Abstract
Polyploidy in plants can enhance stress resistance and secondary metabolite production, offering potential benefits for Clausena lansium (L.) Skeel, a medicinally valuable species. However, systematic studies of polyploidy-induced morphological, anatomical, and metabolic changes in this species are lacking. This study aimed to induce and characterize polyploid C. lansium lines, assess ploidy-dependent variations, and evaluate their impact on bioactive metabolite accumulation. Three cultivars were hybridized, treated with colchicine, and bred, yielding 13 stable polyploid lines confirmed by flow cytometry and chromosome counting. The polyploids exhibited distinct traits, including larger pollen grains, altered leaf margins, increased leaflet numbers, enlarged guard cells with reduced stomatal density, and thicker leaf tissues. Metabolomic analysis revealed that tetraploids accumulated significantly higher levels of flavonoids, alkaloids, and phenolic acids compared to diploids, while triploids showed moderate increases. These findings demonstrate that polyploidization, particularly tetraploidy, enhances C. lansium’s medicinal potential by boosting pharmacologically active compounds. The study expands germplasm resources and supports the development of high-quality cultivars for pharmaceutical applications.
1. Introduction
Clausena lansium (Lour.) Skeels, also known as wampee or wampi, is an evergreen subtropical small tree that belongs to the Rutaceae family. Wampee is a fruit crop and traditional medicinal plant that is native to southern China, Thailand, and Vietnam. It is usually distributed in South China and Southeast Asia. C. lansium receives large amounts of popularity among customers for its nutritional value and pharmacological uses. Wampee fruit ripens from May to July in southern China; it tastes sweet or slightly acidic; it is similar to grapefruit when ripe, resembles a diminutive lemon, and is about 2.0–4.0 cm in diameter with 0–5 seeds [1]. Almost the whole plant of C. lansium contains significant amounts of nutrients and bioactive substances, including its leaves, stems, roots, and fruits, which are applied as the traditional Chinese medicine for various diseases, such as cough, allergies, asthma, etc. [2].
Recent studies also found its various uses: The geranylated carbazole alkaloids extracted from C. lansium are good for Parkinson’s disease [3]; amides isolated from C. lansium seeds possessed the potential to be exploited as botanical fungicides for commercial application [4]; the volatile compound (E)-2-hexenal extracted from C. lansium enhances the Penicillium italicum resistance of postharvest citrus fruit [5]; etc. Recently, the reference genome of C. lansium was constructed which would facilitate the identification of some useful medicinal compounds from wampee resources and reveal their biosynthetic pathways [6]. However, the germplasm of C. lansium is extremely limited, and creating new germplasm is of great significance for the breeding of new cultivars.
Polyploidy is the genome that has more than two full sets of chromosomes, which would cause the genome multiplication and an increase in gene content [7]. Polyploidization is a very common phenomenon in angiosperms plants [8,9]. Some of the obvious advantages of plant polyploidy breeding include the increment in plant organs (“gigas” effect) with higher yield levels, the improvement in the product quality, and the enhancement of both biotic and abiotic stresses [10,11,12]. Moreover, polyploidy often caused fertility failure by meiotic errors and produced seedless varieties [11]. Polyploidy is of great significance in the production practice and theoretical research of variety breeding, genetic diversity, genomics, and mutants [13,14,15]. There are several ways to induce polyploid seedlings, such as hybridization and chemical induction. Among them, chemicals are the most commonly used and effective approach; colchicine, oryzalin, and trifluralin are all applied to induce polyploids, especially colchicine. There are lots of colchicine-induced polyploids reported, such as watermelon [16], pear [17], citrus [18], and so on.
Most of the C. lansium is diploidy, as a medical uses plant; in this study, the polyploidy plants of C. lansium were induced in several ways (seedlings, hybridization, and colchicine-induced). To date, unlike other medically important tree species, polyploid breeding in C. lansium remains unexplored, with no reported studies on its polyploid induction or characterization. To find out the polyploidization effect of C. lansium, morphological and anatomical structure observation, physiological analysis, and metabolomics analysis, were carried out. The relation between the effective substances content and the polyploidy was also revealed. These results improve our understanding of C. lansium’s effective substance synthesis mechanism and provide useful information for enhancing the medicinal substances in C. lansium. This study of the polyploidization of C. lansium is not only beneficial for the enrichment of the germplasm resources; it is also good for the improvement of the extraction of medicinal substances.
2. Material and Methods
2.1. Materials
Three cultivars of C. lansium were used in this research (Figure 1); they were all planted in the orchard of South China Agricultural University:
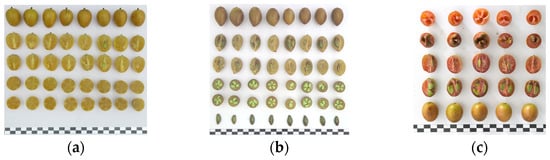
Figure 1.
Three cultivars used in this study. Note: (a) ‘Wuhe’ wampee; (b) ‘Huami No. 3’ wampee; (c) ‘Zirou’ wampee. Bar = 1 cm.
- ‘Wuhe’ wampee (WH): A seedless cultivar selected from Yunan country, Yunfu city, of Guangdong Province in China. The fruits are oval-cordate in shape with brownish skin at a mature stage. It tastes sour and sweet with a 95% seedless rate; it has been reported as a mixoploid [19].
- ‘Huami No. 3’ wampee (HM3): A cultivar developed by the College of Horticulture at South China Agricultural University and Guangzhou Institute of Fruit Tree Science from a cross between ‘Baitang’ × ‘Jinjixin’. Notable features include excellent flavor, superior fruit quality, and early and abundant fruiting, as well as strong stress resistance; diploidy.
- ‘Zirou’ wampee (ZR): This cultivar is found in Guanxi Province, the fruit has distinctive pale purple skin and flesh; it has nearly round-shaped fruits; diploidy. It is reported to have a high content of anthocyanins [20].
2.2. The Induction of Polyploid C. lansium Germplasm
2.2.1. Hybridization of ‘ZR × WH’ to Create Polyploid Plants
Pollen Collection and Observation of WH
Fresh flower buds of WH were collected before blooming. The anthers were peeled off in a culture dish and put in a 30 °C oven (Shanghai Yiheng, DHG-9070A, Shanghai, China) and dried for 12–24 h. Then they were gently stacked in a 200-mesh stainless steel sieve to release the pollen. The pollen was loaded into a 1.5 mL centrifuge tube with silicone beads, sealed with sealing film, and stored at −80 °C (Thermo Fisher Scientific Inc., 88600 V, CA, USA) for later use.
A small amount of pollen sample was dipped with a toothpick onto a conductive carbon film with double-sided adhesive and placed on an ion sputtering instrument (Hitachi, Ltd., MC1000, Tokyo, Japan) sample stage for about 30 s to spray gold. A high-power scanning electron microscope (Zeiss Group, EVO MA15, Oberkochen, Germany) was used to observe the morphology and surface structure of the pollen grains. The pollen of ZR and HM3 was taken as the CK (control check).
Hybridization Between ZR and WH
On the afternoon of the day before pollination, before the flowering of ZR (when the flowers were about to bloom before the pollen matured), the stamens were removed and bagged. The next day, in the morning from 7 to 9 o’clock, we removed the bag, brushed the WH pollen on the ZR style, then bagged and marked it.
SCoT (Start Codon Targeted Polymorphism) Marker Analysis for Hybrid Identification
SCoT is a PCR-based molecular marker technique that uses primers targeting the conserved region flanking the start codon (ATG) in plant genes. This method generates reproducible, dominant markers suitable for genetic diversity analysis, cultivar identification, and hybrid verification [21].
We referred to the SCoT reaction system and procedure proposed by Nugroho [22]. SCoT primers (Table 1) [21] were synthesized by Beijing Qingke Biotechnology Co., Ltd. (Guangzhou, China).

Table 1.
