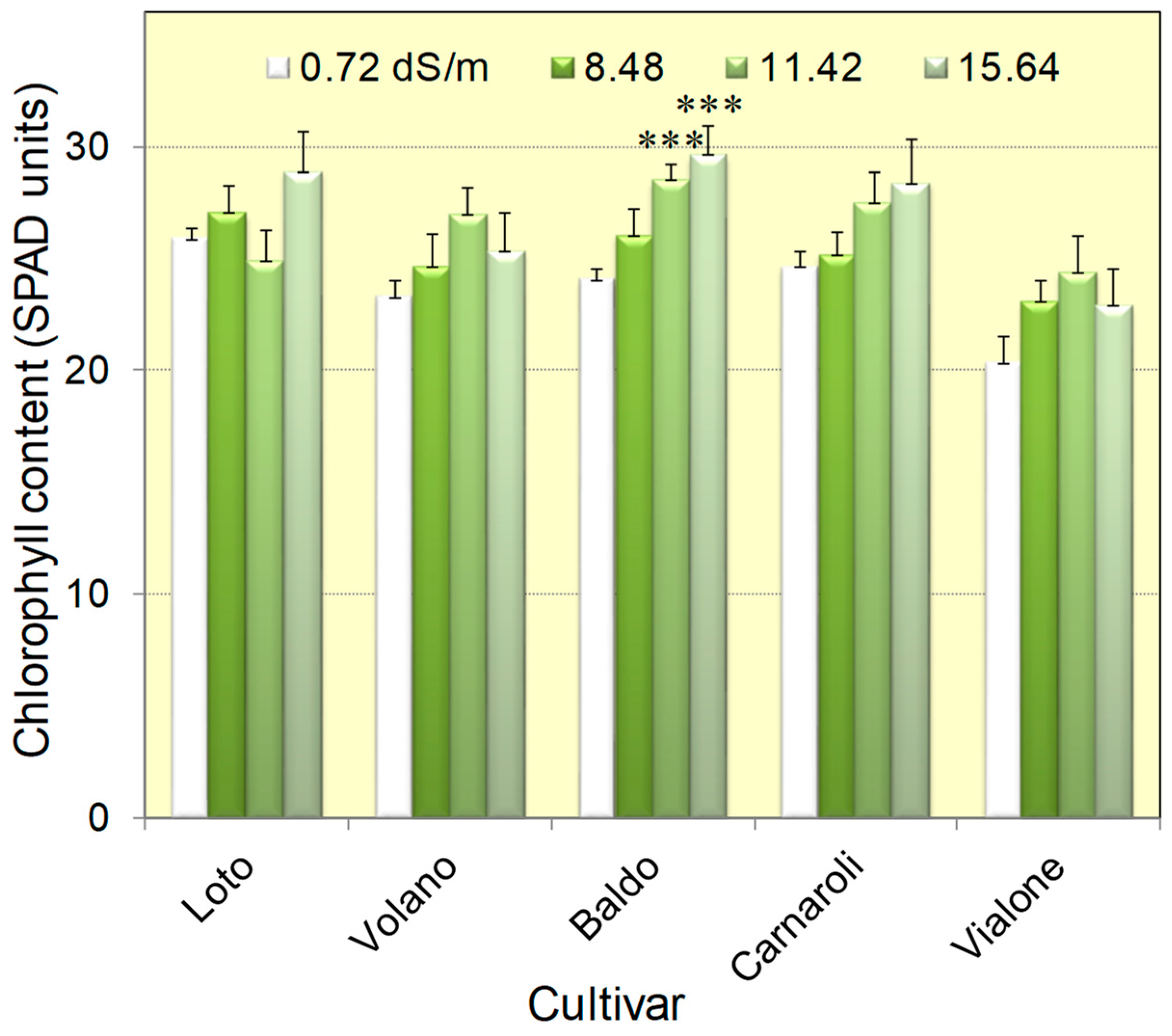
3.1. Seedlings of Five Italian Rice Genotypes Were Able to Cope with Salt Concentrations as High as 16 dS/m
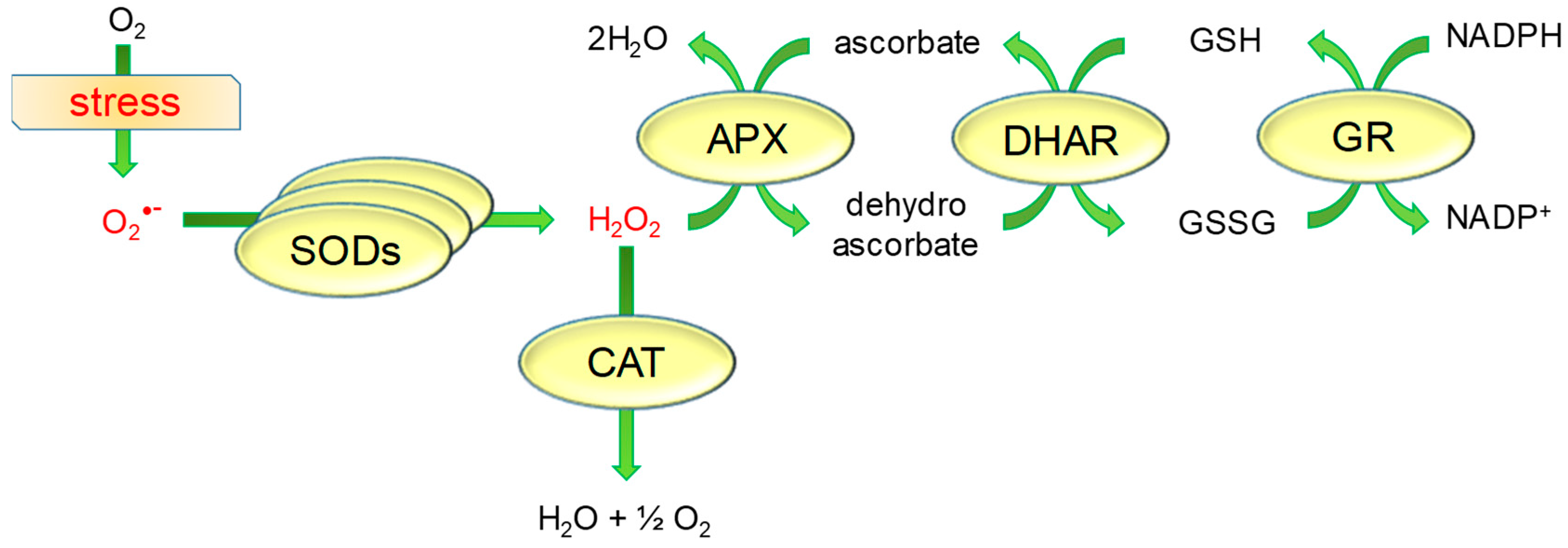
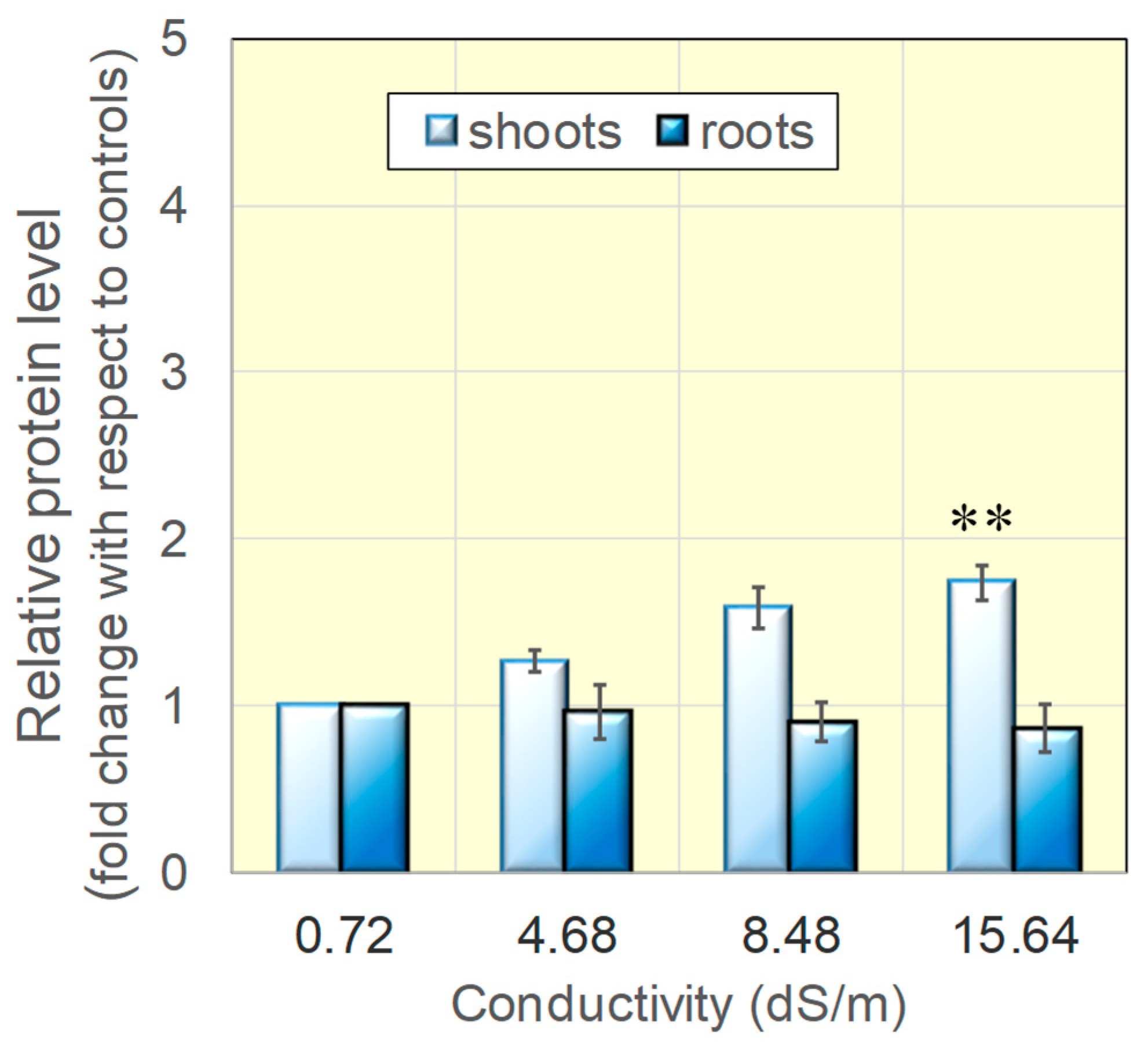
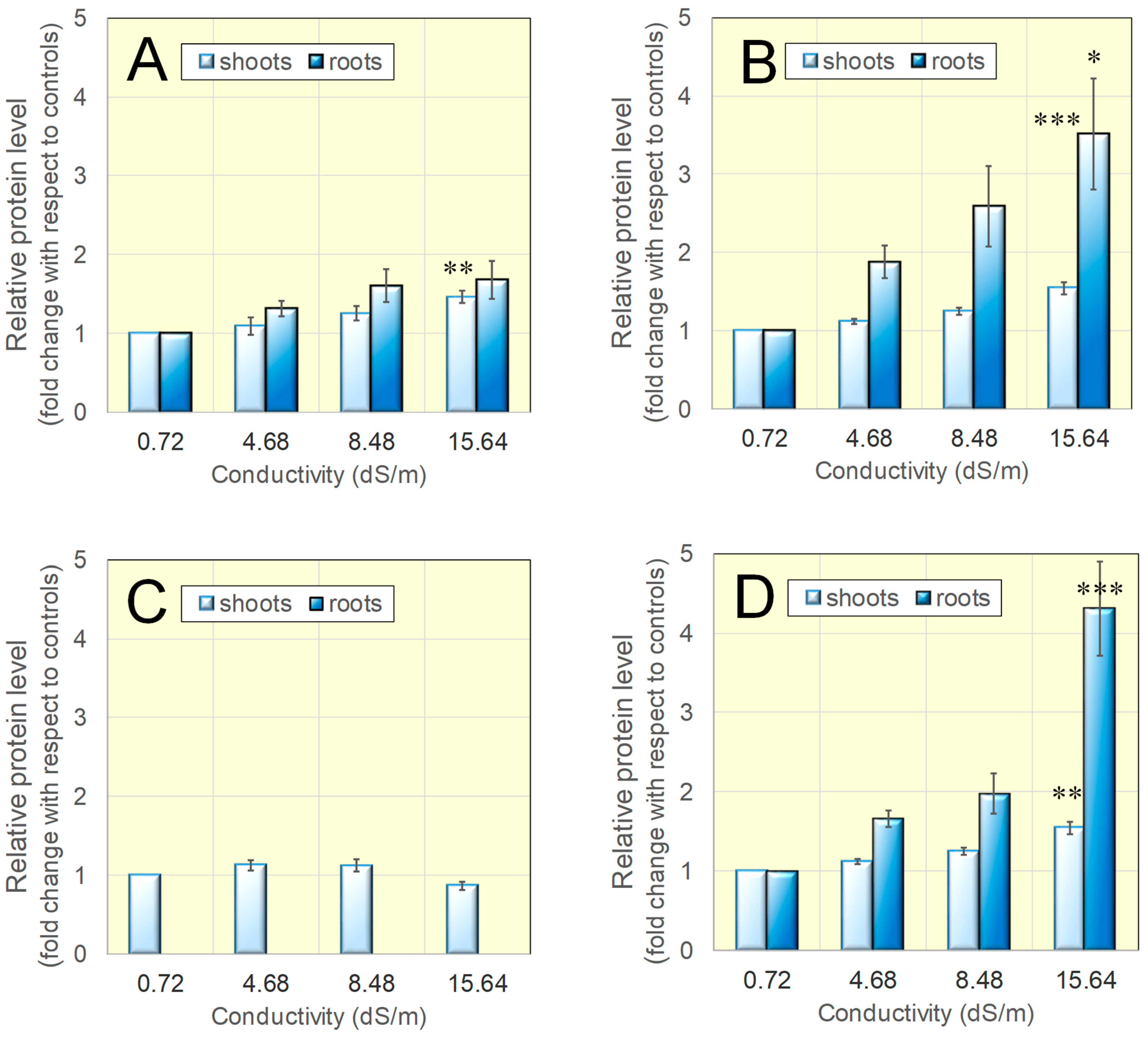
3.2. Protein Levels for Catalase, Mn- and Cu/Zn-Dependent Superoxide Dismutases Were Found Increased in Salt-Grown Rice Seedlings
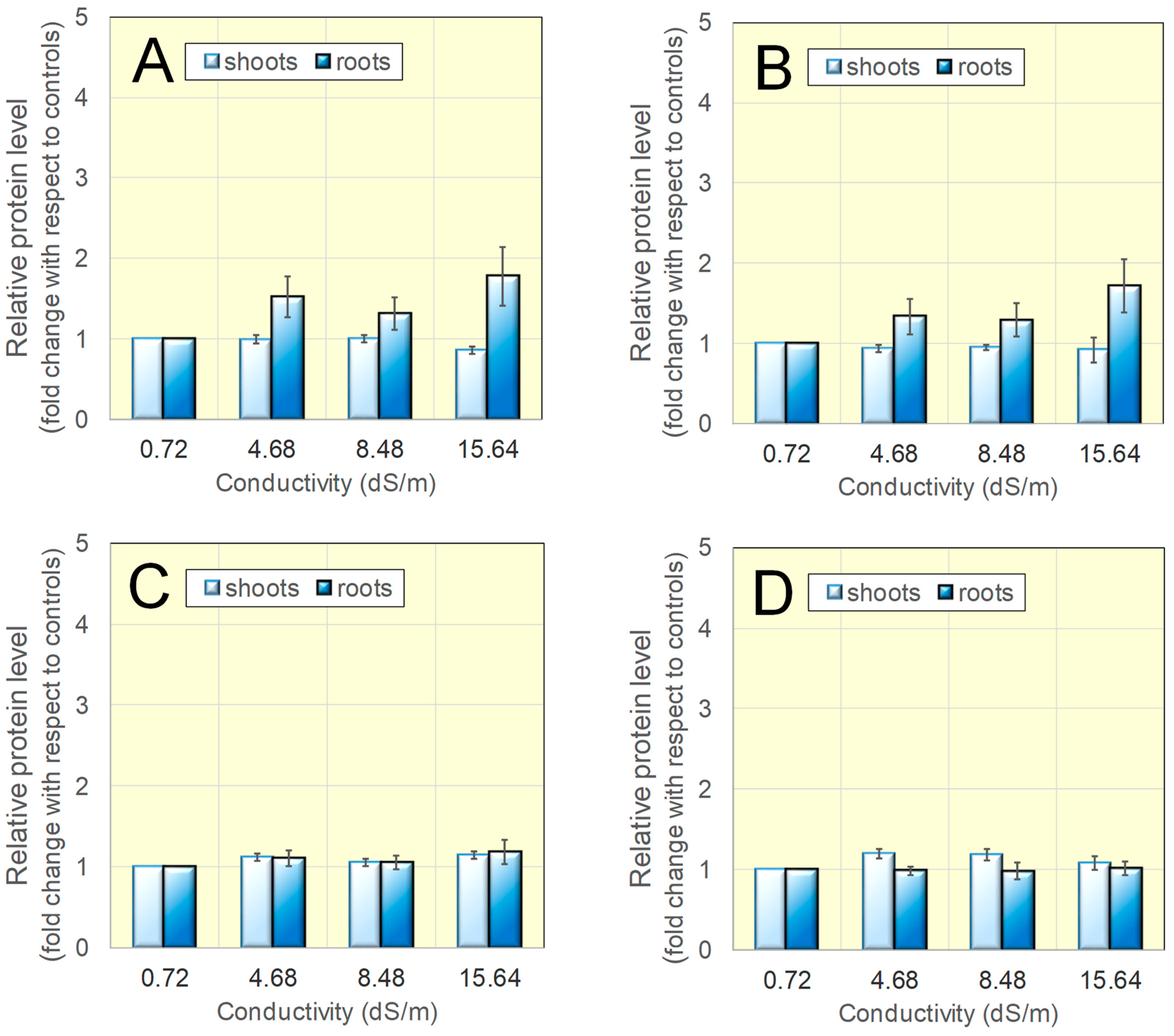
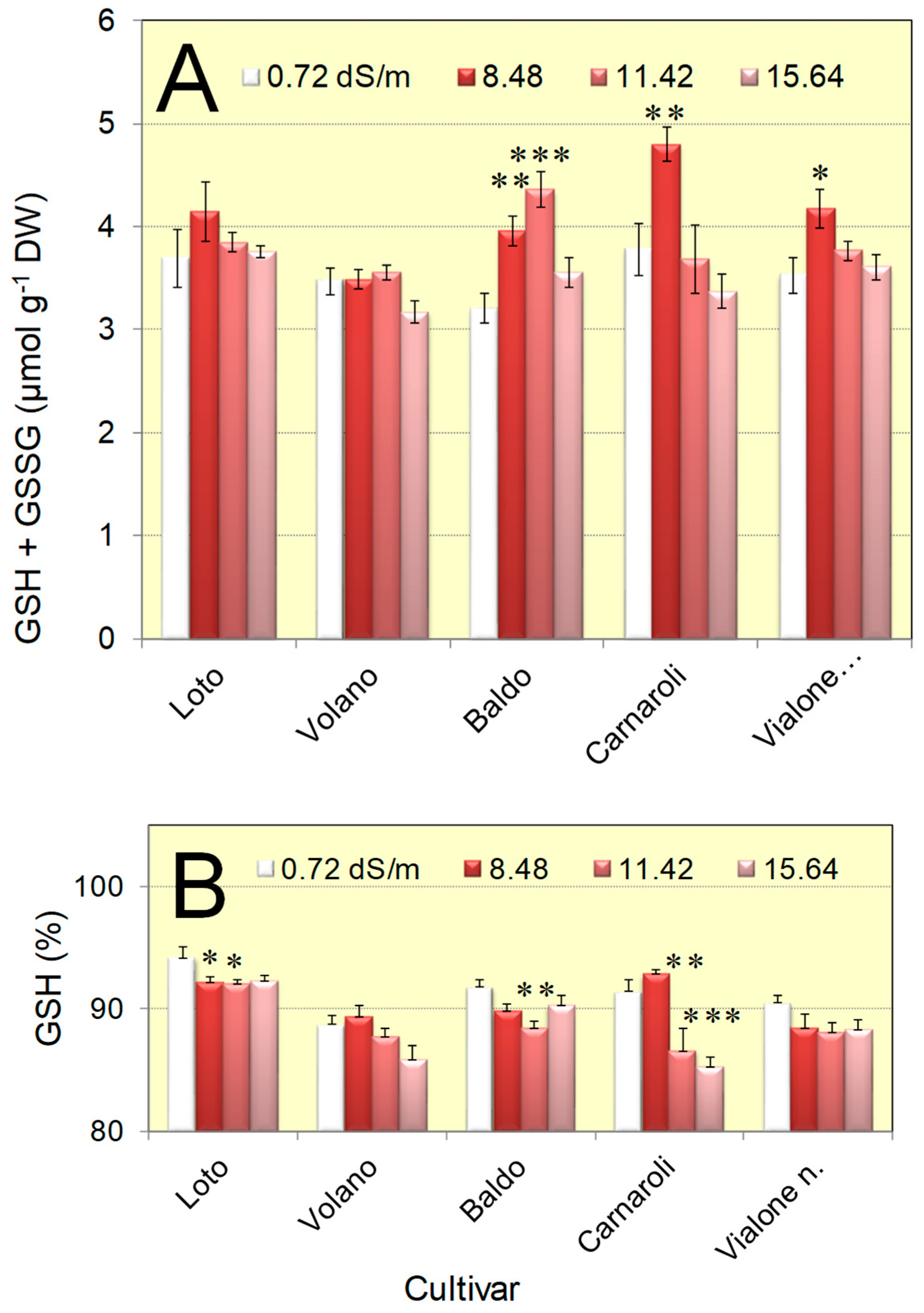
3.3. The Concentration of Glutathione and Its Reduction Level Vary Only Slightly in Salt-Adapted Seedlings
4. Discussion
4.1. Limitations of This Study
4.2. Future Directions
4.3. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| APX | ascorbate peroxidase |
| CAT | catalase |
| DHA | dehydroascorbate |
| DHAR | dehydroascorbate reductase |
| DTNB | 2,2′-dinitro–5,5′-dithio-dibenzoic acid |
| DW/FW | dry-to-fresh weight ratio |
| GR | glutathione reductase |
| GSH | glutathione, reduced |
| GSSG | glutathione, oxidized |
| MVP | 1-methyl-2-vinylpyridinium |
| ROS | reactive oxygen species |
| SOD | superoxide dismutase |
| TBARS | thiobarbituric acid reactive substances |
References
- Munns, R.; Gilliham, M. Salinity tolerance of crops—What is the cost? New Phytol. 2015, 208, 668–673. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Camac, J.; Robinson, A.; Kompas, T. Predicting changes in agricultural yields under climate change scenarios and their implications for global food security. Sci. Rep. 2025, 15, 2858. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Grattan, S.; Zeng, L.; Shannon, M.; Roberts, S. Rice is more sensitive to salinity than previously thought. Calif. Agric. 2002, 56, 189–198. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef] [PubMed]
- Yancey, P.H. Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J. Exp. Biol. 2005, 208, 2819–2830. [Google Scholar] [CrossRef]
