Identification and Comparative Analysis of Genetic Effects of 2Ns Chromosome Introgression from Psathyrostachys huashanica and Leymus mollis into Common Wheat
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Chromosome Preparation and Observation
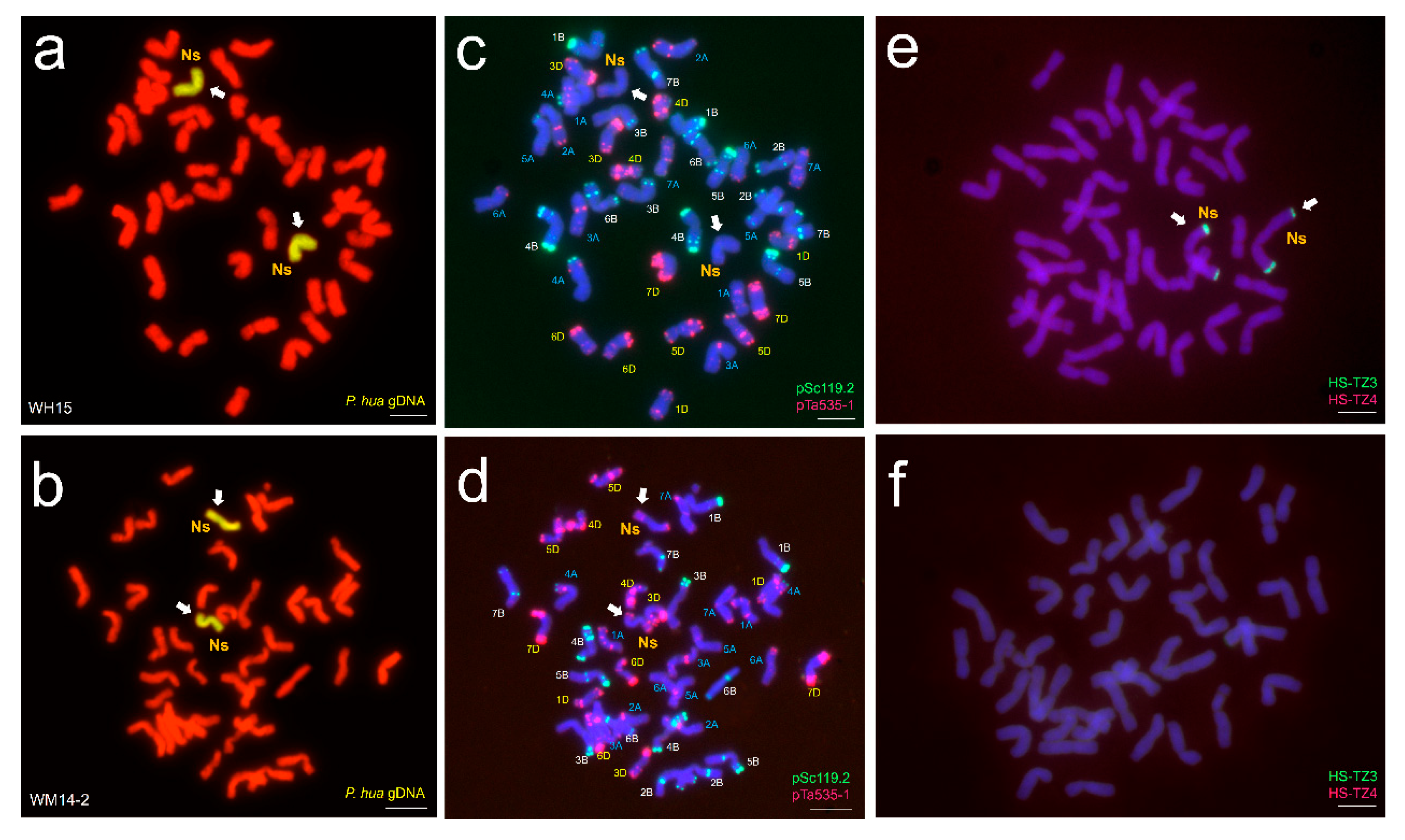
2.3. In Situ Hybridization (FISH/GISH) Analysis
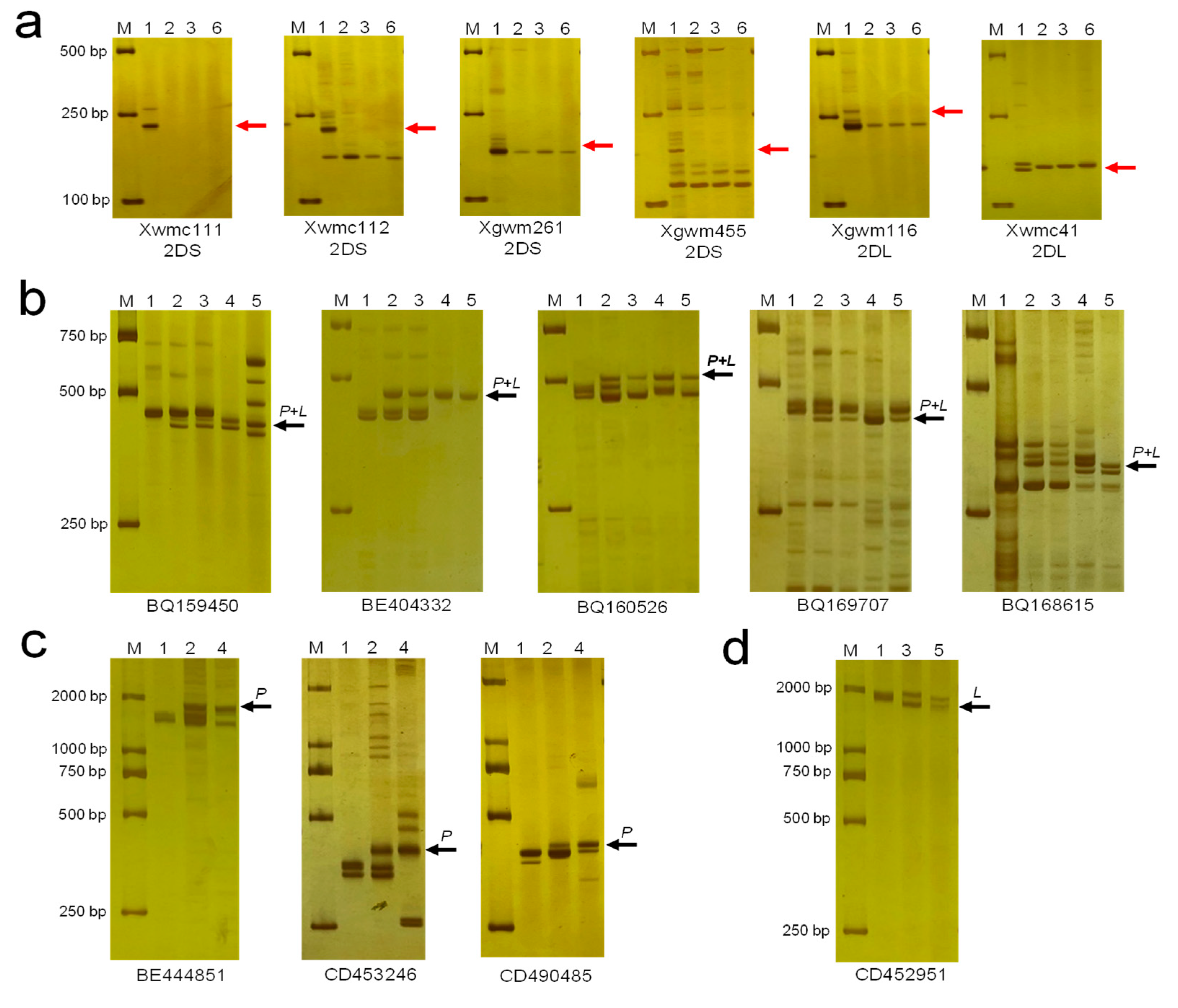
2.4. Molecular Marker Analysis
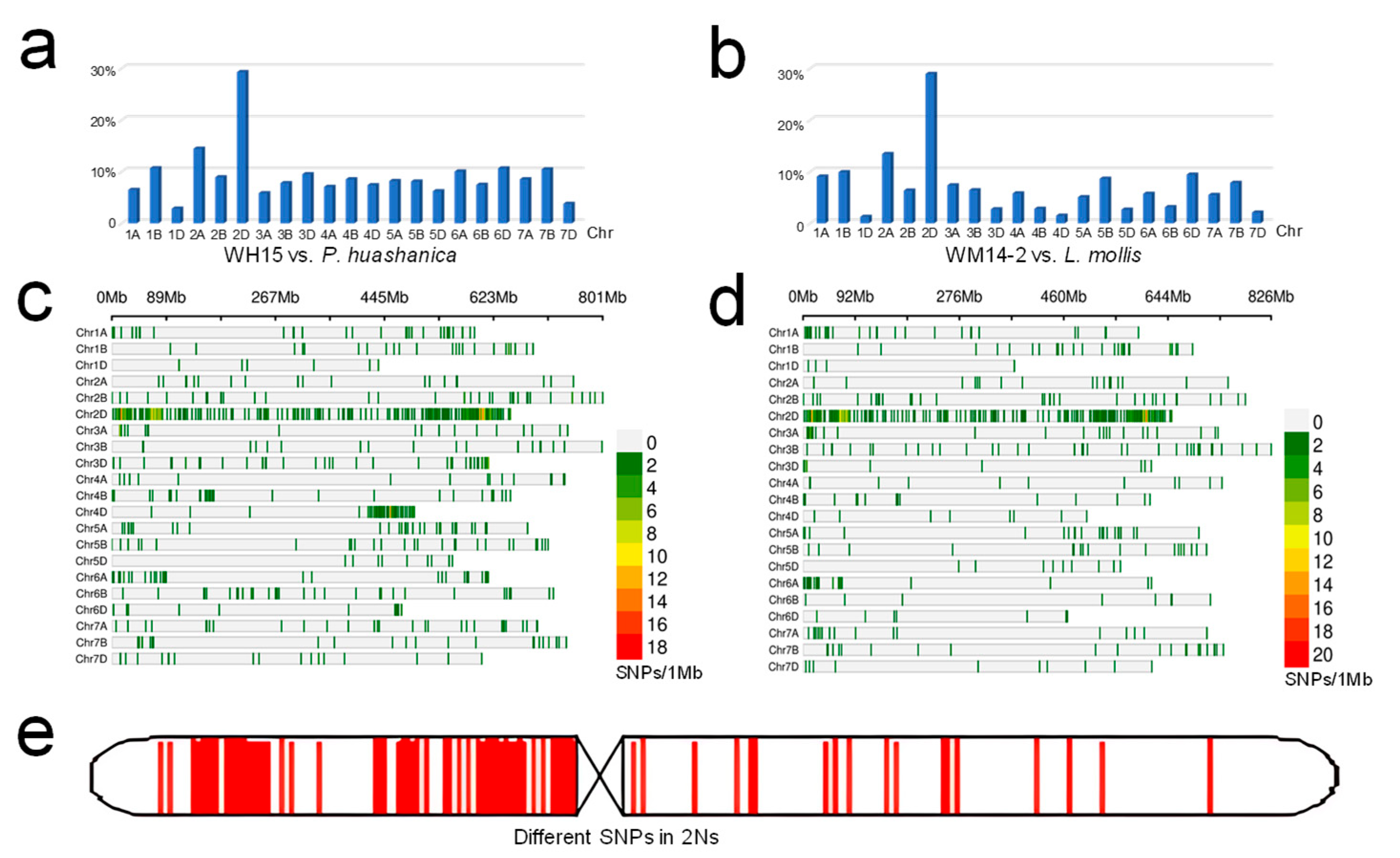
2.5. K SNP Array Analysis
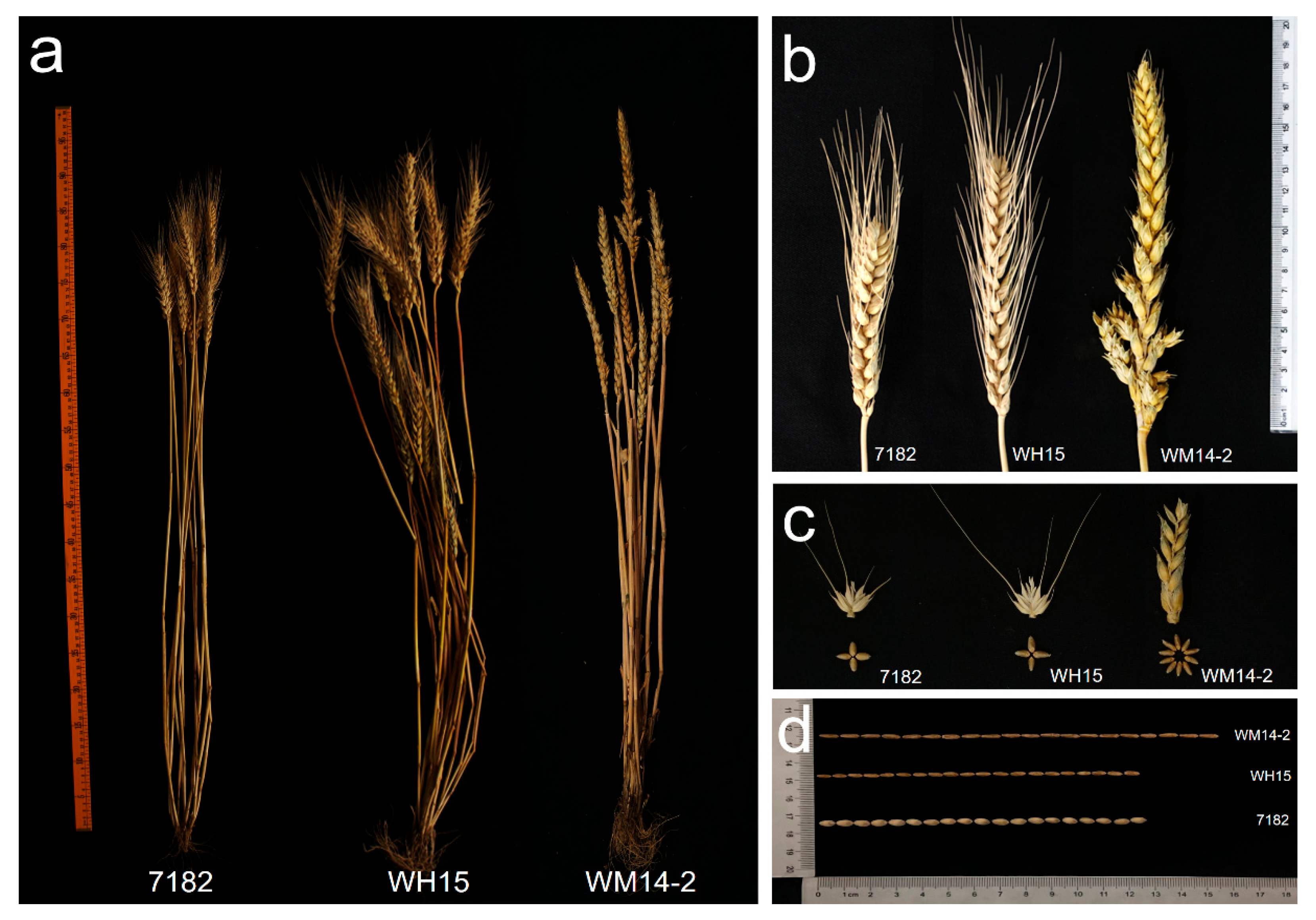
2.6. Agronomic Trait and Disease Resistance Evaluation
2.7. Development of Species-Specific SCAR Markers
3. Results
3.1. In Situ Hybridization
3.2. Molecular Marker Analysis
3.3. K SNP Array Genotyping
3.4. Comparison of Agronomic Traits and Disease Resistance
3.5. Analysis of Genetic Stability
3.6. Development of 2Ns-Specific Molecular Markers
4. Discussion
4.1. Evolutionary Analysis of the Origin of the Ns Genome in Leymus
4.2. Sequence Divergence in Ns Chromosomes Between the Two Genera
4.3. Introducing 2Ns Chromosome-Mediated Trait Improvement in Wheat
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| bp | Base pairs |
| CS | Chinese Spring |
| DAPI | 4′,6-diamidino-2-phenylindole |
| EST-STS | Expressed sequence tag–sequence tagged site |
| FISH | Fluorescence in situ hybridization |
| GISH | Genomic in situ hybridization |
| HR | Highly resistant |
| HS | Highly susceptible |
| IM | Near-immune |
| IT | Immunity type |
| L. mollis | Leymus mollis |
| MR | Moderately resistant |
| MS | Moderately susceptible |
| PCR | Polymerase chain reaction |
| P. huashanica | Psathyrostachys huashanica |
| PI | Propidium iodide |
| PLUG | PCR-based landmark unique gene |
| VHS | Very highly susceptible |
| SCAR | Sequence-characterized amplified region |
| SLAF-seq | Simplified genome sequencing |
| SNP | Single nucleotide polymorphism |
| SSR | Simple sequence repeat |
References
- Xiao, J.; Liu, B.; Yao, Y.Y.; Guo, Z.F.; Jia, H.Y.; Kong, L.R.; Zhang, A.M.; Ma, W.J.; Ni, Z.F.; Xu, S.B.; et al. Wheat genomic study for genetic improvement of traits in China. Sci. China Life Sci. 2022, 65, 1718–1775. [Google Scholar] [CrossRef]