Five SCoT primer sequences [21].
2.2.2. Induction Polyploid Plants of HM3 by Colchicine Treatment
To induce polyploidy in HM3, mature seeds were surface-sterilized (thoroughly washed to remove surface mucilage) and air-dried at 25 °C. Then the seeds were subjected to colchicine treatments in dark conditions at concentrations of 0.1%, 0.2%, and 0.3% for durations of 12, 24, 48, and 72 h, respectively, with water-treated seeds serving as controls (CKs). Each treatment consisted of 30 seeds with three biological replicates. Following treatment, seeds were sown in trays under controlled conditions, with root emergence monitored at 7 days after sowing (DAS) and germination rates recorded at 10-day intervals until stabilization.
Germination rate (%) = number of seedlings emergence/number of
seeds sown × 100%
seeds sown × 100%
2.2.3. Screen Polyploid Plants of the Seedlings of WH
As a mixoploid, WH contains both diploid and non-diploid (e.g., tetraploid) cell populations, enabling its seedling progeny to exhibit ploidy segregation.
In this study, we took the seeds of the WH mature fruit and cleaned and dried them at 25 °C. We sowed the seeds separately in the tray (peat soil to perlite = 3:1), and observations of the plantlet were carried out after the seedlings had grown.
The polyploid seedling of the WH genotype was selected for downstream analysis, due to its genetic uniformity, which provided a consistent genetic background.
2.3. Identification of Polyploid Plants
2.3.1. Identification by Flow Cytometry
We referred to the flow cytometry method which was introduced by Weaver [23]. Fresh young leaf tissues (0.5 cm2) were excised and placed in sterile dishes containing 1 mL of LB01 nuclear extraction buffer. The leaves were rapidly chopped and allowed to stand for 1 min to ensure complete nuclear release. The resulting suspension was then pipetted and filtered through a 30 μm cell strainer. Subsequently, 10 μL of 10 mg/mL propidium iodide (PI) staining solution was added and thoroughly mixed. The samples were incubated in darkness at room temperature for 30 m for nuclear staining. Ploidy analysis was performed using a flow cytometer (BD Biosciences, FACSMelody, San Jose, CA, USA) equipped with 488 nm and 633 nm excitation lasers, with diploid plants serving as control materials.
2.3.2. Chromosome Counting Analysis
The chromosome preparation protocol involved collecting fresh apical leaf tissues (avoiding midveins) between 9:00 and 10:00 AM, followed by 3 h pretreatment in 8-hydroxyquinoline at room temperature. After fixation in methanol/glacial acetic acid (3:1) for 24 h, the samples were washed with distilled water and subjected to 20 m hypotonic treatment. Enzymatic digestion was performed using a 1:1 mixture of 0.6% cellulase and 0.25% pectinase (Macklin, P816736-10g, Shanghai, China) at 37 °C for 3 h, followed by additional hypotonic treatment (10 m) and fixation (1 h). Chromosome spreads were prepared by dropping the fixed material onto pre-chilled slides, dispersing cells by gentle tapping, staining with Giemsa for 30 m, and air-drying before observation under 100× magnification using oil immersion microscopy.
2.4. The Morphological Observation of the Polyploid Plants
Using the diploid seedling of WH as the CK, the morphological characters and leaf anatomic structure of the polyploid plantlets selected from WH were analyzed. The plantlet height, the leaf morphology, the stomata, the thickness of the upper and lower epidermis of the leaves, and the palisade mesophyll were evaluated.
2.5. Stomata and Anatomical Observation of Polyploid Leaves
2.5.1. Stomata Observation and Measurement
Stomatal characteristics were examined using the nail polish imprint technique [24]. Briefly, a thin layer of clear nail polish was applied to the abaxial leaf surface, allowed to dry, and then gently peeled off using adhesive tape. The resulting impressions were mounted on glass slides and observed under a light microscope. Stomata density (number/mm2) = Stomata number in each view field/area of view field (mm2).
2.5.2. Observation of Leaf Anatomical Structure
Leaf segments adjacent to the midrib were excised and fixed in FAA solution. Following conventional paraffin embedding and sectioning, the samples were double-stained with safranin and fast green to prepare permanent slides. Cross-sectional structures were examined under an optical microscope, with images captured at 100× and 200× magnification for target regions. Using ImageJ (v2024) software, quantitative measurements were performed to determine the dimensions, including the thickness of lower/upper epidermis, palisade mesophyll, and spongy mesophyll.
2.6. Determination of Physiological Index
2.6.1. Photosynthetic Parameter Measurement
Photosynthetic indices were measured using a portable photosynthesis system (PP Systems, CIRAS-3, Amesbury, MA, USA) under consistent environmental conditions, with diploid plants serving as controls. Between 8:00 and 11:00 a.m., four–five fully expanded, healthy leaves per plant were selected for analysis. Key parameters recorded included: net photosynthetic rate (Pn), stomatal conductivity (Gs), intercellular CO2 concentration (Ci), and transpiration rate (E).
Determination of chlorophyll content: 0.1 g of leaf powder was weighed and transferred into a centrifuge tube containing 5 mL of absolute ethanol. The mixture was extracted in darkness using an orbital shaker until the leaf powder turned white. Absorbance was measured at 663 nm and 645 nm using a microplate reader (BioTek Cytation5, Winooski, VT, USA). Three technical replicates were performed per sample, and pigment concentrations were calculated using the following equations:
Chlorophyll a (mg/g) = 12.72 × OD 663 − 2.59 × OD645
Chlorophyll b (mg/g) = 22.28 × OD645 − 4.67 × OD663
Total chlorophyll (mg/g) = Chlorophyll a + Chlorophyll b
OD663 and OD645 represented the optical density values at wavelengths of 663 nm and 645 nm, respectively.
2.6.2. Flavonoids
Total flavonoid content was quantified using a commercial assay kit (LHT-1-G, Suzhou Comin Biotechnology Co., Ltd., Suzhou, China) following the manufacturer’s protocol; the distilled water was used as a control. In an alkaline nitrite solution, flavonoids form red complexes with aluminum ions exhibiting a characteristic absorption peak at 502 nm. The flavonoid content of sample extracts can be quantified by measuring absorbance at this specific wavelength.
2.7. Statistical Analysis of Physiological and Ploidy Analysis Data
Data were subjected to one-way analysis of variance (ANOVA) followed by Duncan’s multiple range test (p < 0.05) using SPSS 25.0 (IBM, USA).
2.8. Metabolomic Profiling and Statistical Analysis of Polyploid C. lansium
Metabolomic profiling was conducted on leaves of diploid and polyploid C. lansium (obtained from WH seedings). Fresh leaf samples were first freeze-dried using a Scientz-100F lyophilizer (Ningbo Scientz Biotechnology Co., Ltd., Ningbo, China), then homogenized into fine powder with a Retsch MM 400 ball mill (Retsch GmbH, Haan, Germany; 30 Hz, 1.5 min). For metabolite extraction, 50 mg of powder (weighed using Mettler Toledo MS105DΜ, Mettler Toledo, Columbus, OH, USA) was mixed with 1200 μL of −20 °C pre-cooled 70% methanolic aqueous solution containing internal standards. The mixture underwent intermittent vortexing (30 s every 30 min, 6 times) followed by centrifugation (12,000 rpm, 3 min), with the resulting supernatant filtered through 0.22 μm microporous membranes.
LC-MS/MS analysis was performed using an ExionLC™ AD UPLC system (SCIEX, Marlborough, MA, USA) (https://sciex.com.cn/, accessed on 22 December 2023) coupled with a tandem mass spectrometer (SCIEX, Marlborough, MA, USA), with chromatographic and mass spectrometric parameters set according to established methods [6,25]. Mass spectrometry data were analyzed using Analyst 1.6.3 software. Multivariate statistical analysis included principal component analysis (PCA) using R (base package, v4.1.2) and hierarchical clustering with heatmap visualization (ComplexHeatmap package, v2.9.4) to evaluate metabolic variations. Differential metabolites were identified based on Pearson correlation coefficients (R (base package, v4.1.2)), with subsequent KEGG pathway annotation and enrichment analysis performed following Sun [17].
