Exploring Genetic Variation in Root Traits and Root–Fungal Associations in Aegilops tauschii
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Experimental Conditions
2.2. Plant Growth and Experiment Setup
2.2.1. Crude Field Inoculum
2.2.2. Quality and Uniformity Controls
2.3. Data Collection
2.3.1. Architectural Traits
2.3.2. Morphological Traits
2.3.3. Physiological Traits
2.3.4. Root Biotic Traits
2.3.5. Shoot System Traits
2.4. Statistical Analysis
3. Results
3.1. Root Trait Variations Among Wheat Genotypes
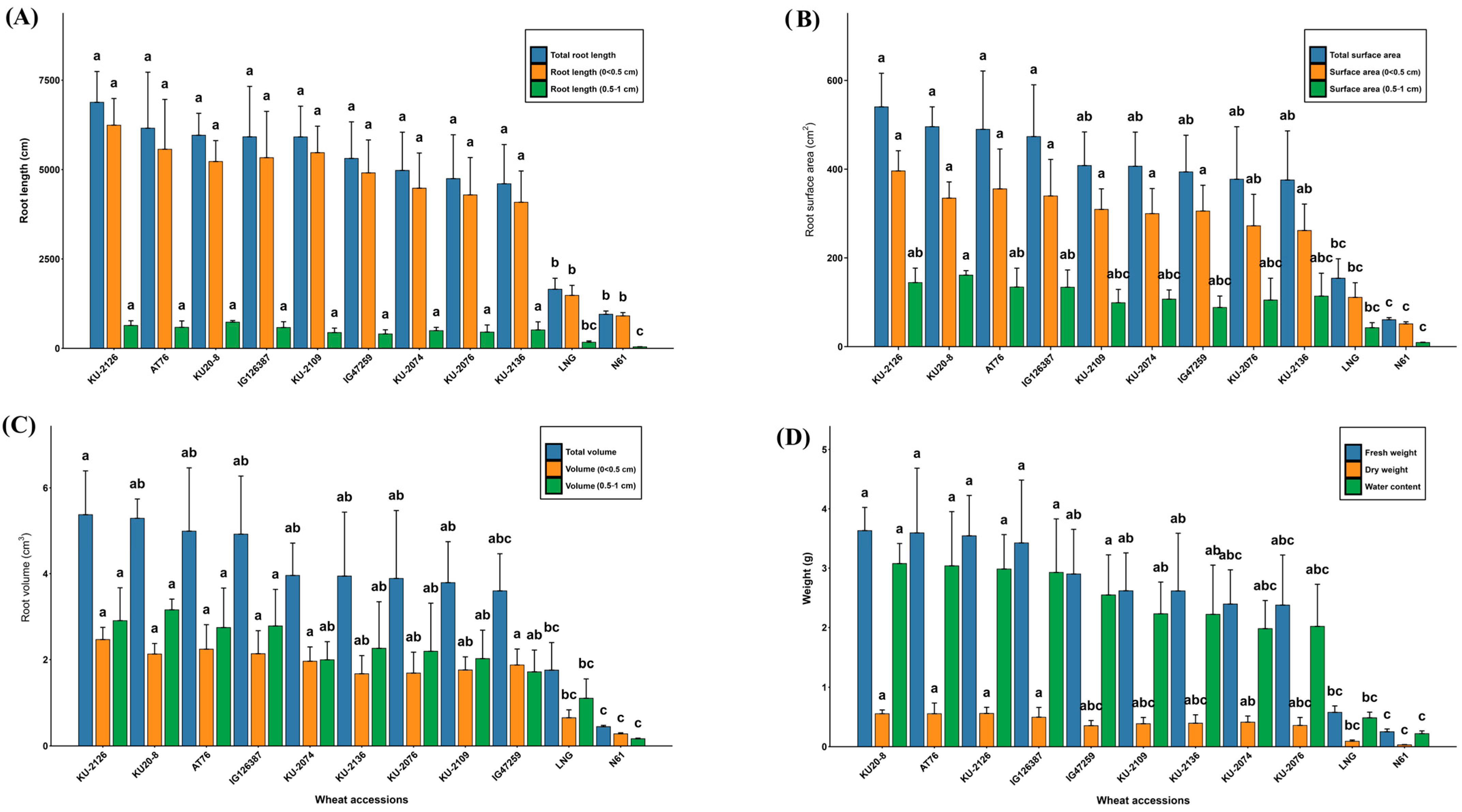
3.1.1. Root Architectural Traits
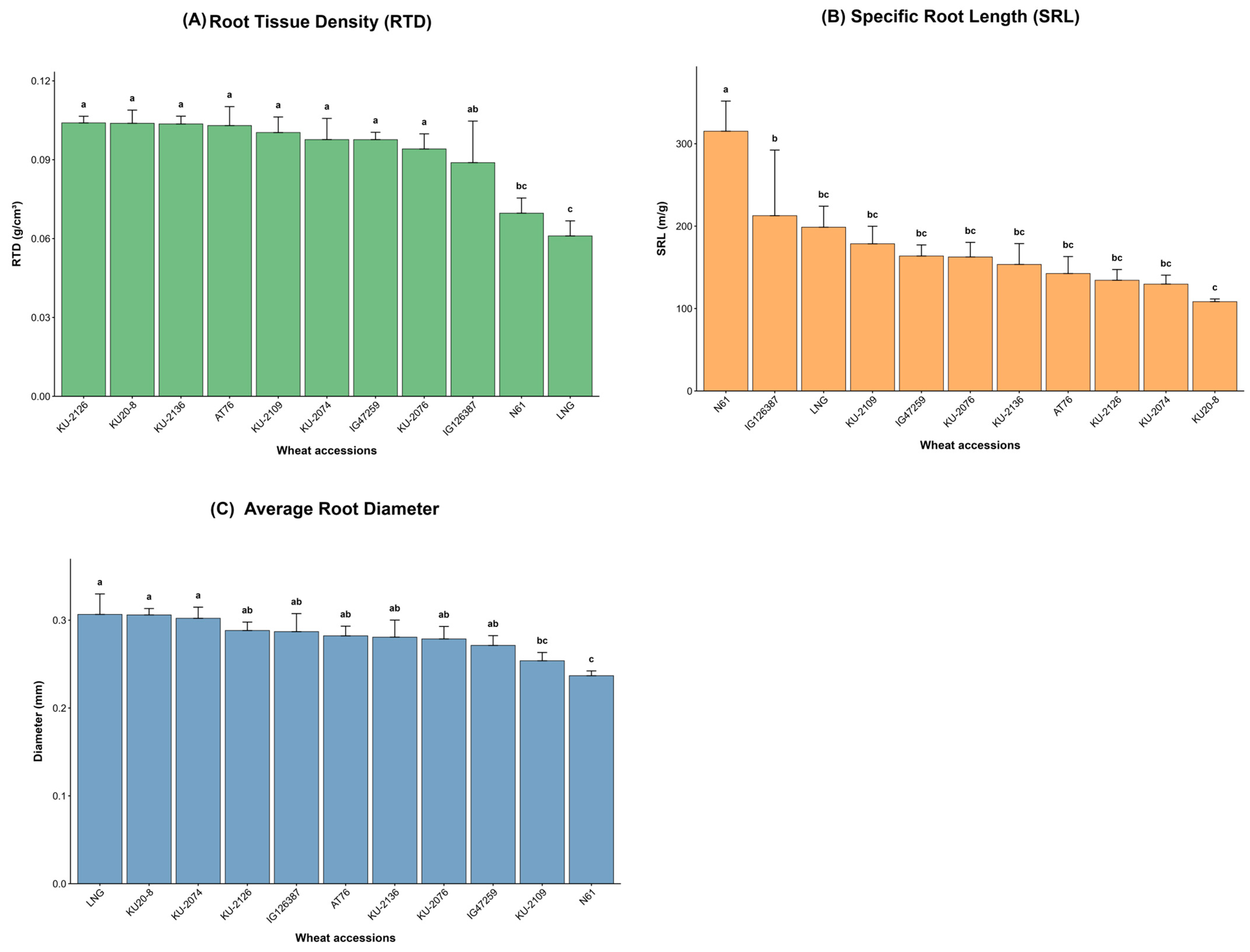
3.1.2. Morphological and Physiological Root Traits
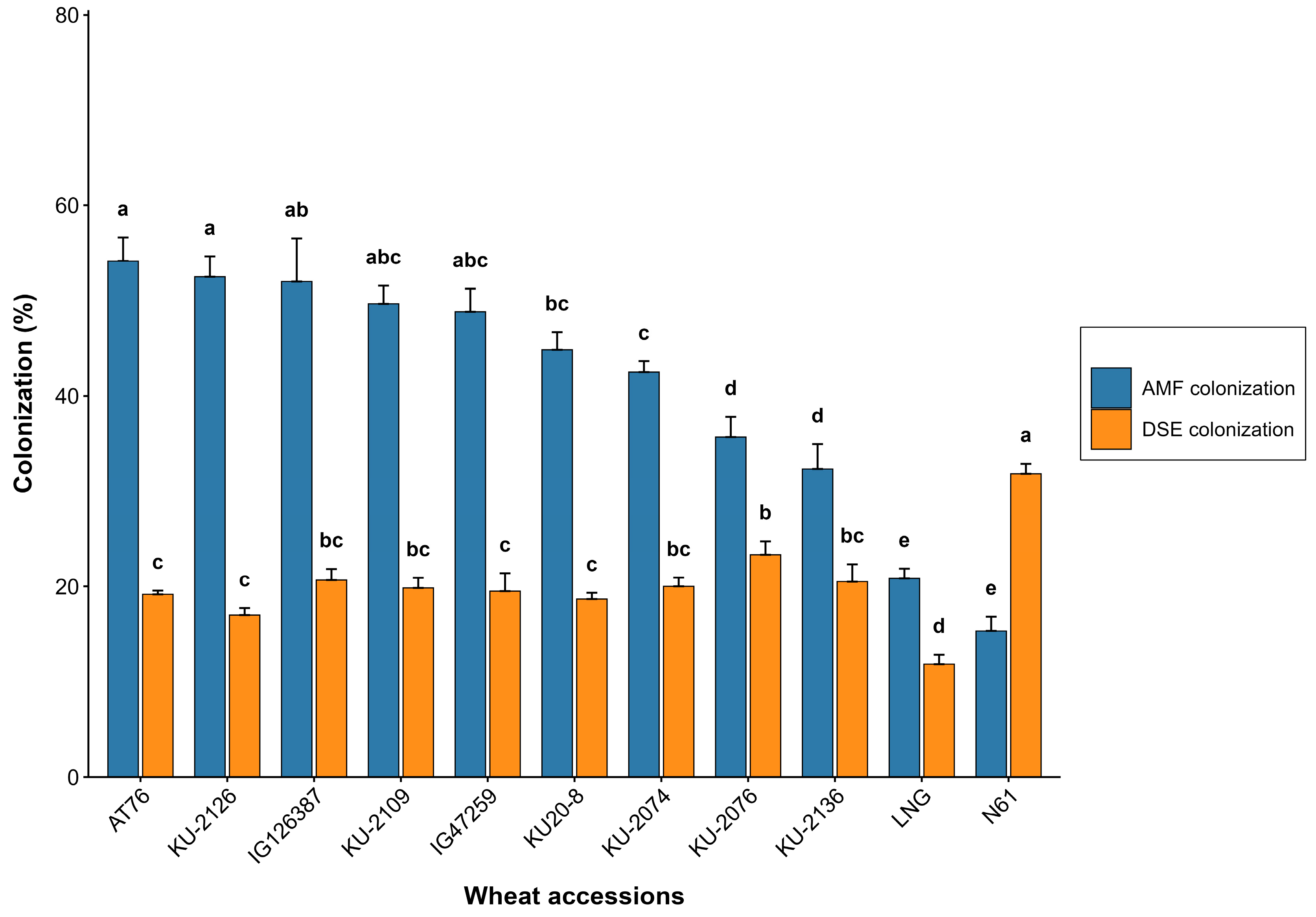
3.1.3. Biotic Traits
3.1.4. Shoot System Traits
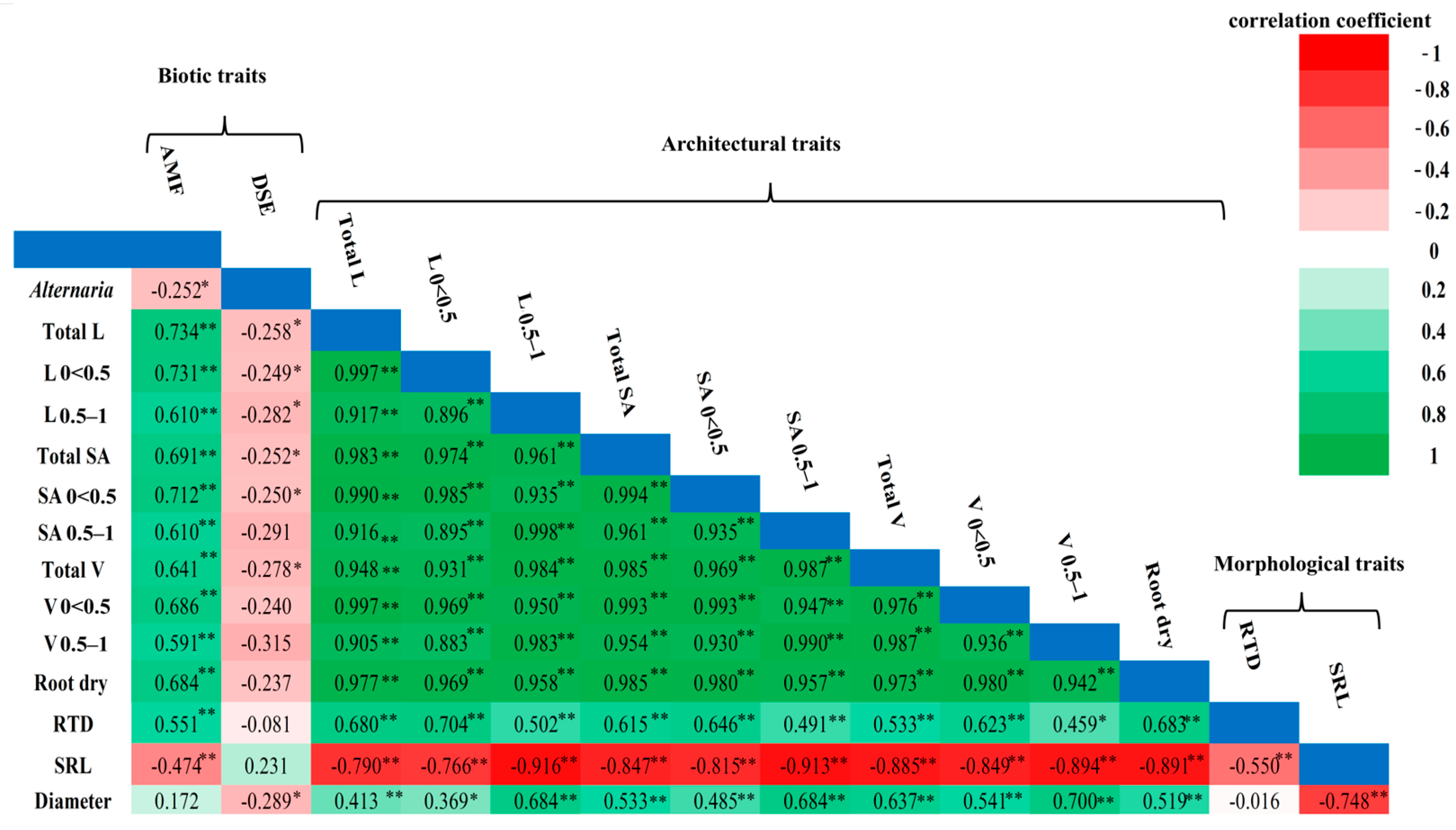
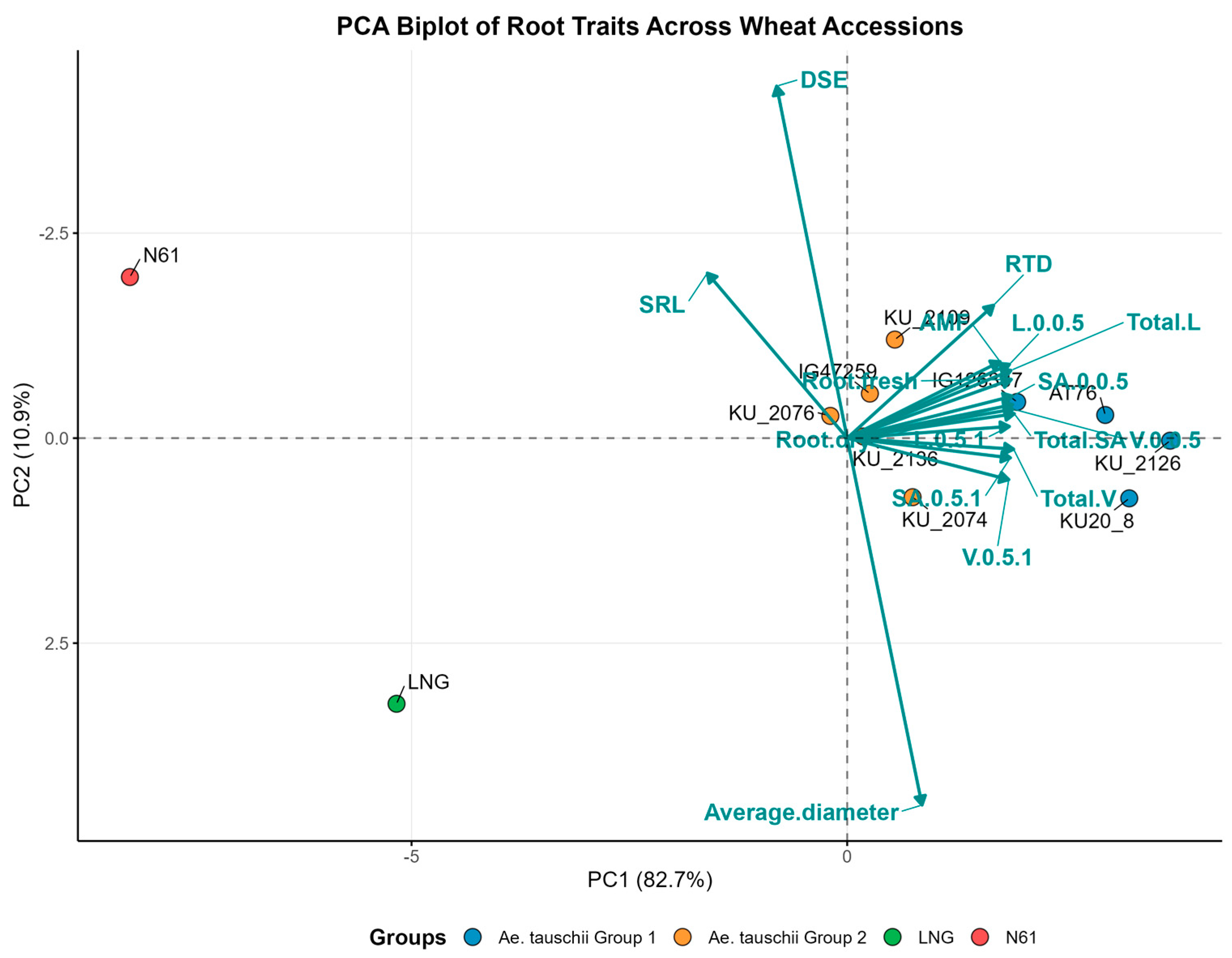
3.2. Association Among Root Traits
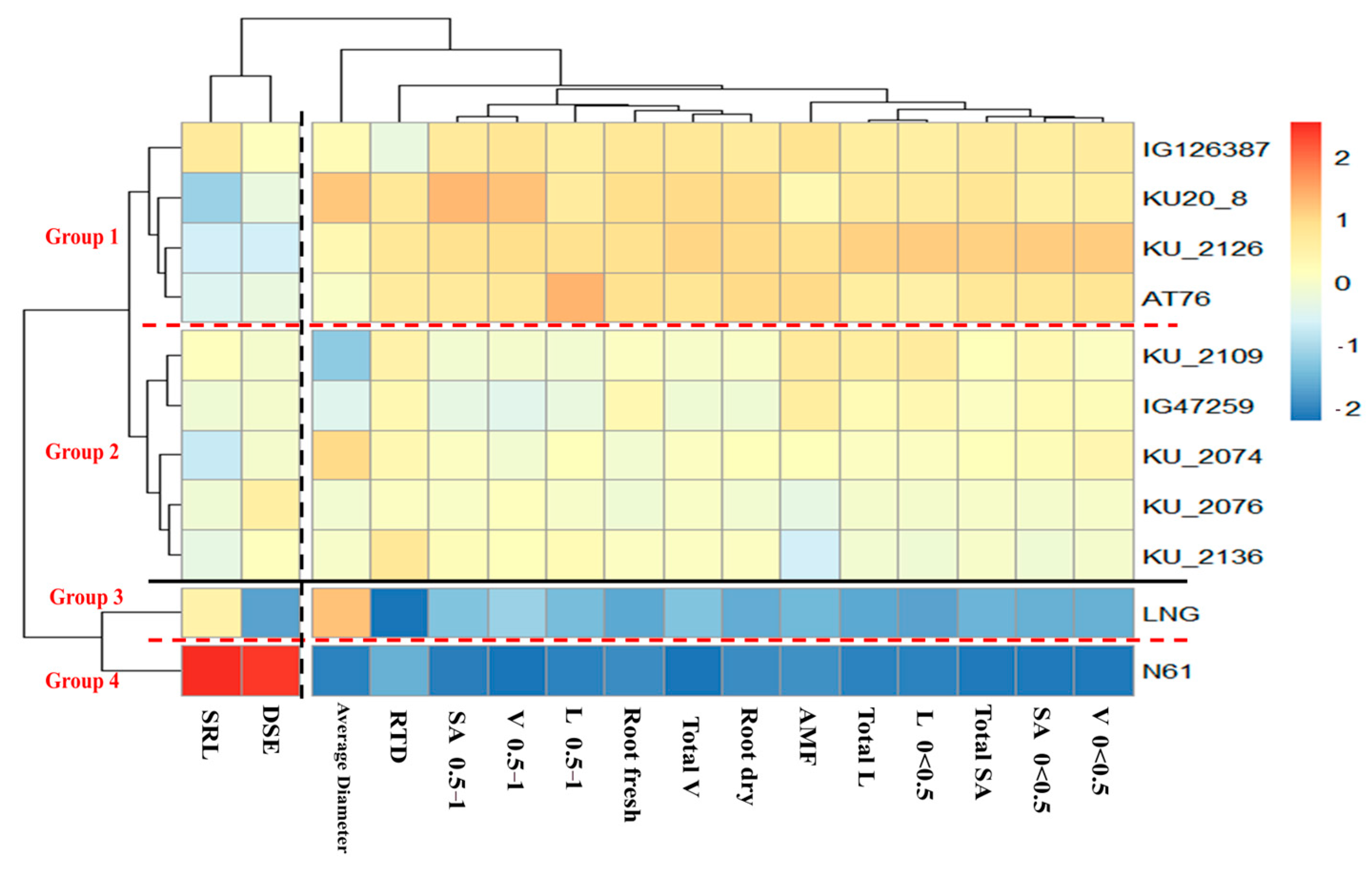
3.3. Wheat Accession Grouping and Association by Root Traits
4. Discussion
5. Conclusions
6. Limitations
7. Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bishopp, A.; Lynch, J.P. The Hidden Half of Crop Yields. Nat. Plants 2015, 1, 15117. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, L.; Reynolds, M.P.; Wang, J.; Chang, X.; Mao, X.; Jing, R. Recognizing the Hidden Half in Wheat: Root System Attributes Associated with Drought Tolerance. J. Exp. Bot. 2021, 72, 5117–5133. [Google Scholar] [CrossRef] [PubMed]
- Ober, E.S.; Alahmad, S.; Cockram, J.; Forestan, C.; Hickey, L.T.; Kant, J.; Maccaferri, M.; Marr, E.; Milner, M.; Pinto, F.; et al. Wheat Root Systems as a Breeding Target for Climate Resilience. Theor. Appl. Genet. 2021, 134, 1645–1662. [Google Scholar] [CrossRef] [PubMed]