- Tian, X.B.; Wang, Z.Y.; Liu, W.X.; Zhao, Y.S. Harness the wild: Progress and perspectives in wheat genetic improvement. J. Genet. Genom. 2025, in press. [Google Scholar] [CrossRef] [PubMed]
- Han, G.H.; Yan, H.W.; Li, L.H.; An, D.G. Advancing wheat breeding using rye: A key contribution to wheat breeding history. Trends Biotechnol. 2025, in press. [CrossRef] [PubMed]
- Yao, Y.Y.; Guo, W.L.; Gou, J.Y.; Hu, Z.R.; Liu, J.; Ma, J.; Zong, Y.; Xin, M.M.; Chen, W.; Li, Q.; et al. Wheat2035: Integrating pan-omics and advanced biotechnology for future wheat design. Mol. Plant 2025, 18, 272–297. [Google Scholar] [CrossRef] [PubMed]
- Li, H.H.; Men, W.Q.; Ma, C.; Liu, Q.W.; Dong, Z.J.; Tian, X.B.; Wang, C.L.; Liu, C.; Gill, H.S.; Ma, P.T.; et al. Wheat powdery mildew resistance gene Pm13 encodes a mixed lineage kinase domain-like protein. Nat. Commun. 2024, 15, 2449. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.H.; Govta, L.b.; Sung, Y.C.; Coaker, G.; Fahima, T. The spectrum of diverse disease-resistance genes cloned and characterized in the Triticeae Tribe. Annu. Rev. Phytopathol. 2025, in press. [Google Scholar] [CrossRef]
- Singh, R.P.; Hodson, D.P.; Singh, P.K.; Lan, C.; He, X.; Lagudah, E.S.; Juliana, P.; Ayliffe, M.; Bhavani, S.; Saunders, D.G.O.; et al. Challenges to wheat disease resistance and current global strategies. Annu. Rev. Phytopathol. 2025, in press. [Google Scholar] [CrossRef]
- Han, G.H.; Liu, H.; Zhu, S.Y.; Gu, T.T.; Cao, L.J.; Yan, H.W.; Jin, Y.; Wang, J.; Liu, S.Y.; Zhou, Y.L.; et al. Two functional CC-NBS-LRR proteins from rye chromosome 6RS confer differential age-related powdery mildew resistance to wheat. Plant Biotechnol. J. 2024, 22, 66–81. [Google Scholar] [CrossRef]
- Liu, C.; Ran, H.; Xiaolu, W.; Gong, W.P.; Cheng, D.G.; Cao, X.Y.; Liu, A.F.; Li, H.S.; Liu, J.J. Research progress of wheat wild hybridization, disease resistance genes transfer and utilization. Sci. Agric. Sin. 2020, 53, 1287–1308. [Google Scholar]