Principles of Metabolite Identification and Quantification
Metabolite identification was performed using an in-house database (MWDB, MetWare Database) based on secondary mass spectrometry (MS/MS) data. During analysis, isotopic signals, redundant adducts (e.g., K+, Na+, ), and fragment ions derived from higher molecular weight compounds were excluded to ensure accurate annotation.
For metabolite quantification, multiple reaction monitoring (MRM) mode on a triple quadrupole mass spectrometer was employed. The MRM analysis is shown in Figure 2. The first quadrupole filters the target precursor ion (parent ion), eliminating interference from other ions with similar mass-to-charge ratios. The selected precursor ion undergoes collision-induced dissociation (CID), generating fragment ions. The third quadrupole then isolates a specific product ion, further reducing background noise and enhancing quantification accuracy and reproducibility. Following MS data acquisition, chromatographic peak areas were integrated for all detected metabolites. Cross-sample peak alignment and integration correction were applied to the same metabolite across different samples [26].
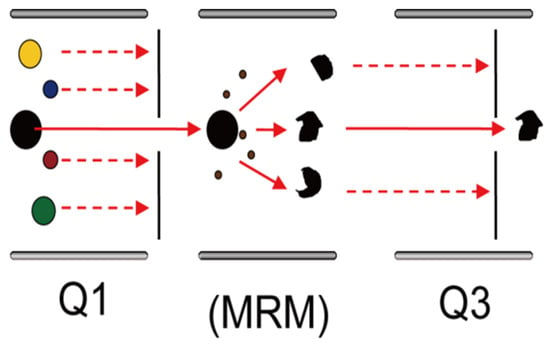
Figure 2.
Schematic diagram of mass spectrometry-based multiple reaction monitoring (MRM) mode. Note: Q1 (Da): precursor ion mass-to-charge ratio after electrospray ionization (ESI); Q3 (Da): characteristic fragment ion; MRM, multiple reaction monitoring.
3. Results and Discussion
3.1. The Pollen Structure Observation
The pollens of WH, ZR, and HM3 are all of the radial symmetrical type (Figure 3); their appearance shapes are similar, the pattern of the pollen wall is mesh carving, the ridge connects as a polygon or irregular mesh, the polar view is nearly round or round with three splits, and the equatorial view is oval. The P/E of ZR is 2.06 and belongs to the extra-long sphere; WH and HM3 belong to the long sphere. Among them, the P/E of HM3 is the largest, and the pollen of WH is the biggest (Table 2). Because WH is mixoploidy, the others are diploid, in the germplasm identification, the size of the pollen grains can be used as a reference to identify the ploidy preliminary.
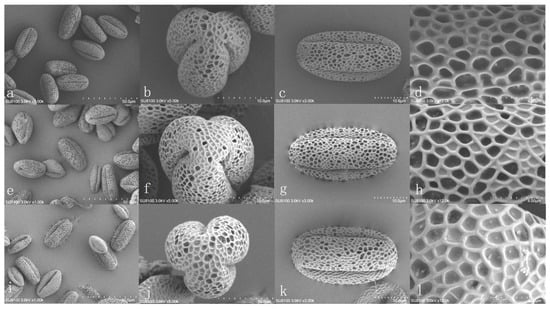
Figure 3.
The electron microscopy of the pollen surface structure of three C. lansium. Note: (a,e,i) display the overall morphology of pollen grains from ‘Zirou’ wampee, ‘Wuhe’ wampee, and ‘Huami No. 3’ wampee, respectively (scale bar = 50 μm). (b,f,j) show the polar views of pollen grains from ‘Zirou’ wampee, ‘Wuhe’ wampee, and ‘Huami No. 3’ wampee, respectively (scale bar = 10 μm). Panels (c,g,k) present the equatorial views of ‘Zirou’ wampee, ‘Wuhe’ wampee, and ‘Huami No. 3’ wampee, respectively (scale bar = 10 μm). Panels (d,h,l) exhibit the exine ornamentation patterns of ‘Zirou’ wampee, ‘Wuhe’ wampee, and ‘Huami No. 3’ wampee, respectively (scale bar = 4 μm).

Table 2.
The pollen structure analysis of three C. lansium.
3.2. The Obtain of Polyploid Plants by Hybridization and the Identification of the Hybrids
A total of 1853 flowers were cross-pollinated between ‘ZR’ and ‘WH’, yielding 202 fruits. From these, 100 interspecific hybrid seedlings were successfully germinated.
3.2.1. The Ploidy Identification of the Hybrids
Ploidy identification was performed using flow cytometry with the diploid offspring of HM3 as the control, followed by chromosome counting verification. Flow cytometry analysis revealed distinct ploidy levels: diploids showed a characteristic single peak (fluorescence intensity 50–55), triploids exhibited 1.5-fold higher intensity (75–85), and tetraploids demonstrated double intensity (100–110), while mixoploids displayed multiple peaks (50–55 and 75–85). Subsequent chromosome counting of newly emerged leaves confirmed these results. Ultimately, we successfully identified two triploids (H1, H2) and one tetraploid (H3) plant through this comprehensive analysis (Figure 4).
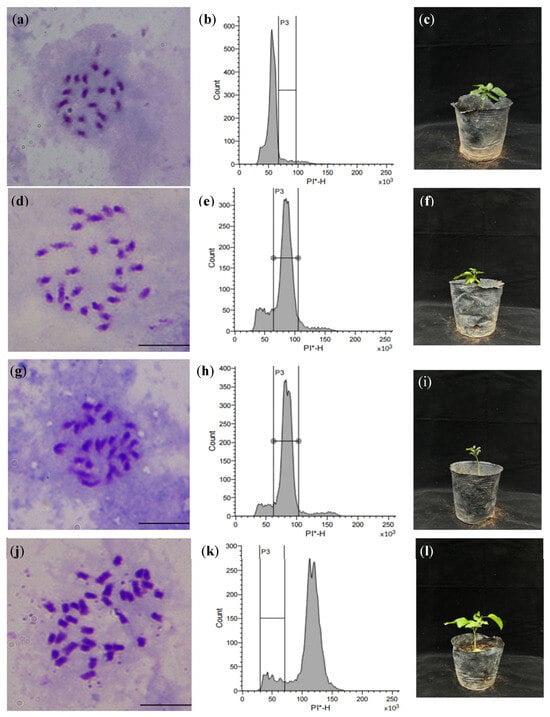
Figure 4.
The polyploid plant identification of seedlings of ZR × WH. Note: (a) the chromosomes of CK (diploid, 2n = 2x = 18); (b) the flow cytometry histogram of CK, PI-H = 50 × 103; (c) the plantlet of CK; (d,g) the chromosomes of H1 and H2 (triploid, 2n = 3x = 27); (e,h) the flow cytometry histogram of H1 and H2, PI-H = 75 × 103; (f,i) the plantlet of H1 and H2; (j) the chromosomes of H3 (tetraploid, 2n = 4x = 36); (k) the flow cytometry histogram of H3, PI-H = 100 × 103; (l) the plantlet of H3. PI-H, The height value of PI fluorescence signal; PI, Propidium iodide; * means PI stained. Bar = 10 μm.
3.2.2. Hybrid Verification by SCoT Markers
The three polyploid plants were genetically authenticated as true hybrids through SCoT polymorphism analysis using two random primers (SCoT12 and SCoT57). All the examined polyploids (H1, H2, H3) exhibited male parent-specific diagnostic bands (Figure 5), confirming their hybrid origin. This molecular evidence substantiates that the polyploid plants are genuine hybrids derived from the ‘ZR’ × ‘WH’ cross, rather than accidental self-pollination products.
