Transcriptome Analysis Elucidates the Mechanism of an Endophytic Fungus Cladosporium sp. ‘BF-F’ in Enhancing the Growth of Sesuvium portulacastrum
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant and Fungal Material
2.2. Fungal Inoculation and Plant Growth Analysis
2.3. RNA Extraction and Sequencing
2.4. Differential Expression Analysis
2.5. Quantitative Real-Time PCR (qRT-PCR)
2.6. Statistical Analysis
3. Results
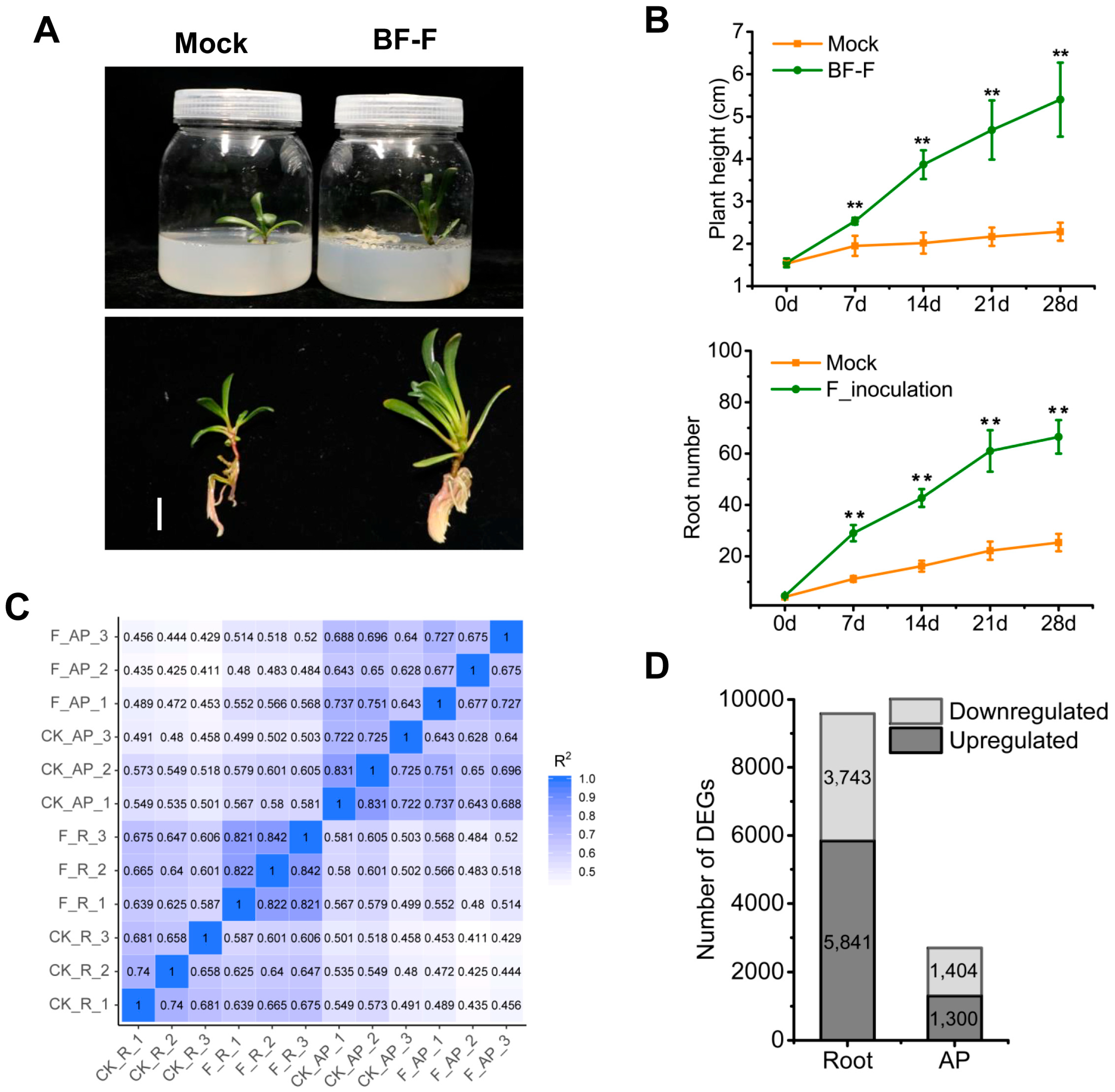
3.1. Cladosporium sp. ‘BF-F’ Promotes the Growth of S. portulacastrum