- Forlani, G.; Trovato, M.; Funck, D.; Signorelli, S. Regulation of proline accumulation and its molecular and physiological functions in stress defence. In Osmoprotectant-Mediated Abiotic Stress Tolerance in Plants: Recent Advances and Future Perspectives; Hossain, M.A., Kumar, V., Burritt, D.J., Fujita, M., Mäkelä, P.S.A., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 73–97. [Google Scholar]
- Gul, Z.; Tang, Z.H.; Arif, M.; Ye, Z. An insight into abiotic stress and influx tolerance mechanisms in plants to cope in saline environments. Biology 2022, 11, 597. [Google Scholar] [CrossRef] [PubMed]
- Shabala, S.; Pottosin, I. Regulation of potassium transport in plants under hostile conditions: Implications for abiotic and biotic stress tolerance. Physiol. Plant 2014, 151, 257–279. [Google Scholar] [CrossRef]
- Teakle, N.L.; Tyerman, S.D. Mechanisms of Cl− transport contributing to salt tolerance. Plant Cell Environ. 2010, 33, 566–589. [Google Scholar] [CrossRef]
- Flowers, T.J.; Munns, R.; Colmer, T.D. Sodium chloride toxicity and the cellular basis of salt tolerance in halophytes. Ann. Bot. 2015, 115, 419–431. [Google Scholar] [CrossRef]
- Soltabayeva, A.; Ongaltay, A.; Omondi, J.O.; Srivastava, S. Morphological, physiological and molecular markers for salt-stressed plants. Plants 2021, 10, 243. [Google Scholar] [CrossRef]
- Assaha, D.V.M.; Ueda, A.; Saneoka, H.; Al-Yahyai, R.; Yaish, M.W. The role of Na+ and K+ transporters in salt stress adaptation in glycophytes. Front. Physiol. 2017, 8, 509. [Google Scholar] [CrossRef]
- Keisham, M.; Mukherjee, S.; Bhatla, S.C. Mechanisms of sodium transport in plants—Progresses and challenges. Int. J. Mol. Sci. 2018, 19, 647. [Google Scholar] [CrossRef] [PubMed]
- Sackey, O.K.; Feng, N.; Mohammed, Y.Z.; Dzou, C.F.; Zheng, D.; Zhao, L.; Shen, X. A comprehensive review on rice responses and tolerance to salt stress. Front. Plant Sci. 2025, 16, 1561280. [Google Scholar] [CrossRef]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 2012, 217037. [Google Scholar] [CrossRef]