- Singer, E.; Vogel, J.P.; Northen, T.; Mungall, C.J.; Juenger, T.E. Novel and Emerging Capabilities That Can Provide a Holistic Understanding of the Plant Root Microbiome. Phytobiomes J. 2021, 5, 122–132. [Google Scholar] [CrossRef]
- Liu, Q.; Cheng, L.; Nian, H.; Jin, J.; Lian, T. Linking Plant Functional Genes to Rhizosphere Microbes: A Review. Plant Biotechnol. J. 2023, 21, 902–917. [Google Scholar] [CrossRef]
- Marcussen, T.; Sandve, S.R.; Heier, L.; Spannagl, M.; Pfeifer, M.; Jakobsen, K.S.; Wulff, B.B.H.; Steuernagel, B.; Mayer, K.F.X.; Olsen, O.-A. Ancient Hybridizations among the Ancestral Genomes of Bread Wheat. Science 2014, 345, 1250092. [Google Scholar] [CrossRef]
- Wang, H.; Hu, Z.; Huang, K.; Han, Y.; Zhao, A.; Han, H.; Song, L.; Fan, C.; Li, R.; Xin, M.; et al. Three Genomes Differentially Contribute to the Seedling Lateral Root Number in Allohexaploid Wheat: Evidence from Phenotype Evolution and Gene Expression. Plant J. 2018, 95, 976–987. [Google Scholar] [CrossRef]
- Gaurav, K.; Arora, S.; Silva, P.; Sánchez-Martín, J.; Horsnell, R.; Gao, L.; Brar, G.S.; Widrig, V.; John Raupp, W.; Singh, N.; et al. Population Genomic Analysis of Aegilops Tauschii Identifies Targets for Bread Wheat Improvement. Nat. Biotechnol. 2022, 40, 422–431. [Google Scholar] [CrossRef]
- Feldman, M.; Levy, A.A. Evolution of Wheat Under Cultivation. In Wheat Evolution and Domestication; Springer InternationalPublishing: Cham, Switzerland, 2023. [Google Scholar]
- Wang, L.; Liu, K.; Mao, S.; Li, Z.; Lu, Y.; Wang, J.; Liu, Y.; Wei, Y.; Zheng, Y. Large-Scale Screening for Aegilops Tauschii Tolerant Genotypes to Phosphorus Deficiency at Seedling Stage. Euphytica 2015, 204, 571–586. [Google Scholar] [CrossRef]