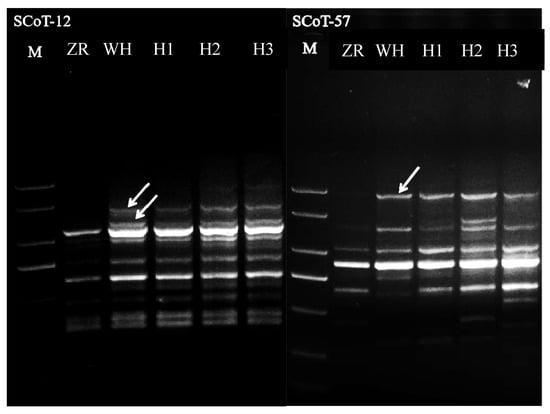
Figure 5.
Polyploid plant hybrid identification by ScoT 12 and ScoT 57. Note: M: 2000 bp Marker; ZR: female parent, WH: male parent; H1, H2, and H3 mean the polyploid plants. The white arrow means the specific band of WH.
3.3. Evaluation of Colchicine-Induced Polyploid Plants in HM3
Following colchicine treatments at varying concentrations, HM3 seedlings were counted 7 d post-sowing (Table 3). The CK demonstrated optimal germination with an average of 84 seedlings, significantly outperforming all treatment groups (p < 0.05). Our results revealed a concentration-dependent phytotoxic effect, where increasing colchicine concentrations (0.1–0.3%) correlated with progressively reduced seedling numbers at equivalent exposure durations, indicating that colchicine solution has a certain toxic effect. On the 27th day of sowing, the 0.2% colchicine treatment for 72 h yielded 49 surviving seedlings, representing a 45.6% mortality rate that approximated the target LD50 threshold. With flow cytometry, 22 mixoploidy plants and one tetraploid plant were obtained. Chromosomal analysis of the tetraploid confirmed its ploidy status through direct microscopic observation of leaf cells (Figure 6), revealing clear tetraploid (4x) chromosomal counts compared to diploid controls.

Table 3.
Seedlings growth of HM3 treated with colchicine.
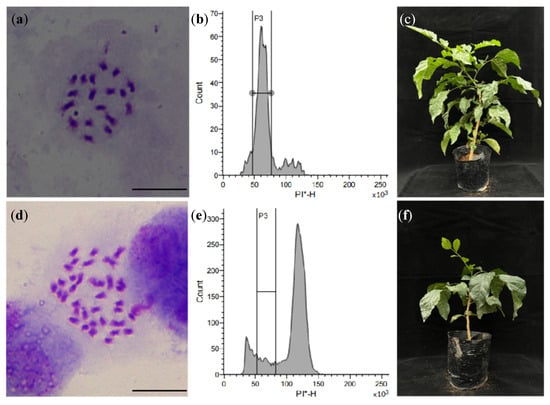
Figure 6.
The polyploid plant identification of seedlings of HM3 by colchicine treatment. Note: (a) the chromosomes of CK (HM3 seeds treated with water, diploid, 2n = 2x = 18); (b) the flow cytometry histogram of CK, PI-H= 50 × 103; (c) the plantlet of CK; (d) the chromosomes of C1 (colchicine concentration 0.3%, 48 h, tetraploid, 2n = 4x = 36); (e) the flow cytometry histogram of H3, PI-H = 100 × 103; (f) the plantlet of C1. PI-H, The height value of PI fluorescence signal; PI, Propidium iodide; * means PI stained. Bar = 10 μm.
3.4. Polyploid Identification, Observation and Analyzation of WH Seedlings
3.4.1. Polyploid Identification of WH Seedlings
A total of 804 WH seeds were sown, yielding 446 seedlings with a germination rate of 55.3%. Initial screening by flow cytometry identified six mixoploid, seven triploid, and two tetraploid plants. Subsequent cytogenetic analysis via chromosome counting of leaf cells revealed that the six initially detected mixoploid plants reverted to diploid status. Thus, the final ploidy distribution consisted of seven triploid and two tetraploid plants (Figure 7).
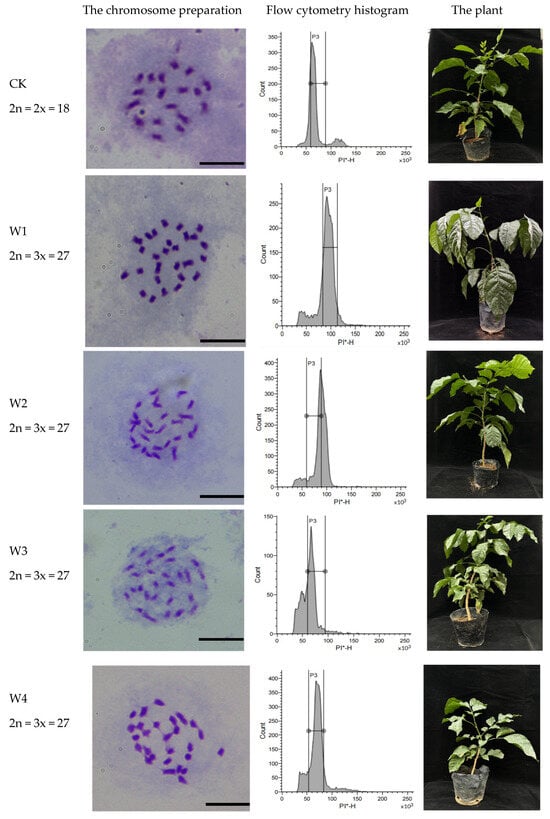

Figure 7.
The polyploid plant identification of WH seedlings. Note: CK: diploid seedling of WH, 2n = 2x = 18, PI-H= 50 × 103; W1–W7: triploid seedlings of WH, 2n = 3x = 27, PI-H = 75 × 103; W8–W9: tetraploid seedlings of WH, 2n = 4x = 36, PI-H = 100 × 103. PI-H, The height value of PI fluorescence signal; PI, Propidium iodide; * means PI stained. Bar = 10 μm.
3.4.2. The Morphological Comparison Between Diploid and Polyploid C. lansium of WH
As shown in Table 4, the leaflets of both the diploid and polyploid plantlets are all broad-ovate, with a shiny dark green surface; the leaf base is cuneate; however, in terms of the leaf margin, the leaflet numbers of one compound leaf, and the leaf surface, the diploids are different from the polyploids.

Table 4.
The morphological comparison between diploid and polyploid C. lansium of WH.
3.4.3. Comparative Analysis of Stomatal Characteristics Between Diploid and Polyploid of WH Offspring
In this study, we compared stomatal traits between diploid and polyploid WH plants grown under identical conditions (Table 5 and Figure 8). As the results showed, the polyploids exhibited significantly larger guard cell length and width compared to the diploids. Conversely, stomatal density was markedly lower in the polyploids than in their diploid counterparts.

Table 5.
The stomata comparison between diploid and polyploid C. lansium of WH.
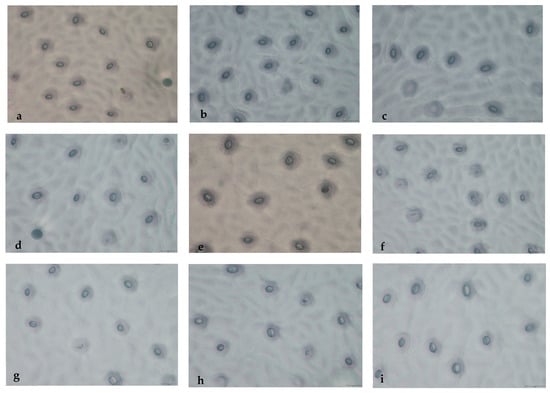
Figure 8.
The stomata observation between diploid and polyploid C. lansium of WH (40×). Note: (a) CK (2n = 2x = 18); (b–h) W1–W7 (2n = 3x = 27); (i) W8 (2n = 4x = 36). Bar = 20 μm.