- Choudhury, S.; Panda, P.; Sahoo, L.; Panda, S.K. Reactive oxygen species signaling in plants under abiotic stress. Plant Signal. Behav. 2013, 8, e23681. [Google Scholar] [CrossRef]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Ma, X.; Wan, P.; Liu, L. Plant salt-tolerance mechanism: A review. Biochem. Biophys. Res. Commun. 2018, 495, 286–291. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Ascorbate and glutathione: The heart of the redox hub. Plant Physiol. 2011, 155, 2–18. [Google Scholar] [CrossRef]
- Garratt, L.C.; Janagoudar, B.S.; Lowe, K.C.; Anthony, P.; Power, J.B.; Davey, M.R. Salinity tolerance and antioxidant status in cotton cultures. Free Radic. Biol. Med. 2002, 33, 502–511. [Google Scholar] [CrossRef] [PubMed]
- Vaidyanathan, H.; Sivakumar, P.; Chakrabarty, R.; Thomas, G. Scavenging of reactive oxygen species in NaCl-stressed rice (Oryza sativa L.)—Differential response in salt-tolerant and sensitive varieties. Plant Sci. 2003, 165, 1411–1418. [Google Scholar] [CrossRef]
- El-Shabrawi, H.; Kumar, B.; Kaul, T.; Reddy, M.K.; Singla-Pareek, S.L.; Sopory, S.K. Redox homeostasis, antioxidant defense, and methylglyoxal detoxification as markers for salt tolerance in Pokkali rice. Protoplasma 2010, 245, 85–96. [Google Scholar] [CrossRef]
- Abogadallah, G.M. Antioxidative defense under salt stress. Plant Signal. Behav. 2010, 5, 369–374. [Google Scholar] [CrossRef]
- Kibria, M.G.; Hossain, M.; Murata, Y.; Hoque, M.A. Antioxidant defense mechanisms of salinity tolerance in rice genotypes. Rice Sci. 2017, 24, 155–162. [Google Scholar] [CrossRef]
- Laus, M.N.; De Santis, M.A.; Flagella, Z.; Soccio, M. Changes in antioxidant defence system in durum wheat under hyperosmotic stress: A concise overview. Plants 2021, 11, 98. [Google Scholar] [CrossRef]
- Muthayya, S.; Sugimoto, J.D.; Montgomery, S.; Maberly, G.F. An overview of global rice production, supply, trade, and consumption. Ann. N. Y. Acad. Sci. 2014, 1324, 7–14. [Google Scholar] [CrossRef]