- Tkacz, A.; Pini, F.; Turner, T.R.; Bestion, E.; Simmonds, J.; Howell, P.; Greenland, A.; Cheema, J.; Emms, D.M.; Uauy, C.; et al. Agricultural Selection of Wheat Has Been Shaped by Plant-Microbe Interactions. Front. Microbiol. 2020, 11, 132. [Google Scholar] [CrossRef]
- Wipf, H.M.L.; Coleman-Derr, D. Evaluating Domestication and Ploidy Effects on the Assembly of the Wheat Bacterial Microbiome. PLoS ONE 2021, 16, e0248030. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Yin, X.; Qiu, K.; Rees, R.M.; Harrison, M.T.; Chen, F.; Wen, X. Wheat Cultivar Replacement Drives Soil Microbiome and Microbial Cooccurrence Patterns. Agric. Ecosyst. Environ. 2024, 360, 108774. [Google Scholar] [CrossRef]
- Kou, H.; Zhang, Z.; Yang, Y.; Wei, C.; Xu, L.; Zhang, G. Advances in the Mining of Disease Resistance Genes from Aegilops Tauschii and the Utilization in Wheat. Plants 2023, 12, 880. [Google Scholar] [CrossRef] [PubMed]
- Bektas, H.; Hohn, C.E.; Waines, J.G. Characteristics of the Root System in the Diploid Genome Donors of Hexaploid Wheat (Triticum aestivum L.). Genet. Resour. Crop Evol. 2017, 64, 1641–1650. [Google Scholar] [CrossRef]
- Feldman, M.; Levy, A.A. Genome Evolution Due to Allopolyploidization in Wheat. Genetics 2012, 192, 763–774. [Google Scholar] [CrossRef]
- Smith, S.E.; Read, D.J. Mycorrhizal Symbiosis; Academic Press: Cambridge, MA, USA, 2010; ISBN 978-0-08-055934-6. [Google Scholar]
- Zhu, X.; Song, F.; Liu, S.; Liu, F. Arbuscular Mycorrhiza Improve Growth, Nitrogen Uptake, and Nitrogen Use Efficiency in Wheat Grown under Elevated CO2. Mycorrhiza 2015, 26, 133–140. [Google Scholar] [CrossRef]