3.4.4. Comparative Leaf Anatomy of Diploid and Polyploid WH Offspring
Comparative anatomical analysis revealed significant differences in leaf structure between the diploid and polyploid offspring of ‘WH’ (Figure 9 and Table 6). The polyploid plants exhibited substantially thicker upper epidermis compared to the diploids, with the tetraploids showing even greater thickness than the triploids, demonstrating a clear ploidy dosage effect. Both the palisade and spongy parenchyma layers were significantly more developed in the polyploids than in the diploids.

Figure 9.
The leaf anatomy structure of diploid and polyploid C. lansium of WH (200×). Note: (a) CK (2n = 2x = 18); (b) W1 (2n = 3x = 27); (c) W8 (2n = 4x = 36). Bar = 100 μm.

Table 6.
The leaf anatomy comparison between diploid and polyploid C. lansium of WH.
3.4.5. Physiology Comparison Between Diploid and Polyploid C. lansium of WH
Physiological comparison between the diploid and polyploid C. lansium of ‘WH’ revealed significant differences in multiple parameters (Table 7). The tetraploids exhibited a significantly higher net photosynthetic rate compared to the diploids. The chlorophyll a content in the diploid and tetraploid seedlings of WH was 1.12 mg/g and 1.28 mg/g, respectively, with the tetraploids exhibiting approximately 1.1 times the content of the diploids. The chlorophyll b content reached 0.84 mg/g in the diploids and 0.97 mg/g in the tetraploids, representing a 1.2-fold increase in the tetraploids. Total chlorophyll content measured 1.96 mg/g for the diploids and 2.66 mg/g for the tetraploids, indicating that the tetraploids contained 1.5 times the total chlorophyll of the diploids.

Table 7.
Physiology comparison between diploid and polyploid C. lansium of WH.
The polyploids generally showed greater stomatal conductance and transpiration rates than the diploids, except for W3 and W6. The intercellular CO2 concentration was significantly elevated in the polyploids, with the exception of W3. Secondary metabolite analysis showed that while the triploids had non-significantly higher flavonoid content than the diploids, the tetraploids accumulated significantly more flavonoids. In contrast, the diploids contained significantly higher total phenolic content than the polyploids. No differences were observed in malondialdehyde (MDA) or soluble protein levels between the ploidy types. Soluble sugar content was similar in the diploids and triploids, but significantly increased in the tetraploids. These findings demonstrate that polyploidization, particularly tetraploidy, optimizes physiological performance in C. lansium, enhancing photosynthetic efficiency, modifying stomatal function, and preferentially accumulating valuable flavonoids. This supports the strategic use of polyploid breeding to develop high-performance cultivars with superior medicinal and agronomic traits.
3.5. Metabolites Analysis Between Diploid and Polyploid C. lansium of WH
3.5.1. Qualitative and Quantitative Analysis of the Metabolites in Diploid and Polyploid Seedlings of C. lansium
Quality control (QC) was performed using mixed samples, and the results were visualized through total ion chromatograms (TIC) and multiple reaction monitoring (MRM) metabolite detection peak maps (extracted ion chromatograms, XIC) (Figure 10). The major metabolites identified in C. lansium leaves are presented in Figure 11. Flavonoids were the most abundant class of metabolites, consistent with their established role as key bioactive compounds in C. lansium with medicinal significance.
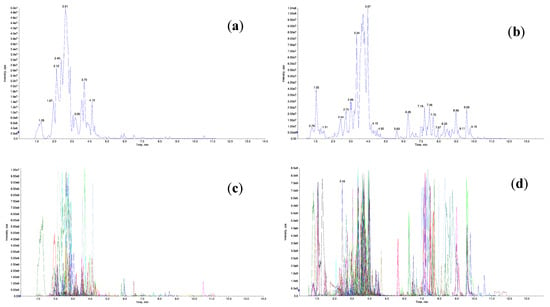
Figure 10.
The TIC and MRM peak map for C. lansium. Note: (a) TIC in negative ion mode; (b) TIC in positive ion mode; (c) MRM in in negative ion mode; (d) MRM in positive ion mode. Each chromatographic peak in different colors represents a detected metabolite. The transverse coordinate for the metabolite detection RT (retention time); the ordinate is the ion flow intensity in cps (count per second).
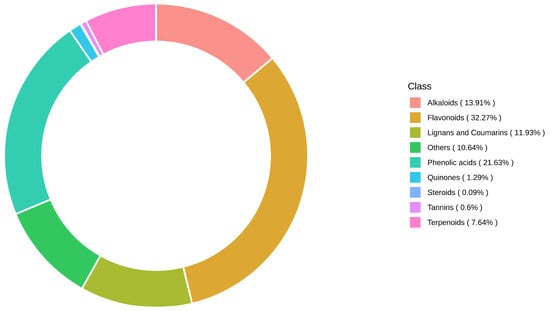
Figure 11.
Circular plot of the metabolite category composition. Note: Each color represents a metabolite category, and the block area represents the proportion. Total is 100%.
3.5.2. The PCA and Orthogonal Partial Least Squares–Discriminant Analysis (OPLS-DA)
Principal component analysis (PCA) was performed to assess overall metabolite differences between groups and intra-group variability (Figure 12a). The results revealed a distinct clustering of samples within the same group, except for Group 4, which exhibited greater dispersion.
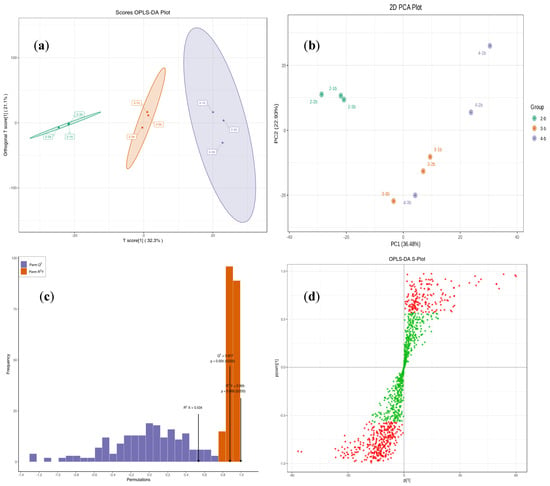
Figure 12.
The PCA and OPLS-DA of the materials. Note: (a) The 2D PCA plot of the materials. PC1, the first principal component; PC2, the second principal component, and the percentage represents the explanatory of this principal component in the dataset. Each point in the figure represents one sample, and samples in the same group are represented by the same color. (b) The chart of OPLS-DA score chart. The abscissa represents the predicted principal component; the ordinate represents the orthogonal principal component; the percentage represents the interpretation rate of the component to the dataset. Each point in the figure represents one sample; samples from the same group use the same color. (c) OPLS-DA verification. The abscissa represents the model R2Y and Q2 value, and the ordinate is the frequency of the model classification effect in 200 random permutation and combination experiments. Orange represents the random grouping model R2Y, purple represents the random grouping model Q2, and the black arrows represent the values for the R2X, R2Y, and Q2 values of the original model. (d) OPLS-DA S-plot. The abscissa represents the covariance between principal components and metabolites, and the ordinate represents the correlation coefficient between principal components and metabolites. Metabolites closer to the top-right and bottom-left corners indicate more significant differences. Red dots represent metabolites with VIP values > 1, while green dots denote those with VIP values ≤ 1.
Since PCA is less sensitive to weakly correlated variables, partial least squares–discriminant analysis (PLS-DA) was employed to enhance variable discrimination. Further refinement was achieved using orthogonal partial least squares–discriminant analysis (OPLS-DA), which integrates orthogonal signal correction (OSC) with PLS-DA to partition the X-matrix into Y-related and Y-orthogonal components, thereby improving the detection of biologically relevant differential metabolites [27]. The OPLS-DA score plot (Figure 12b) highlighted clear inter-group metabolic distinctions, while model validation (Figure 12c) and the S-plot (Figure 12d) confirmed its robustness and identified key contributing variables.