- Subbarao, G.V.; Kishii, M.; Bozal-Leorri, A.; Ortiz-Monasterio, I.; Gao, X.; Ibba, M.I.; Karwat, H.; Gonzalez-Moro, M.B.; Gonzalez-Murua, C.; Yoshihashi, T.; et al. Enlisting wild grass genes to combat nitrification in wheat farming: A nature-based solution. Proc. Natl. Acad. Sci. 2021, 118, e2106595118. [Google Scholar] [CrossRef]
- Wang, H.W.; Sun, S.L.; Ge, W.Y.; Zhao, L.F.; Kong, L.R. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 2020, 368, eaba5435. [Google Scholar] [CrossRef]
- Li, Y.H.; Tan, B.W.; Yang, J.Y.; Zhang, H.; Zhu, W.; Xu, L.L.; Cheng, Y.R.; Wang, Y.; Zeng, J.; Sha, L.; et al. Integrated review of Psathyrostachy huashanica: From phylogenetic research to wheat breeding application. Mol. Breed. 2025, 45, 42. [Google Scholar] [CrossRef]
- Tan, B.W.; Zhao, L.; Li, L.Y.; Zhang, H.; Zhu, W.; Xu, L.L.; Wang, Y.; Zeng, J.; Fan, X.; Sha, L.N.; et al. Identification of a wheat-Psathyrostachys huashanica 7Ns ditelosomic addition line conferring early maturation by cytological analysis and newly developed molecular and FISH markers. Front. Plant Sci. 2021, 12, 784001. [Google Scholar] [CrossRef]
- Liu, Y.X.; Huang, S.H.; Han, J.; Hou, C.C.; Zheng, D.S.; Zhang, Z.M.; Wu, J. Development and molecular cytogenetic identification of a new wheat–Psathyrostachys huashanica Keng translocation line resistant to powdery mildew. Front. Plant Sci. 2021, 12, 1127. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Tang, S.J.; Li, W.; Zhang, S.B.; Wang, J.L.; Pan, D.F.; Lin, Z.L.; Ma, X.; Chang, Y.N.; Liu, B.; et al. Genome evolution and initial breeding of the Triticeae grass Leymus chinensis dominating the Eurasian Steppe. Proc. Natl. Acad. Sci. 2023, 120, e2308984120. [Google Scholar] [CrossRef]
- Du, X.; Feng, X.B.; Li, R.X.; Jin, Y.L.; Shang, L.H.; Zhao, J.X.; Wang, C.Y.; Li, T.D.; Chen, C.H.; Tian, Z.R.; et al. Cytogenetic identification and molecular marker development of a novel wheat-Leymus mollis 4Ns(4D) alien disomic substitution line with resistance to stripe rust and Fusarium head blight. Front. Plant Sci. 2022, 13, 1012939. [Google Scholar] [CrossRef]
- Dewey, D.R. Genome relations among diploid Elymus junceus and certain tetraploid and octoploid Elymus species. Am. J. Bot. 1970, 57, 633–639. [Google Scholar] [CrossRef]