- Singh, R.K.; Kota, S.; Flowers, T.J. Salt tolerance in rice: Seedling and reproductive stage QTL mapping come of age. Theor. Appl. Genet. 2021, 134, 3495–3533. [Google Scholar] [CrossRef] [PubMed]
- Reddy, I.N.B.L.; Kim, B.-K.; Yoon, I.-S.; Kim, K.-H.; Kwon, T.-R. Salt tolerance in rice: Focus on mechanisms and approaches. Rice Sci. 2017, 24, 123–144. [Google Scholar] [CrossRef]
- Rajkumari, N.; Chowrasia, S.; Nishad, J.; Ganie, S.A.; Mondal, T.K. Metabolomics-mediated elucidation of rice responses to salt stress. Planta 2023, 258, 111. [Google Scholar] [CrossRef]
- Reddy, I.N.B.L.; Kim, S.-M.; Kim, B.-K.; Yoon, I.-S.; Kwon, T.-R. Identification of rice accessions associated with K+/Na+ ratio and salt tolerance based on physiological and molecular responses. Rice Sci. 2017, 24, 360–364. [Google Scholar] [CrossRef]
- Li, Q.; Zhu, P.; Yu, X.; Xu, J.; Liu, G. Physiological and molecular mechanisms of rice tolerance to salt and drought stress: Advances and future directions. Int. J. Mol. Sci. 2024, 25, 9404. [Google Scholar] [CrossRef]
- Kumar, V.; Srivastava, A.K.; Sharma, D.; Pandey, S.P.; Pandey, M.; Dudwadkar, A.; Parab, H.J.; Suprasanna, P.; Das, B.K. Antioxidant defense and ionic homeostasis govern stage-specific response of salinity stress in contrasting rice varieties. Plants 2024, 13, 778. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, R.P.; Prashar, V.; Jawali, N.; Sunkar, R.; Misra, H.S.; Saini, A. Molecular and biochemical analysis of duplicated cytosolic CuZn superoxide dismutases of rice and in silico analysis in plants. Front. Plant Sci. 2022, 13, 864330. [Google Scholar] [CrossRef]
- Nath, K.; Kumar, S.; Poudyal, R.S.; Yang, Y.N.; Timilsina, R.; Park, Y.S.; Nath, J.; Chauhan, P.S.; Pant, B.; Lee, C.-H. Developmental stage-dependent differential gene expression of superoxide dismutase isoenzymes and their localization and physical interaction network in rice (Oryza sativa L.). Genes Genom. 2014, 36, 45–55. [Google Scholar] [CrossRef]