Differential metabolites were screened based on variable importance in projection (VIP) scores; the top 20 metabolites with the highest VIP values (Figure 13) represent key bioactive compounds that contribute substantially to C. lansium’s medicinal and physiological properties. These included six phenolic acids, four flavonoids, five terpenoids, three alkaloids, and two lignans/coumarins. These metabolites collectively enhance C. lansium’s medicinal value through antioxidant, antimicrobial, and anticancer properties while improving plant defense and stress adaptation. For instance, the most significant metabolite, shizukolidol, is a traditional medicine Chinese component with antibacterial activity [28]; santalal is kind of anti-tumor active compound which exhibits tumor-selective cytotoxicity [29]; Kaempferol-7-O-rhamnoside and Kaempferol-3-7-O-robinoside show aldose reductase inhibition with the potential to develop the treatment of diabetes [30], and the novel combination with MTX mildly improved the reduction in inflammation in experimental arthritis [31].
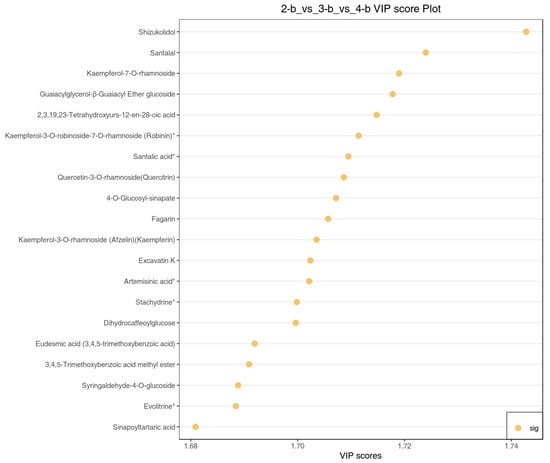
Figure 13.
Plots of the differential metabolite VIP values. Note: The abscissa represents the VIP values, the ordinate indicates the differential metabolites, and the yellow represents the value of the metabolites that are significantly different in the three differential comparison groups. * means presence of isomeric forms of this compound, though the exact configuration remains undetermined, and the specific isomer cannot be identified.
These compounds serve as key targets for quality control in herbal products and future breeding programs aimed at optimizing bioactive content.
3.5.3. Differential Metabolites of C. lansium in Different Ploidy Leaves
As shown in Figure 14A, triploid and tetraploid C. lansium plants exhibited a significantly higher accumulation of flavonoids, alkaloids, phenolic acids, and other bioactive metabolites compared to their diploid counterparts. Moreover, these metabolites were extremely high in the tetraploid, suggesting a positive correlation between ploidy level and medicinal compound content, particularly for flavonoids. These findings align with previous physiological studies comparing diploid and polyploid C. lansium.
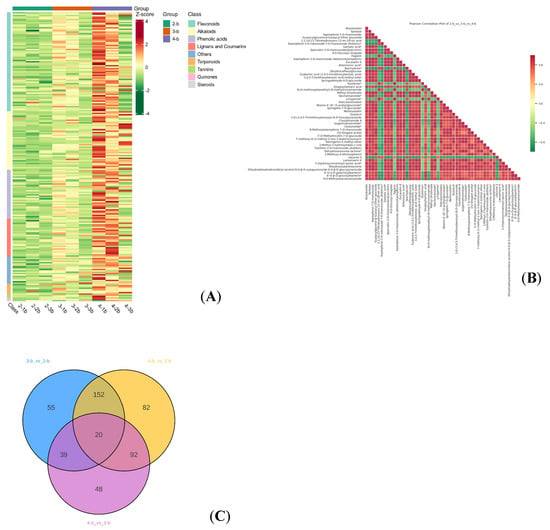
Figure 14.
Differential metabolites of different ploidy materials of C. lansium. Note: (A) Heatmap of the differential metabolite clusters: The horizontal is the sample name, the vertical is the differential metabolite information; different colors are different values obtained after different relative content standardization treatments (red represents high content, green represents low content). Heatmap_class: Classification by the metabolite class; class is the first class of substances; heatmap_col-row_cluster: Both metabolites and samples are cluster analyzed; the cluster line on the left is the metabolite cluster line, and the cluster line above the graph is the sample; cluster.line.heatmap_row_cluster: Cluster analysis is performed only for the metabolites, and the cluster line on the left side of the figure is the metabolite cluster line. (B) Heatmap of differential metabolite correlation; transverse and longitudinal axle are both the name of different metabolites; different colors represent the Pearson correlation coefficient r; the legend on the right shows the correlation coefficient and color between groups; red is strong positive correlation, green is strong negative correlation; the deeper color represents the absolute value of the correlation coefficient between samples. Plots for all differential metabolites by default, showing the top 50 differential metabolites with the largest VIP value when the number of differential metabolites exceeds 50. (C) Venn of differential metabolite: Each circle in the figure represents a comparison group; the numbers in the overlapping circles represent the number of differential metabolites shared between the comparison groups, and the numbers without the overlap represent the number of differential metabolites unique to the comparison group. * means presence of isomeric forms of this compound, though the exact configuration remains undetermined, and the specific isomer cannot be identified.
Metabolites often exhibit synergistic or antagonistic relationships, and correlation analysis can reveal metabolic proximity and potential regulatory interactions during biological state transitions. We performed Pearson correlation coefficient analysis on the differentially accumulated metabolites (Figure 14B), which helped identify coordinated or mutually exclusive metabolic patterns.
Furthermore, Venn diagram analysis (Figure 14C) identified 20 conserved metabolites present across all three ploidy groups (Table 8), suggesting that these compounds may represent essential biochemical components of C. lansium. Notably, the majority of these conserved metabolites were alkaloids and flavonoids, further supporting the pharmaceutical value of this species.

Table 8.
Top 20 most abundant metabolites identified in C. lansium.
3.5.4. KEGG Pathway Enrichment Analysis of Differential Metabolites
Based on the differential metabolite results, KEGG pathway enrichment analysis was performed. ‘Rich Factor’ is calculated as the ratio of the number of differentially abundant metabolites in a given pathway to the total number of metabolites annotated to that pathway. A higher Rich Factor indicates a greater degree of enrichment (i.e., more significant overrepresentation of differential metabolites in the pathway). p-value means the p-value of the hypergeometric test, and the calculation formula of the hypergeometric distribution is as follows:
‘N’ represents the number of metabolites with KEGG annotation in the total metabolites; ‘n’ represents the number of differential metabolites in N; ‘M’ represents the number of metabolites in a KEGG pathway in N; and ‘m’ represents the number of differential metabolites for a KEGG pathway in M. The closer the p-value is to 0, the more significant the enrichment. The size of the points in the plot represents the number of differentially significant metabolites enriched to the corresponding pathway. After selecting the top 20 pathways of p-values from small to large to analyze, as shown in Figure 15, only 13 pathways met the criteria in practice. The highest differential metabolite enrichment is in the ‘Flavonoid biosynthesis’ and the ‘Biosynthesis of secondary metabolites’ pathways, which are the most critical pathways for the medicinal component synthesis; this also indicated that the ploidy would affect the medicinal component accumulation.
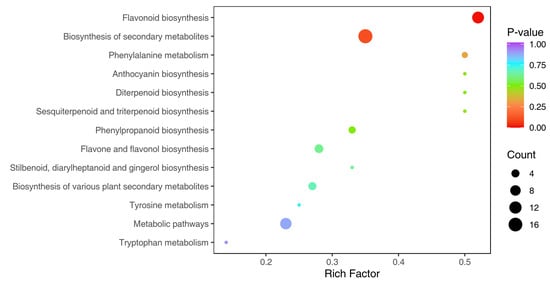
Figure 15.