- Savary, R.; Masclaux, F.G.; Wyss, T.; Droh, G.; Cruz Corella, J.; Machado, A.P.; Morton, J.B.; Sanders, I.R. A Population Genomics Approach Shows Widespread Geographical Distribution of Cryptic Genomic Forms of the Symbiotic Fungus Rhizophagus Irregularis. ISME J. 2018, 12, 17–30. [Google Scholar] [CrossRef]
- Li, M.; Wang, R.; Tian, H.; Gao, Y. Transcriptome Responses in Wheat Roots to Colonization by the Arbuscular Mycorrhizal Fungus Rhizophagus Irregularis. Mycorrhiza 2018, 28, 747–759. [Google Scholar] [CrossRef]
- Inbaraj, M.P. Plant-Microbe Interactions in Alleviating Abiotic Stress—A Mini Review. Front. Agron. 2021, 3, 667903. [Google Scholar] [CrossRef]
- Grebenikova, N.; Korshunov, A.; Rud’, V.; Savchenko, I.; Marques, M. Root Rot Grain Crops on Cereals Caused by the Phytopathogenic Fungi. MATEC Web Conf. 2018, 245, 11006. [Google Scholar] [CrossRef]
- Fiorilli, V.; Vannini, C.; Ortolani, F.; Garcia-Seco, D.; Chiapello, M.; Novero, M.; Domingo, G.; Terzi, V.; Morcia, C.; Bagnaresi, P.; et al. Omics Approaches Revealed How Arbuscular Mycorrhizal Symbiosis Enhances Yield and Resistance to Leaf Pathogen in Wheat. Sci. Rep. 2018, 8, 9625. [Google Scholar] [CrossRef]
- Dowarah, B.; Gill, S.S.; Agarwala, N. Arbuscular Mycorrhizal Fungi in Conferring Tolerance to Biotic Stresses in Plants. J. Plant Growth Regul. 2022, 41, 1429–1444. [Google Scholar] [CrossRef]
- Mahjoob, M.M.M.; Chen, T.-S.; Gorafi, Y.S.A.; Yamasaki, Y.; Kamal, N.M.; Abdelrahman, M.; Iwata, H.; Matsuoka, Y.; Tahir, I.S.A.; Tsujimoto, H. Traits to Differentiate Lineages and Subspecies of Aegilops tauschii, the D Genome Progenitor Species of Bread Wheat. Diversity 2021, 13, 217. [Google Scholar] [CrossRef]