- Kruppa, K.; Türkösi, E.; Holušová, K.; Kalapos, B.; Szakács, É.; Cséplő, M.; Farkas, A.; Ivanizs, L.; Szőke-Pázsi, K.; Mikó, P.; et al. Genotyping-by-sequencing uncovers a Thinopyrum 4StS·1JvsS Robertsonian translocation linked to multiple stress tolerances in bread wheat. Theor. Appl. Genet. 2024, 138, 13. [Google Scholar] [CrossRef]
- Dolezel, J.; Lucretti, S.; Molnár, I.; Cápal, P.; Giorgi, D. Chromosome analysis and sorting. Cytom. Part A 2021, 99, 328–342. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.E.A.H.P.; Rodrigues, M.S.; Siqueira, W.J.; Marques, M.O.M.; Mondego, J.M.C. Comparative analysis indicates a simple protocol for DNA extraction of the aromatic plant Lippia alba. Anal. Biochem. 2023, 675, 115225. [Google Scholar] [CrossRef]
- Kuang, Z.Y.; Ji, X.J.; Xu, S.R.; Han, H.M.; Zhang, J.P.; Zhou, S.H.; Yang, X.M.; Li, X.Q.; Li, L.H.; Liu, W.H. Generation and identification of a wheat–Agropyron cristatum (L.) Gaertn. 3P chromosome addition line and substitution line. Euphytica 2023, 219, 23. [Google Scholar] [CrossRef]
- Li, J.C.; Li, J.J.; Zhao, L.; Zhao, J.X.; Wu, J.; Chen, X.H.; Zhang, L.Y.; Dong, P.H.; Wang, L.M.; Zhao, D.H.; et al. Rapid identification of Psathyrostachys huashanica Keng chromosomes in wheat background based on ND-FISH and SNP array methods. J. Integr. Agric. 2023, 22, 2–16. [Google Scholar] [CrossRef]
- Tang, Z.X.; Yang, Z.J.; Fu, S.L. Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa–535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genet. 2014, 55, 313–318. [Google Scholar] [CrossRef]
- Ren, P.X.; Zhao, D.H.; Zeng, Z.K.; Yan, X.F.; Zhao, Y.; Lan, C.X.; Wang, C.P. Pleiotropic effect analysis and marker development for grain zinc and iron concentrations in spring wheat. Mol. Breed. 2022, 42, 49. [Google Scholar] [CrossRef]
- An, D.G.; Ma, P.T.; Zheng, Q.; Fu, S.L.; Li, L.H.; Han, F.P.; Han, G.H.; Wang, J.; Xu, Y.F.; Jin, Y.L.; et al. Development and molecular cytogenetic identification of a new wheat-rye 4R chromosome disomic addition line with resistances to powdery mildew, stripe rust and sharp eyespot. Theor. Appl. Genet. 2019, 132, 257–272. [Google Scholar] [CrossRef] [PubMed]