Differential metabolite pathway enrichment map by KEGG analysis. Note: The abscissa represents the corresponding ‘Rich Factor’ of each pathway; the ordinate indicated the pathway’s name (sorted by p-value), the color of the points reflects the size of p-value, and the redder color means more significant enrichment. The size of the dot represents the enriched number of differential metabolites; the bigger dot indicates more differential metabolites.
4. Discussion
4.1. Comparative Evaluation of Polyploid Induction Methods in C. lansium
This study successfully induced polyploidy in C. lansium through three distinct approaches: hybridization, colchicine treatment, and seedling selection. Our results yielded nine triploid and four tetraploid plants, with hybridization proving to be the most effective method (3% success rate), significantly outperforming colchicine induction (0.1‰ success rate). These findings align with previous reports that both colchicine concentration and exposure duration critically influence polyploid induction efficiency [32], suggesting the need for further optimization of chemical induction protocols for C. lansium. The superior performance of hybridization can be attributed to the mixed-ploidy nature of ‘Wuhe’, which exhibits high inherent potential for polyploid production.
4.2. Morphological, Anatomical Structure and Physiology Modified of Polyploid C. lansium
Polyploidization induced significant modifications in the stomatal characteristics and leaf anatomy of C. lansium. Our observations of enlarged guard cells and reduced stomatal density in polyploids corroborate findings in watermelon [16]. The wampee polyploid plants also developed significantly thicker upper epidermal layers, palisade, and spongy parenchyma tissues. These findings are consistent with previous reports in atemoya [33], where polyploidization resulted in wider xylem vessels and improved drought tolerance. The reinforced epidermal layers and expanded mesophyll tissue suggest polyploid plants may have evolved superior adaptive traits. Specifically, these structural changes likely contribute to superior drought resistance and pest defense mechanisms compared to their diploid counterparts, potentially due to improved water retention capacity and reduced transpirational water loss. The consistent pattern of leaf architecture modification through polyploidization demonstrates its potential as a mechanism for developing plants with improved stress resilience. These structural adaptations may explain the ecological advantages frequently observed in polyploid plants across various species. Importantly, these morphological changes correlate with enhanced production of bioactive compounds, particularly flavonoids and polyphenols—valuable phytochemicals with demonstrated antitussive, expectorant, antimicrobial, anti-inflammatory, and hypolipidemic properties [34]. Beyond genomic changes, polyploidization in C. lansium also confers physiological advantages, including heightened photosynthetic activity and altered carbon metabolism. These adjustments in photoassimilate production and distribution may represent an adaptive strategy to improve environmental stress resilience [9].
4.3. Metabolomic Profiling of Diploid and Polyploid C. lansium
Modern metabolomics has emerged as a powerful tool for plant studies [35]. Recent studies found that the ploidy is positively correlated with most metabolites’ biosynthesis, especially flavonoids. The integrated metabolomics and transcriptome analysis of the leaves of diploid and tetraploid Fagopyrum tataricum showed that polyploidization up-regulated flavonoid biosynthesis pathway genes and promoted flavone biosynthesis [36]. The study of Cyclocarya paliurus found that the flavonoid glucuronides were significantly accumulated in the tetraploid [37]. Our findings are consistent with these previous reports and support the growing consensus that ploidy level positively correlates with enhanced biosynthesis of secondary metabolites, particularly flavonoids. The comprehensive metabolite profiling conducted in this study provides new insights into the pharmaceutical potential of polyploid C. lansium.
5. Conclusions
This study demonstrates significant advancements in C. lansium polyploidy research, successfully establishing 13 polyploid lines that enrich the species’ genetic resources. The polyploid plants exhibit distinct structural modifications, including larger pollen grains, increased guard cell dimensions, reduced stomatal density, and thickened leaf tissues (epidermis, palisade, and spongy parenchyma). Metabolomic analysis reveals that triploid and tetraploid plants accumulate substantially higher concentrations of these medicinally important metabolites compared to diploids, with flavonoid content showing particularly strong ploidy-dependent enhancement. These findings establish polyploid C. lansium as a superior source material for pharmaceutical applications and provide a foundation for developing high-value cultivars through targeted polyploid breeding programs focused on optimizing bioactive compound production. The observed positive correlation between ploidy level and phytochemical content suggests intentional ploidy manipulation and represents a promising strategy for enhancing the medicinal value of this species. This study indicated that the ploidy is positively related with the content of bioactivity components. Compared with the diploidy, the polyploidy of C. lansium significantly upregulated the medical component, which means they are the better source materials for the extraction of metabolites with high pharmaceutical value.
Author Contributions
Y.D.: Writing—original draft, Methodology, Investigation, Formal analysis; L.W.: Software, Methodology, Investigation, Formal analysis; H.W.: Investigation, Formal analysis; Z.Z. (Zhichun Zhang): Formal analysis, Data curation; J.Z. Conceptualization, Resources; G.H.: Supervision, Project administration; Y.Q.: Supervision, Resources, Funding acquisition; Z.Z. (Zhike Zhang): Writing—review and editing, Supervision, Project administration, Funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the Science and Technology Planning Project of Guangdong Province and Yunan County (h20240204); Yonggen Sci-Tech Station Project (202308).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Zhao, Z.C.; Hu, G.B.; Ouyang, R.; Liu, Y.C.; Yi, Y.; Chen, Y.Y. The earlier identification of the seedless characteristic of the wampee [Clausena lansium (Lour.) Skeels] hybrid by a random amplified polymorphic DNA (RAPD) marker. Afr. J. Biotechnol. 2010, 9, 8578–8582. [Google Scholar]
- Chokeprasert, P.; Charles, A.L.; Sue, K.H.; Huang, T.C. Volatile components of the leaves: Fruits and seeds of wampee Clausena lansium (Lour.) Skeels. J. Food Compos. Anal. 2007, 20, 52–56. [Google Scholar] [CrossRef]
- Liu, Y.P.; Guo, J.M.; Wang, X.P.; Liu, Y.Y.; Zhang, W.; Wang, T.; Qiang, L.; Fu, Y.H. Geranylated carbazole alkaloids with potential neuroprotective activities from the stems and leaves of Clausena lansium. Bioorg. Chem. 2019, 92, 103278. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Xiong, Z.; Xie, N.; Liu, S.Z.; Zhang, L.L.; Xu, F.; Guo, W.H.; Feng, J.T. Bioassay-guided isolation of antifungal amides against Sclerotinia sclerotiorum from the seeds of Clausena lansium. Ind. Crops Prod. 2018, 121, 352–359. [Google Scholar] [CrossRef]