- Yang, Q.; Zheng, Y.; Li, X. Integrative RNA-seq and ATAC-seq analysis unveils antioxidant defense mechanisms in salt-tolerant rice variety Pokkali. BMC Plant Biol. 2025, 25, 364. [Google Scholar] [CrossRef]
- Guan, Q.; Liao, X.; He, M.; Li, X.; Wang, Z.; Ma, H.; Yu, S.; Liu, S. Tolerance analysis of chloroplast OsCu/Zn-SOD overexpressing rice under NaCl and NaHCO3 stress. PLoS ONE 2017, 12, e0186052. [Google Scholar] [CrossRef] [PubMed]
- Motohashi, T.N.K.; Prodhan, S.H.; Nakao, K.; Shishido, T.; Yamamoto, Y.; Moriwaki, T.; Hattori, E.; Asada, M.; Morishima, H.; Hirose, S.; et al. Production of salt stress tolerant rice by overexpression of the catalase gene, katE, derived from Escherichia coli. Asia-Pac. J. Mol. Biol. Biotechnol. 2010, 18, 37–42. [Google Scholar]
- Lu, Z.; Liu, D.; Liu, S. Two rice cytosolic ascorbate peroxidases differentially improve salt tolerance in transgenic Arabidopsis. Plant Cell Rep. 2007, 26, 1909–1917. [Google Scholar] [CrossRef]
- Zheng, X.; Yuan, Z.; Yu, Y.; Yu, S.; He, H. OsCSD2 and OsCSD3 Enhance seed storability by modulating antioxidant enzymes and abscisic acid in rice. Plants 2024, 13, 310. [Google Scholar] [CrossRef]
- Faivre-Rampant, O.; Bruschi, G.; Abbruscato, P.; Cavigiolo, S.; Picco, A.M.; Borgo, L.; Lupotto, E.; Piffanelli, P. Assessment of genetic diversity in Italian rice germplasm related to agronomic traits and blast resistance (Magnaporthe oryzae). Mol. Breed. 2011, 27, 233–246. [Google Scholar] [CrossRef]
- Forlani, G.; Bertazzini, M.; Cagnano, G. Stress-driven increase in proline levels, and not proline levels themselves, correlates with the ability to withstand excess salt in a group of 17 Italian rice genotypes. Plant Biol. 2019, 21, 336–342. [Google Scholar] [CrossRef]