- Giancaspro, A.; Lionetti, V.; Giove, S.L.; Zito, D.; Fabri, E.; Reem, N.; Zabotina, O.A.; De Angelis, E.; Monaci, L.; Bellincampi, D.; et al. Cell wall features transferred from common into durum wheat to improve Fusarium Head Blight resistance. Plant Sci. 2018, 274, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Wood, R.; Liu, A.G.; Bowyer, F.; Wilby, P.R.; Dunn, F.S.; Kenchington, C.G.; Cuthill, J.F.H.; Mitchell, E.G.; Penny, A. Integrated records of environmental change and evolution challenge the Cambrian Explosion. Nat. Ecol. Evol. 2019, 3, 528–538. [Google Scholar] [CrossRef] [PubMed]
- McColgan, Á.; DiFrisco, J. Understanding developmental system drift. Development 2024, 151, 203054. [Google Scholar] [CrossRef]
- Soucy, S.M.; Huang, J.L.; Gogarten, J.P. Horizontal gene transfer: Building the web of life. Nat. Rev. Genet. 2015, 16, 472–482. [Google Scholar] [CrossRef]
- Guan, J.T.; Garcia, D.F.; Zhou, Y.; Appels, R.; Li, A.L.; Mao, L. The battle to sequence the bread wheat genome: A tale of the three kingdoms. Genom. Proteom. Bioinf. 2020, 18, 221–229. [Google Scholar] [CrossRef]
- Zhao, L.B.; Xie, D.; Huang, L.; Zhang, S.J.; Luo, J.T.; Jiang, B.; Ning, S.Z.; Zhang, L.Q.; Yuan, Z.W.; Wang, J.R.; et al. Integrating the physical and genetic map of bread wheat facilitates the detection of chromosomal rearrangements. J. Integr. Agric. 2021, 20, 2333–2342. [Google Scholar] [CrossRef]
- Farooq, M.; Frei, M.; Zeibig, F.; Pantha, S.; Özkan, H.; Kilian, B.; Siddique, K.H.M. Back into the wild: Harnessing the power of wheat wild relatives for future crop and food security. J. Exp. Bot. 2025, in press. [Google Scholar] [CrossRef]
- Bödvarsdóttir, S.K.; Anamthawat-Jónsson, K. Isolation, characterization, and analysis of Leymus-specific DNA sequences. Genome 2003, 46, 673–682. [Google Scholar] [CrossRef]
- Wang, R.R.C.; Zhang, J.Y.; Lee, B.S.; Jensen, K.B.; Kishii, M.; Tsujimoto, H. Variations in abundance of 2 repetitive sequences in Leymus and Psathyrostachys species. Genome 2006, 49, 511–519. [Google Scholar] [CrossRef]