- Yang, C.; Lin, Z.Z.; Luo, Z.; Wang, Z.Q.; Liu, P.; Xu, R.W.; Zhu, F.; Cheng, Y.J. The volatile compound (E)-2-hexenal in wampee (Clausena lansium) represses the development of Penicillium italicum and enhances the disease resistance of postharvest citrus fruit. Postharvest Biol. Technol. 2025, 219, 113241. [Google Scholar] [CrossRef]
- Wei, Y.Z.; Wang, Y.; Hu, F.C.; Wang, W.; Wei, C.B.; Xu, B.Q.; Liu, L.Q.; Li, H.Y.; Wang, C.; Zhang, H.N.; et al. The Clausena lansium genome provides new insights into alkaloid diversity and the evolution of the methyltransferase family. J. Integr. Agric. 2024, 23, 3537–3553. [Google Scholar] [CrossRef]
- Moghe, G.D.; Shiu, S.H. The causes and molecular consequences of polyploidy in flowering plants. Ann. N. Y. Acad. Sci. 2014, 1320, 16–34. [Google Scholar] [CrossRef] [PubMed]
- Husband, B.C.; Baldwin, S.J.; Suda, J. The incidence of polyploidy in natural plant populations: Major patterns and evolutionary processes. In Plant Genome Diversity; Leitch, I.J., Greilhuber, J., Dolezel, J., Wendel, J.F., Eds.; Springer Inc.: Vienna, Austria, 2013; Volume 2, pp. 255–276. [Google Scholar]
- Van de Peer, Y.; Ashman, T.L.; Soltis, P.S.; Soltis, D.E. Polyploidy: An evolutionary and ecological force in stressful times. Plant Cell 2021, 33, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Stebbins, G.L. Variation and Evolution in Plants; Columbia University Press: New York, NY, USA, 1950. [Google Scholar]
- Sattler, M.C.; Carvalho, C.R.; Clarindo, W.R. The polyploidy and its key role in plant breeding. Planta 2016, 243, 281–296. [Google Scholar] [CrossRef] [PubMed]
- Hagen, E.R.; Mason, C.M. Differences in pathogen resistance between diploid and polyploid plants: A systematic review and meta-analysis. Oikos 2024, 5, e09908. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, Y.; Tang, W.H.; Zhang, G.; Wang, Y.H.; Yan, Z.M. Genetic variation, polyploidy, hybridization influencing the aroma profiles of Rosaceae family. Genes 2024, 15, 1339. [Google Scholar] [CrossRef] [PubMed]
- Germanà, M. Doubled haploid production in fruit crops. Plant Cell Tissue Organ Cult. 2006, 86, 131–146. [Google Scholar] [CrossRef]
- Kulkarni, R.N.; Baskaran, K.; Jhang, T. Breeding medicinal plant, periwinkle [Catharanthus roseus (L) G. Don]: A review. Plant Genet. Resour. 2016, 14, 283–302. [Google Scholar] [CrossRef]
- Khan, M.N.E.A.; Hassan, J.; Biswas, M.S.; Khan, H.I.; Sultana, H.; Suborna, M.N.; Rajib, M.M.R.; Akter, J.; Gomasta, J.; Anik, A.A. Morphological and anatomical characterization of colchicine-induced polyploids in watermelon. Hortic. Environ. Biotechnol. 2023, 61, 461–474. [Google Scholar] [CrossRef]
- Sun, Q.; Sun, H.; Li, L.; Bell, R.L. In vitro colchicine-induced polyploid plantlet production and regeneration from leaf explants of the diploid pear (Pyrus communis L.) cultivar, ‘Fertility’. J. Hortic. Sci. Biotechnol. 2009, 84, 548–552. [Google Scholar] [CrossRef]
- Bora, L.; Vijayakumar, R.M.; Ganga, M.; Ganesan, N.M.; Sarkar, M.; Kundu, M. Induction of polyploids in acid lime (Citrus aurantifolia) through colchicine treatment of in vitro derived shoot tip explants. Plant Breed. 2023, 142, 118–127. [Google Scholar] [CrossRef]
- Feng, L.; Liu, R. Preliminary report on chromosomal ploidy variation in ‘Yunan Wuhe’ wampee. Chin. Agric. Sci. Bull. 2007, 9, 115–118. (In Chinese) [Google Scholar]
- Peng, S.J.; Zhu, X.Y.; Chen, J.X.; Chen, J.Y.; Hu, X.L.; Wu, J.L.; Zhang, Z.K.; Zhao, J.T.; Hu, G.B.; Sabir, I.A.; et al. Integrated analyses of the transcriptome and metabolome revealed the coloring mechanism of red-pericarp wampee (Clausena lansium). Plant Physiol. Biochem. 2025, 224, 109959. [Google Scholar] [CrossRef] [PubMed]
- Collard, B.C.; Mackill, D.J. Start codon targeted (SCoT) polymorphism: A simple, novel DNA marker technique for generating gene-targeted markers in plants. Plant Mol. Biol. Rep. 2009, 27, 86–93. [Google Scholar] [CrossRef]
- Nugroho, K.; Santoso, T.J.; Kosmiatin, M.; Sukma, D.; Purwito, A.; Husni, A.; Reflinur, R.; Lestari, P. Genetic diversity and population structure of Indonesia’s mandarin citrus genotypes using simple sequence repeat and start codon targeted markers. Genet. Resour. Crop Evol. 2025, 72, 2939–2957. [Google Scholar] [CrossRef]
- Weaver, J.L. Flow cytometry: Measuring cell populations and studying cell physiology—Introduction to flow cytometry. Methods 2000, 21, 199–201. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Leng, Q. A new method to prepare clean cuticular membrane from fossil leaves with thin and fragile cuticles. Sci. China-Earth Sci. 2011, 54, 223–227. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, W.; Li, C.; Wang, Z.; Lu, C.; Cheng, J.; Wei, S.; Yang, J.; Yang, Q. Integrated transcriptomic and metabolomic analyses elucidate the mechanism of flavonoid biosynthesis in the regulation of mulberry seed germination under salt stress. BMC Plant Biol. 2024, 24, 132. [Google Scholar] [CrossRef] [PubMed]
- Fraga, C.G.; Clowers, B.H.; Moore, R.J.; Zink, E.M. Signature-discovery approach for sample matching of a nerve-agent precursor using liquid chromatography-mass spectrometry, XCMS, and chemometrics. Anal. Chem. 2010, 82, 4165–4173. [Google Scholar] [CrossRef] [PubMed]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Reyes, T.; Monribot-Villanueva, J.L.; Jiménez-Martínez, O.D.; Aguilar-Colorado, A.S.; Bonilla-Landa, I.; Flores-Estévez, N.; Luna-Rodríguez, M.; Guerrero-Analco, J.A. Sesquiterpene lactones and phenols from polyfollicles of Magnolia vovidessi and their antimicrobial activity. Nat. Prod. Commun. 2018, 13, 521–525. [Google Scholar] [CrossRef]
- Matsuo, Y.; Mimaki, Y. α-Santalol derivatives from Santalum album and their cytotoxic activities. Phytochemistry 2012, 77, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Quilantang, N.G.; Choi, K.; Lee, B.H.; Lee, S. Kaempferol Rhamnosides from Geranium sibiricum as aldose reductase inhibitors and their content by HPLC analysis. Processes 2020, 8, 694. [Google Scholar] [CrossRef]
- Tsiklauri, L.; Švík, K.; Chrastina, M.; Poništ, S.; Dráfi, F.; Slovák, L.; Alania, M.; Kemertelidze, E.; Bauerova, K. Bioflavonoid Robinin from Astragalus falcatus Lam. mildly improves the effect of metothrexate in rats with adjuvant arthritis. Nutrients 2021, 13, 1268. [Google Scholar] [CrossRef] [PubMed]
- Emsweller, S.L. Colchicine-induced polyploidy in Lilium longiflorum. Am. J. Bot. 1949, 36, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Losada, J.M.; Blanco-Monre, N.; Fonollá, A.; Martínez-Ferrí, E.; Hormaza, J.I. Hydraulic tradeoffs underlie enhanced performance of polyploid trees under soil water deficit. Plant Physiol. 2023, 192, 1821–1835. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhou, J.; Zhang, X.; Hou, H.; Liu, X.; Yang, C.; Shen, S.; Luo, J. Insights into tissue-specific specialized metabolism in wampee (Clausena lansium (Lour.) Skeels) varieties. Foods 2024, 13, 3092. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luo, J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.D.; Qi, Y.; Lin, L.Z.; Wang, J.; Qin, X.B.; Niu, B. Comparative analysis of the transcriptome and metabolome in leaves of diploid and tetraploid Fagopyrum tataricum. Phyton-Int. J. Exp. Bot. 2023, 92, 3149–3162. [Google Scholar] [CrossRef]
- Yu, Y.H.; Qu, Y.Q.; Wang, S.Y.; Wang, Q.; Shang, X.L.; Fu, X.X. An integrative analysis of metabolome and transcriptome reveals the molecular regulatory mechanism of the accumulation of flavonoid glycosides in different Cyclocarya paliurus ploidies. Forests 2023, 14, 770. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).