- Bertazzini, M.; Sacchi, G.A.; Forlani, G. A differential tolerance to mild salt stress conditions among six Italian rice genotypes does not rely on Na+ exclusion from shoots. J. Plant Physiol. 2018, 226, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Cao, Q.; Zhang, K.; Ata-Ul-Karim, S.T.; Tian, Y.; Zhu, Y.; Cao, W.; Liu, X. Optimal leaf positions for SPAD meter measurement in rice. Front. Plant Sci. 2016, 7, 719. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Queval, G.; Noctor, G. A plate reader method for the measurement of NAD, NADP, glutathione, and ascorbate in tissue extracts: Application to redox profiling during Arabidopsis rosette development. Anal. Biochem. 2007, 363, 58–69. [Google Scholar] [CrossRef]
- Chaves, M.M.; Flexas, J.; Pinheiro, C. Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2009, 103, 551–560. [Google Scholar] [CrossRef]
- Zhang, J.; Che, J.; Ouyang, Y. Engineering rice genomes towards green super rice. Curr. Opin. Plant Biol. 2024, 82, 102664. [Google Scholar] [CrossRef]
- Raghuvanshi, R.; Srivastava, A.K.; Verulkar, S.; Suprasanna, P. Unlocking allelic diversity for sustainable development of salinity stress tolerance in rice. Curr. Genomics 2021, 22, 393–403. [Google Scholar] [CrossRef]
- Padmavathi, G.; Bangale, U.; Rao, K.N.; Balakrishnan, D.; Arun, M.N.; Singh, R.K.; Sundaram, R.M. Progress and prospects in harnessing wild relatives for genetic enhancement of salt tolerance in rice. Front. Plant Sci. 2023, 14, 1253726. [Google Scholar] [CrossRef]
- Ahmadi, N.; Negrão, S.; Katsantonis, D.; Frouin, J.; Ploux, J.; Letourmy, P.; Droc, G.; Babo, P.; Trindade, H.; Bruschi, G.; et al. Targeted association analysis identified japonica rice varieties achieving Na+/K+ homeostasis without the allelic make-up of the salt tolerant indica variety Nona Bokra. Theor. Appl. Genet. 2011, 123, 881–895. [Google Scholar] [CrossRef]
- Joshi, S.; Nath, J.; Singh, A.K.; Pareek, A.; Joshi, R. Ion transporters and their regulatory signal transduction mechanisms for salinity tolerance in plants. Physiol. Plant. 2022, 174, e13702. [Google Scholar] [CrossRef]