- Martín, M.; Rubio, A.; Remesal, E.; Cano, C.; Bago, A. Application of the Ultimate Arbuscular Mycorrhizal Inoculant MYCOGEL® in Japan: Results and Prospects. Ph.D. Thesis, Tohoku University, Sendai, Japan, 2018. [Google Scholar]
- Bardgett, R.D.; Mommer, L.; De Vries, F.T. Going Underground: Root Traits as Drivers of Ecosystem Processes. Trends Ecol. Evol. 2014, 29, 692–699. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.M.; Hayman, D.S. Improved Procedures for Clearing Roots and Staining Parasitic and Vesicular-Arbuscular Mycorrhizal Fungi for Rapid Assessment of Infection. Trans. Br. Mycol. Soc. 1970, 55, 158–161. [Google Scholar] [CrossRef]
- McGONIGLE, T.P.; Miller, M.H.; Evans, D.G.; Fairchild, G.L.; Swan, J.A. A New Method Which Gives an Objective Measure of Colonization of Roots by Vesicular—Arbuscular Mycorrhizal Fungi. New Phytol. 1990, 115, 495–501. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2024. [Google Scholar]
- Mohammedali, A.K.H.; Kamal, N.M.; Gorafi, Y.S.A.; Tahir, I.S.A.; Tsujimoto, H.; Taniguchi, T. Variation in Root Traits and Root-Endophyte Interactions in Primary Synthetic Wheat Derived from Aegilops Tauschii Collected from Diverse Soil Types. Agronomy 2025, 15, 1443. [Google Scholar] [CrossRef]
- Barkaoui, K.; Roumet, C.; Volaire, F. Mean Root Trait More than Root Trait Diversity Determines Drought Resilience in Native and Cultivated Mediterranean Grass Mixtures. Agric. Ecosyst. Environ. 2016, 231, 122–132. [Google Scholar] [CrossRef]
- Freschet, G.T.; Roumet, C.; Comas, L.H.; Weemstra, M.; Bengough, A.G.; Rewald, B.; Bardgett, R.D.; De Deyn, G.B.; Johnson, D.; Klimešová, J.; et al. Root Traits as Drivers of Plant and Ecosystem Functioning: Current Understanding, Pitfalls and Future Research Needs. New Phytol. 2021, 232, 1123–1158. [Google Scholar] [CrossRef]