- Orgaard, M.; Heslop-Harrison, J.S. Relationships between species of Leymus, Psathyrostachys, and Hordeum (Poaceae, Triticeae) inferred from Southern hybridization of genomic and cloned DNA probes. Plant Syst. Evol. 1994, 189, 217–231. [Google Scholar] [CrossRef]
- Xiao, K.Z.; Sun, Y.N.; Zhou, S.J. Revealing the abnormal meiosis and the variation of the functional female gametes of aneuploid lily (Lilium) using genomic in situ hybridization (GISH). Euphytica 2023, 219, 108. [Google Scholar] [CrossRef]
- Han, G.H.; Li, H.W.; Cao, L.J.; Liu, S.Y.; Yan, H.W.; Wang, J.; Zhou, Y.L.; An, D.G. A novel wheat-rye 2R (2D) disomic substitution line pyramids two types of resistance to powdery mildew. Plant Dis. 2022, 106, 2433–2440. [Google Scholar] [CrossRef]
- Wang, J.; Liu, C.; Guo, X.R.; Wang, K.; Du, L.P.; Lin, Z.S.; Ye, X.G. Development and genetic analysis of wheat double substitution lines carrying Hordeum vulgare 2H and Thinopyrum intermedium 2Ai#2 chromosomes. Crop J. 2019, 7, 163–175. [Google Scholar]
- Song, Z.P.; Dai, S.F.; Bao, T.Y.; Zuo, Y.Y.; Yan, Z.H. Analysis of structural genomic diversity in Aegilops umbellulata, Ae. markgrafii, Ae. comosa, and Ae. uniaristata by fluorescence in situ hybridization karyotyping. Front. Plant Sci. 2020, 11, 710. [Google Scholar] [CrossRef]
- Zhu, S.Y.; Du, H.N.; Su, F.Y.; Wang, J.; Meng, Q.F.; Liu, T.L.; Guo, R.; Chen, Z.Z.; Li, H.H.; Liu, W.X.; et al. Molecular cytogenetic analyses of two new wheat-rye 6RL translocation lines with resistance to wheat powdery mildew. Crop J. 2023, 11, 584–592. [Google Scholar] [CrossRef]
- Fan, W.; Sun, M.Q.; Zheng, Y.B.; Song, S.W.; Zhang, Z.Y.; Bian, Y. Karyotypic and phenotypic condensation in allotetraploid wheats accompanied with reproductive strategy transformation: From natural evolution to domestication. Planta 2024, 260, 83. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.W.; Wang, Z.; Heng, Y.F.; Chen, Z.; Wang, K.; Xiao, Q.M.; Li, J.; Hu, Z.R.; He, H.; Cao, Y.; et al. Selection of dysfunctional alleles of bHLH1 and MYB1 has produced white grain in the tribe Triticeae. Plant Commun. 2025, 6, 101265. [Google Scholar] [CrossRef]