- Geilfus, C.-M. Chloride in soil: From nutrient to soil pollutant. Environ. Exp. Bot. 2019, 157, 299–309. [Google Scholar] [CrossRef]
- Liu, X.X.; Zhu, Y.X.; Fang, X.Z.; Ye, J.Y.; Du, W.X.; Zhu, Q.Y.; Lin, X.Y.; Jin, C.W. Ammonium aggravates salt stress in plants by entrapping them in a chloride over-accumulation state in an NRT1.1-dependent manner. Sci. Total Environ. 2020, 746, 141244. [Google Scholar] [CrossRef] [PubMed]
- Formentin, E.; Sudiro, C.; Perin, G.; Riccadonna, S.; Barizza, E.; Baldoni, E.; Lavezzo, E.; Stevanato, P.; Sacchi, G.A.; Fontana, P.; et al. Transcriptome and cell physiological analyses in different rice cultivars provide new insights into adaptive and salinity stress responses. Front. Plant Sci. 2018, 9, 204. [Google Scholar] [CrossRef]
- Formentin, E.; Sudiro, C.; Ronci, M.B.; Locato, V.; Barizza, E.; Stevanato, P.; Ijaz, B.; Zottini, M.; De Gara, L.; Lo Schiavo, F. H2O2 signature and innate antioxidative profile make the difference between sensitivity and tolerance to salt in rice cells. Front. Plant Sci. 2018, 9, 1549. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Han, B.; Wang, T.; Chen, S.; Li, H.; Zhang, Y.; Dai, S. Mechanisms of plant salt response: Insights from proteomics. J. Proteome Res. 2012, 11, 49–67. [Google Scholar] [CrossRef]
- Hualpa-Ramirez, E.; Carrasco-Lozano, E.C.; Madrid-Espinoza, J.; Tejos, R.; Ruiz-Lara, S.; Stange, C.; Norambuena, L. Stress salinity in plants: New strategies to cope with in the foreseeable scenario. Plant Physiol. Biochem. 2024, 208, 108507. [Google Scholar] [CrossRef]
- Martins, T.S.; Da-Silva, C.J.; Shabala, S.; Striker, G.G.; Carvalho, I.R.; de Oliveira, A.C.B.; do Amarante, L. Understanding plant responses to saline waterlogging: Insights from halophytes and implications for crop tolerance. Planta 2023, 259, 24. [Google Scholar] [CrossRef]