- Hashem, A.; Akhter, A.; Alqarawi, A.A.; Singh, G.; Almutairi, K.F.; Abd_Allah, E.F. Mycorrhizal Fungi Induced Activation of Tomato Defense System Mitigates Fusarium Wilt Stress. Saudi J. Biol. Sci. 2021, 28, 5442–5450. [Google Scholar] [CrossRef]
- Enebe, M.C.; Erasmus, M. Frontiers|Susceptibility and Plant Immune Control—A Case of Mycorrhizal Strategy for Plant Colonization, Symbiosis, and Plant Immune Suppression. Front. Agron. 2023, 14, 1178258. [Google Scholar] [CrossRef]
- Bennett, A.E.; Groten, K. The Costs and Benefits of Plant–Arbuscular Mycorrhizal Fungal Interactions. Annu. Rev. Plant Biol. 2022, 73, 649–672. [Google Scholar] [CrossRef]
- Mostofa, M.G.; Li, W.; Nguyen, K.H.; Fujita, M.; Tran, L.-S.P. Strigolactones in Plant Adaptation to Abiotic Stresses: An Emerging Avenue of Plant Research. Plant Cell Environ. 2018, 41, 2227–2243. [Google Scholar] [CrossRef]
- Mahjoob, M.M.M.; Kamal, N.M.; Gorafi, Y.S.A.; Tsujimoto, H. Genome-Wide Association Study Reveals Distinct Genetic Associations Related to Leaf Hair Density in Two Lineages of Wheat-Wild Relative Aegilops Tauschii. Sci. Rep. 2022, 12, 17486. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.I.Y.; Kamal, N.M.; Gorafi, Y.S.A.; Abdalla, M.G.A.; Tahir, I.S.A.; Tsujimoto, H. Heat Stress-Tolerant Quantitative Trait Loci Identified Using Backcrossed Recombinant Inbred Lines Derived from Intra-Specifically Diverse Aegilops Tauschii Accessions. Plants 2024, 13, 347. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhu, L.; Fan, R.; Li, Z.; Liu, Y.; Shaheen, A.; Nie, F.; Li, C.; Liu, X.; Li, Y.; et al. A Platform for Whole-Genome Speed Introgression from Aegilops Tauschii to Wheat for Breeding Future Crops. Nat. Protoc. 2024, 19, 281–312. [Google Scholar] [CrossRef] [PubMed]