- Guo, L.; Yu, L.W.; Tong, J.Y.; Zhao, Y.Y.; Yang, Y.; Ma, Y.R.; Cui, L.; Hu, Y.G.; Wang, Z.H.; Gao, X. Addition of Aegilops geniculata 1Ug chromosome improves the dough rheological properties by changing the composition and micro-structure of gluten. Food Chem. 2021, 358, 129850. [Google Scholar] [CrossRef]
- Wan, W.T.; Zhao, R.H.; Chen, T.T.; Wang, L.; Zhang, X.; Li, H.F.; Wang, X.E.; Bie, T.D. Rapid development of wheat-Dasypyrum villosum compensating translocations resistant to powdery mildew using a triple marker strategy conducted on a large ph1b-induced population. Theor. Appl. Genet. 2023, 136, 148. [Google Scholar] [CrossRef] [PubMed]
- Beying, N.; Schmidt, C.; Pacher, M.; Houben, A.; Puchta, H. CRISPR–Cas9-mediated induction of heritable chromosomal translocations in Arabidopsis. Nat. Plants 2020, 6, 638–645. [Google Scholar] [CrossRef] [PubMed]

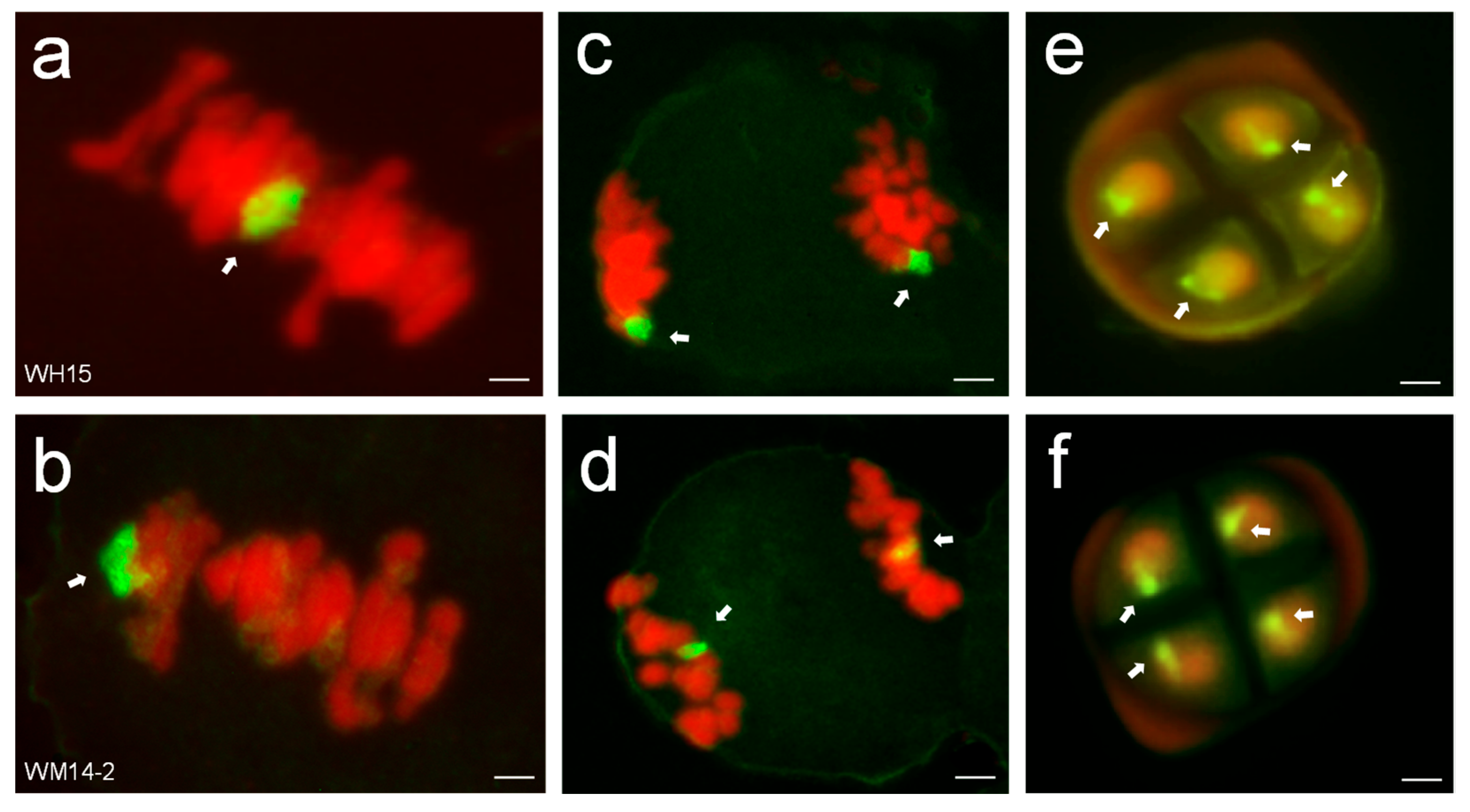
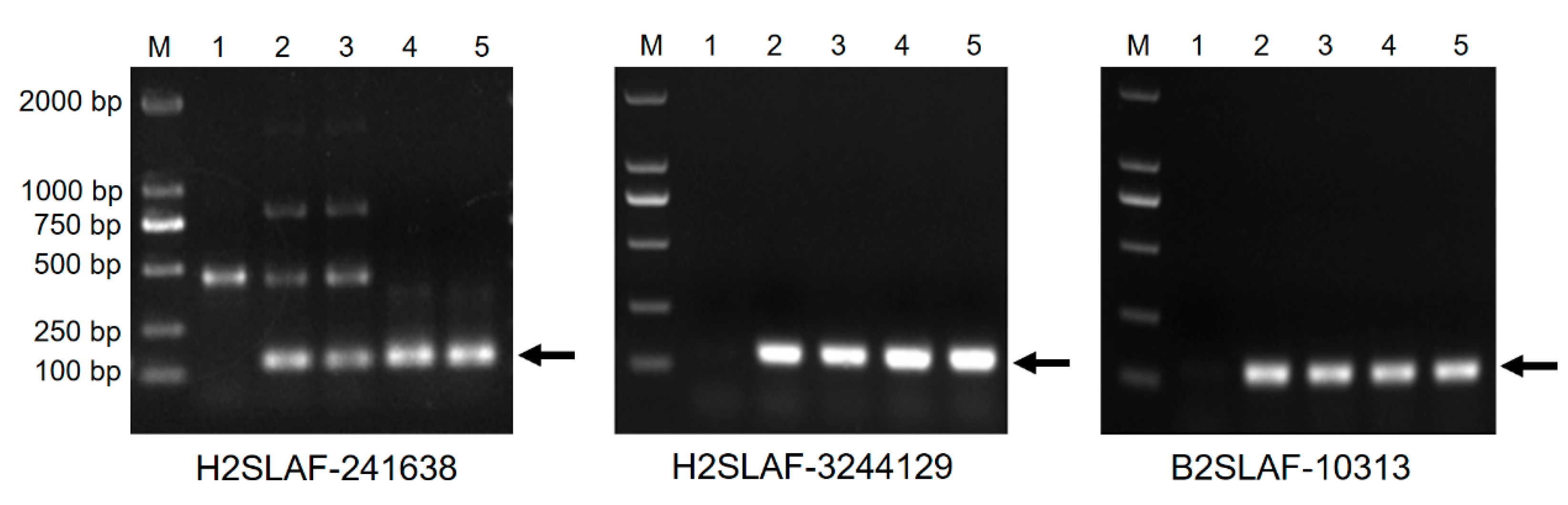
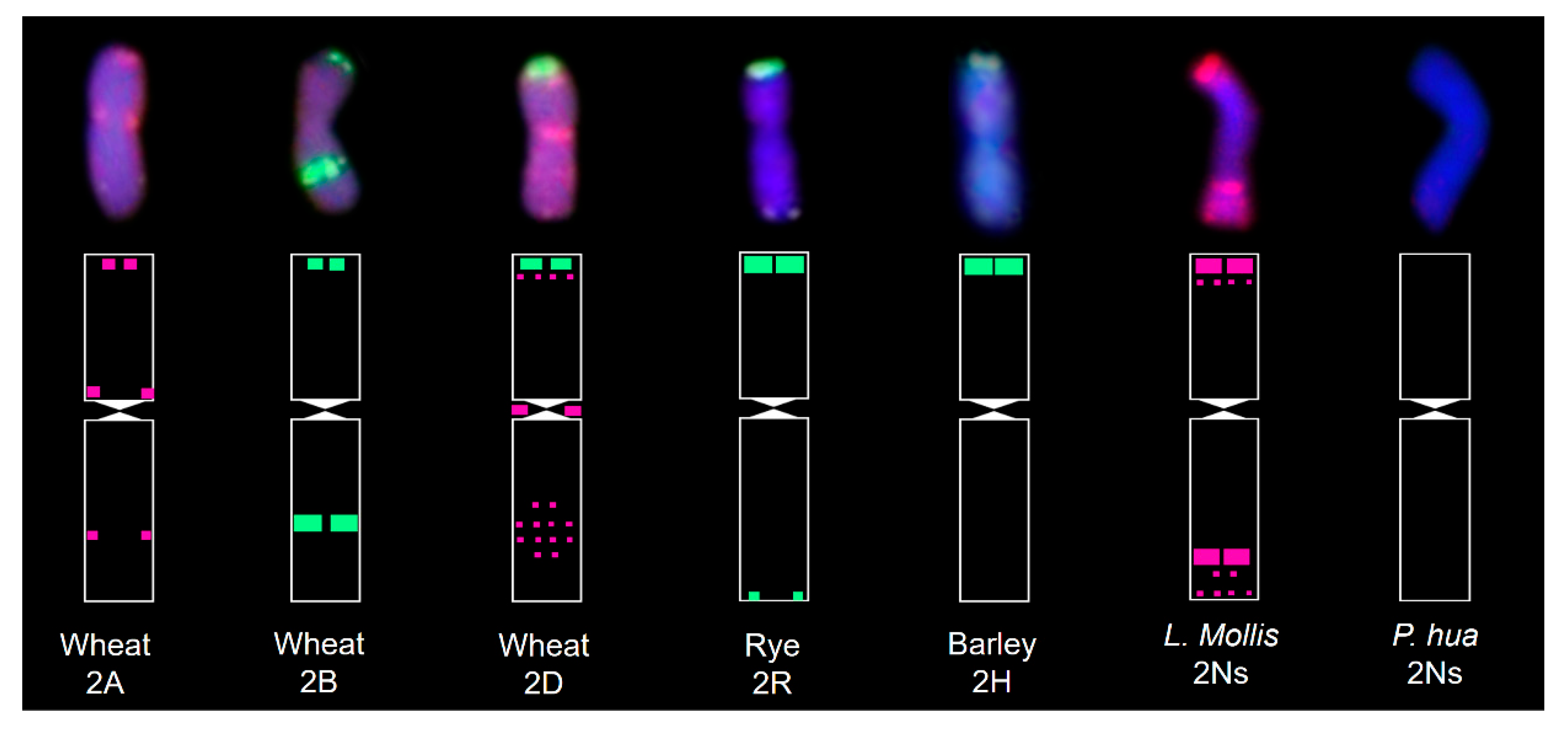
| Material | Plant Height (cm) | Tiller Number | Spike Length (cm) | Spikelets Per Spike | Kernels Per Spike | Thousand Kernel Weight (g) |
|---|---|---|---|---|---|---|
| 7182 | 75.6 ± 4.3a | 11 ± 2b | 9.04 ± 0.64b | 18 ± 2b | 48 ± 4c | 30.71 ± 0.13b |
| WH15 | 77.5 ± 3.1a | 15 ± 2a | 13.5 ± 0.47a | 24 ± 2b | 55 ± 6b | 30.33 ± 0.14b |
| WM14-2 | 80.2 ± 5.2a | 10 ± 2b | 18.46 ± 1.41a | 28 ± 3a | 68 ± 7a | 36.79 ± 0.44a |
| Materials | Protein Content (%) | Crude Protein Content (%) | Gluten Protein Content (%) | Starch Content (%) | Subsidence Value | Dough Stability Time (min) |
|---|---|---|---|---|---|---|
| 7182 | 15.14 ± 0.36b | 61.22 ± 0.27a | 33.01 ± 0.24b | 74.43 ± 0.85a | 40.35 ± 0.64a | 3.77 ± 0.44a |
| WH15 | 14.35 ± 0.42b | 60.02 ± 0.23a | 33.92 ± 0.34b | 73.48 ± 0.64a | 38.44 ± 0.53a | 4.23 ± 0.63a |
| WM14-2 | 17.51 ± 0.25a | 58.38 ± 0.31a | 36.24 ± 0.47a | 72.78 ± 0.83a | 42.03 ± 0.72a | 5.25 ± 0.25a |
| Material | Cu (mg/kg) | Fe (mg/kg) | K (mg/kg) | Mg (mg/kg) | Mn (mg/kg) | P (mg/kg) | Zn (mg/kg) |
|---|---|---|---|---|---|---|---|