- Türkan, I.; Demiral, T. Recent developments in understanding salinity tolerance. Environ. Exp. Bot. 2009, 67, 2–9. [Google Scholar] [CrossRef]
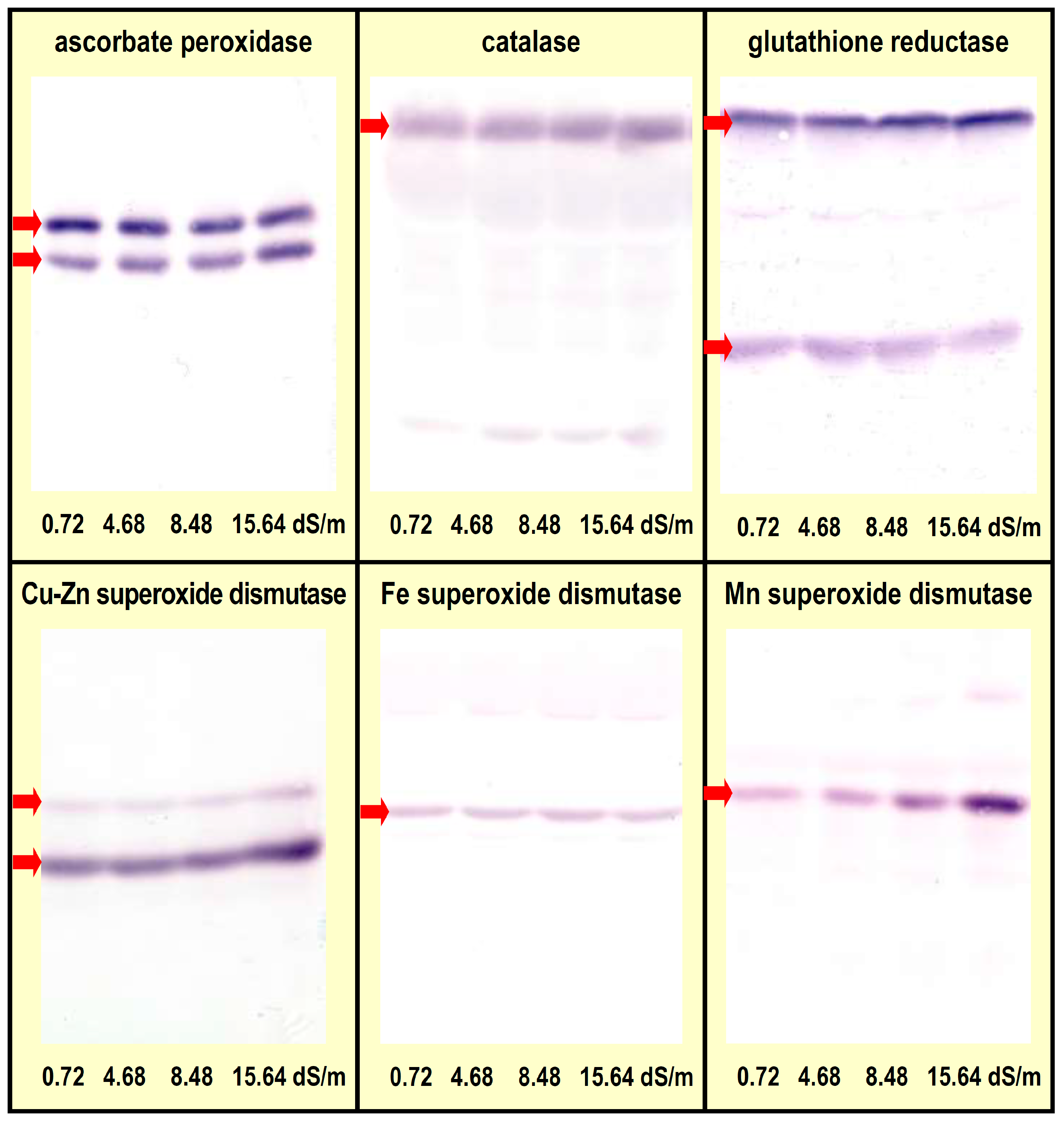
| Agrisera Code | Target | Immunogen | Acrylamide in Separating Gel | Expected Band(s) (kDa) |
|---|---|---|---|---|
| AS08 368 | ascorbate peroxidase | synthetic peptide derived from Arabidopsis thaliana tAPX (At1g77490) and sAPX (At4g08390) | 12% | 27 (cytosolic) 33–37 (plastidial) |
| AS09 501 | catalase | KLH-conjugated peptide chosen from know plant catalase sequences including A. thaliana | 10% | 57 |
| AS06 181 | glutathione reductase | maltose binding protein (MBP) fusion of Zea mays glutathione reductase, O64409 | 10% | 42 (plastidial) 60 (cytosolic) |
| AS06 170 | Cu/Zn superoxide dismutase | heterologously expressed Arabidopsis thaliana Cu/ZnSOD O78310 | 12% | 15 (cytosolic) 21 (plastidial) |
| AS06 125 | Fe superoxide dismutase | overexpressed Chlamydomonas reinhardtii thioredoxin fusion protein A8IGH1, FeSOD | 12% | 29 |
| AS09 524 | Mn superoxide dismutase | KLH-conjugated synthetic peptide derived from MnSOD sequences in dicots and monocots including A. thaliana O81235 | 12% | 25 (21–29) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Forlani, G. Enhancement of Antioxidant Enzymatic Defenses in Salt-Adapted Rice Seedlings. Agriculture 2025, 15, 1234. https://doi.org/10.3390/agriculture15121234
Forlani G. Enhancement of Antioxidant Enzymatic Defenses in Salt-Adapted Rice Seedlings. Agriculture. 2025; 15(12):1234. https://doi.org/10.3390/agriculture15121234
Chicago/Turabian StyleForlani, Giuseppe. 2025. "Enhancement of Antioxidant Enzymatic Defenses in Salt-Adapted Rice Seedlings" Agriculture 15, no. 12: 1234. https://doi.org/10.3390/agriculture15121234
APA StyleForlani, G. (2025). Enhancement of Antioxidant Enzymatic Defenses in Salt-Adapted Rice Seedlings. Agriculture, 15(12), 1234. https://doi.org/10.3390/agriculture15121234