| Genebank Accession | Origin | Taxon | Genome | Lineage |
|---|---|---|---|---|
| KU-2126 | Iran | Aegilops tauschii | D | TauL1 a |
| KU20-8 | Iran | Aegilops tauschii | D | TauL2 a |
| AT76 | China | Aegilops tauschii | D | TauL1 |
| IG126387 | Turkmenistan | Aegilops tauschii | D | TauL1 |
| KU-2109 | Iran | Aegilops tauschii | D | TauL1 |
| IG47259 | Syria | Aegilops tauschii | D | TauL1 |
| KU-2074 | Iran | Aegilops tauschii | D | TauL2 |
| KU-2076 | Iran | Aegilops tauschii | D | TauL2 |
| KU-2136 | Turkey | Aegilops tauschii | D | TauL1 |
| LPGKU2196 (LNG) | American cultivar | Triticum durum | AB | – |
| KT020-032 (N61) | Japanese cultivar | Triticum aestivum | ABD | – |
| Variable | Mean ± SE | F-Statistic a | df1 | df2 | p-Value |
|---|---|---|---|---|---|
| Root architectural trait | |||||
| Total L (cm) | 4825 ± 358 | 16.106 | 10 | 20.543 | ≤0.0001 *** |
| L 0 < 0.5 (cm) | 4363 ± 319 | 15.691 | 10 | 20.623 | ≤0.0001 *** |
| L 0.5–1.0 (cm) | 462 ± 45 | 25.957 | 10 | 20.073 | ≤0.0001 *** |
| Total SA (cm2) | 380 ± 30 | 17.487 | 10 | 20.174 | ≤0.0001 *** |
| SA 0 < 0.5 (cm2) | 276 ± 20 | 16.997 | 10 | 20.300 | ≤0.0001 *** |
| SA 0.5–1.0 (cm2) | 104 ± 10 | 24.166 | 10 | 20.049 | ≤0.0001 *** |
| Total V (cm3) | 3.82 ± 0.35 | 16.868 | 10 | 20 < 0.51 | ≤0.0001 *** |
| V 0 < 0.5 (cm3) | 1.72 ± 0.13 | 17.204 | 10 | 20.210 | ≤0.0001 *** |
| V 0.5–1.0 (cm3) | 2.10 ± 0.22 | 16.976 | 10 | 20.029 | ≤0.0001 *** |
| Root fresh weight (g) | 2.54 ± 0.25 | 12.808 | 10 | 20.635 | ≤0.0001 *** |
| Root dry weight (g) | 0.38 ± 0.04 | 12.902 | 10 | 20.190 | ≤0.0001 *** |
| Root morphological trait | |||||
| RTD (g/cm3) | 0.093 ± 0.003 | 6.106 | 10 | 21.814 | ≤0.0001 *** |