| 7182 | 4.45 ± 0.18b | 37.32 ± 0.82b | 4163.80 ± 5.33c | 1806.70 ± 7.24c | 39.28 ± 0.83b | 3078.80 ± 6.86c | 20.04 ± 1.21c |
| WH15 | 7.90 ± 0.23a | 79.46 ± 0.43a | 8499.05 ± 7.64a | 2752.67 ± 7.54a | 63.81 ± 1.45a | 5500.63 ± 9.23a | 50.63 ± 0.88a |
| WM14-2 | 6.41 ± 0.12a | 82.42 ± 0.56a | 6111.69 ± 6.42b | 2214.00 ± 8.23b | 63.05 ± 1.21a | 4417.08 ± 7.87b | 44.97 ± 1.32b |
| Marker | Primer Sequence (5′–3′) | Tm (°C) | Location |
|---|---|---|---|
| H2SLAF241638 | CCATCTGGCATCTGTGATGTACT GTGCTCGGATGAACGCGC | 60 | 2Ns |
| H2SLAF3244129 | CCGAAGATGAATACGAGTATCGG AGCTTCAGCTGTCTCGGACATG | 60 | 2NsS |
| B2SLAF10313 | CCCGAAGTAATTTGGTCCACC GCCACCCTCAGCTCGATATC | 60 | 2Ns |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pang, Y.; Li, J.; Huo, W.; Hua, X.; Yuan, J.; Tang, X.; Yang, H.; Jia, C.; Li, J.; Zhao, J. Identification and Comparative Analysis of Genetic Effects of 2Ns Chromosome Introgression from Psathyrostachys huashanica and Leymus mollis into Common Wheat. Agriculture 2025, 15, 1887. https://doi.org/10.3390/agriculture15171887
Pang Y, Li J, Huo W, Hua X, Yuan J, Tang X, Yang H, Jia C, Li J, Zhao J. Identification and Comparative Analysis of Genetic Effects of 2Ns Chromosome Introgression from Psathyrostachys huashanica and Leymus mollis into Common Wheat. Agriculture. 2025; 15(17):1887. https://doi.org/10.3390/agriculture15171887
Chicago/Turabian StylePang, Yuhui, Jiaojiao Li, Wenjie Huo, Xueyou Hua, Jiayi Yuan, Xicheng Tang, Huanhuan Yang, Chongyang Jia, Jiachuang Li, and Jixin Zhao. 2025. "Identification and Comparative Analysis of Genetic Effects of 2Ns Chromosome Introgression from Psathyrostachys huashanica and Leymus mollis into Common Wheat" Agriculture 15, no. 17: 1887. https://doi.org/10.3390/agriculture15171887
APA StylePang, Y., Li, J., Huo, W., Hua, X., Yuan, J., Tang, X., Yang, H., Jia, C., Li, J., & Zhao, J. (2025). Identification and Comparative Analysis of Genetic Effects of 2Ns Chromosome Introgression from Psathyrostachys huashanica and Leymus mollis into Common Wheat. Agriculture, 15(17), 1887. https://doi.org/10.3390/agriculture15171887