| SRL (cm/g) | 173 ± 11 | 6.296 | 10 | 20.890 | ≤0.0001 *** |
| Average diameter (cm) | 0.28 ± 0.01 | 6.251 | 10 | 21.826 | ≤0.0001 *** |
| Root physiological trait | |||||
| Carbon (%) | 45.6 ± 0.2 | 1.867 | 10 | 21.718 | 0.108 |
| Nitrogen (%) | 1.43 ± 0.04 | 1.282 | 10 | 21.865 | 0.299 |
| C/N ratio | 33 ± 0.8 | 1.540 | 10 | 21.909 | 0.191 |
| Root biotic traits | |||||
| AMF colonization rate (%) | 41 ± 2.0 | 51.725 | 10 | 21.867 | ≤0.0001 *** |
| DSE colonization rate (%) | 20 ± 1.0 | 17.178 | 10 | 21.735 | ≤0.0001 *** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohammedali, A.K.H.; Gorafi, Y.S.A.; Kamal, N.M.; Tahir, I.S.A.; Tsujimoto, H.; Taniguchi, T. Exploring Genetic Variation in Root Traits and Root–Fungal Associations in Aegilops tauschii. Agriculture 2025, 15, 1889. https://doi.org/10.3390/agriculture15171889
Mohammedali AKH, Gorafi YSA, Kamal NM, Tahir ISA, Tsujimoto H, Taniguchi T. Exploring Genetic Variation in Root Traits and Root–Fungal Associations in Aegilops tauschii. Agriculture. 2025; 15(17):1889. https://doi.org/10.3390/agriculture15171889
Chicago/Turabian StyleMohammedali, Ahmed Khaled Hassan, Yasir Serag Alnor Gorafi, Nasrein Mohamed Kamal, Izzat Sidahmed Ali Tahir, Hisashi Tsujimoto, and Takeshi Taniguchi. 2025. "Exploring Genetic Variation in Root Traits and Root–Fungal Associations in Aegilops tauschii" Agriculture 15, no. 17: 1889. https://doi.org/10.3390/agriculture15171889
APA StyleMohammedali, A. K. H., Gorafi, Y. S. A., Kamal, N. M., Tahir, I. S. A., Tsujimoto, H., & Taniguchi, T. (2025). Exploring Genetic Variation in Root Traits and Root–Fungal Associations in Aegilops tauschii. Agriculture, 15(17), 1889. https://doi.org/10.3390/agriculture15171889