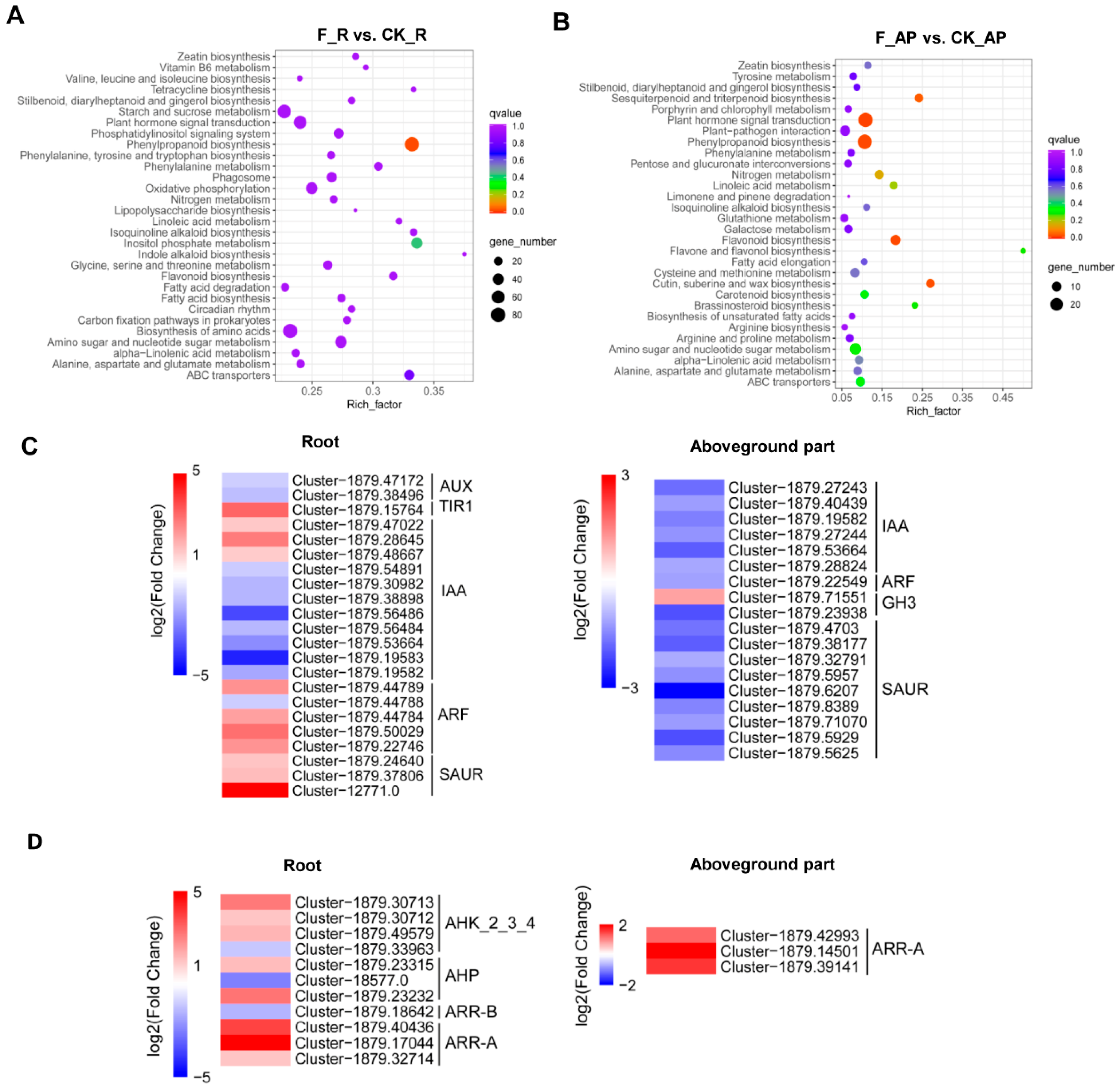
3.2. Inoculation of ‘BF-F’ Affected the Expression of Genes in S. portulacastrum
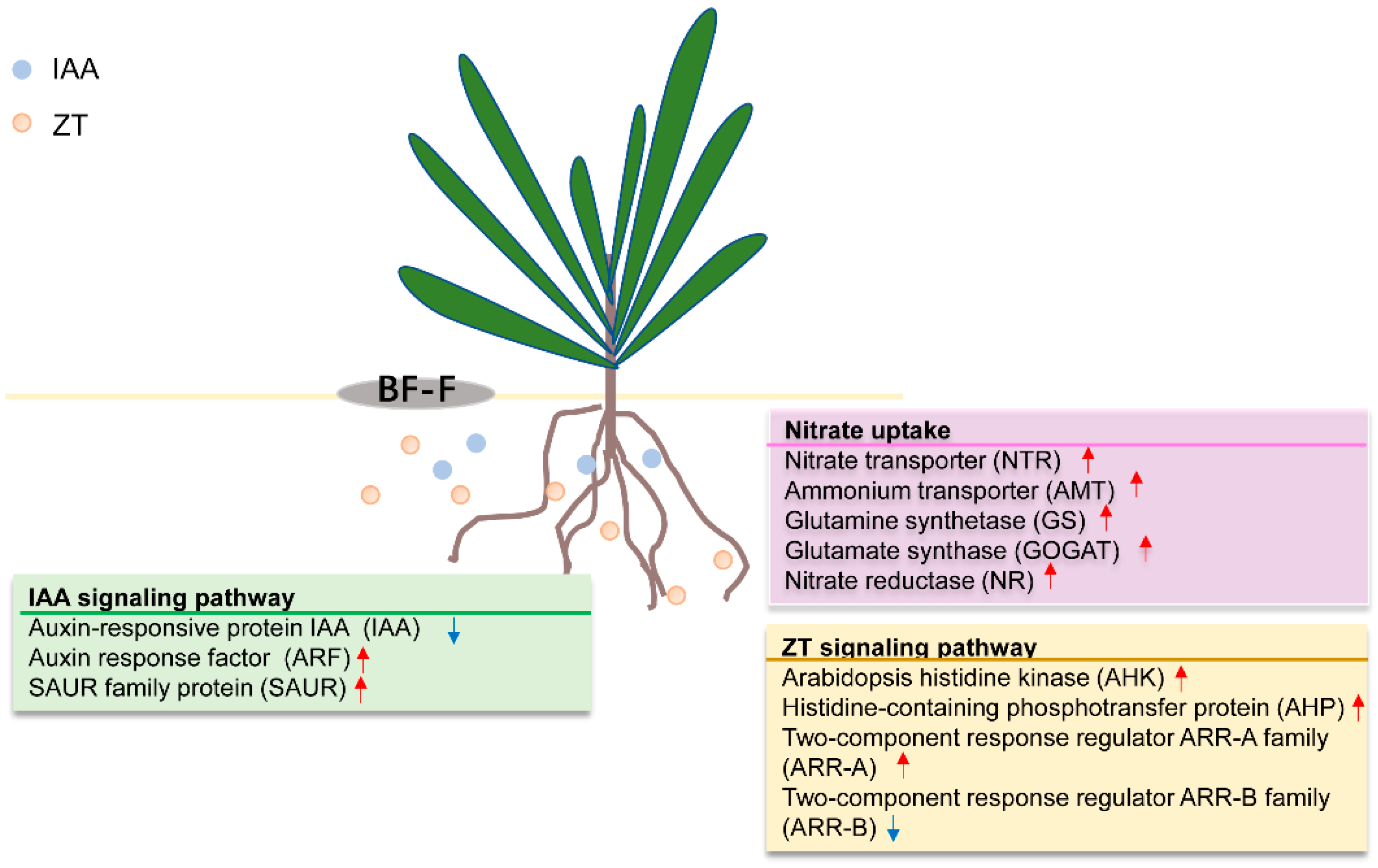
3.3. Inoculation of “BF-F” Affected Hormone Signaling Transduction in S. portulacastrum
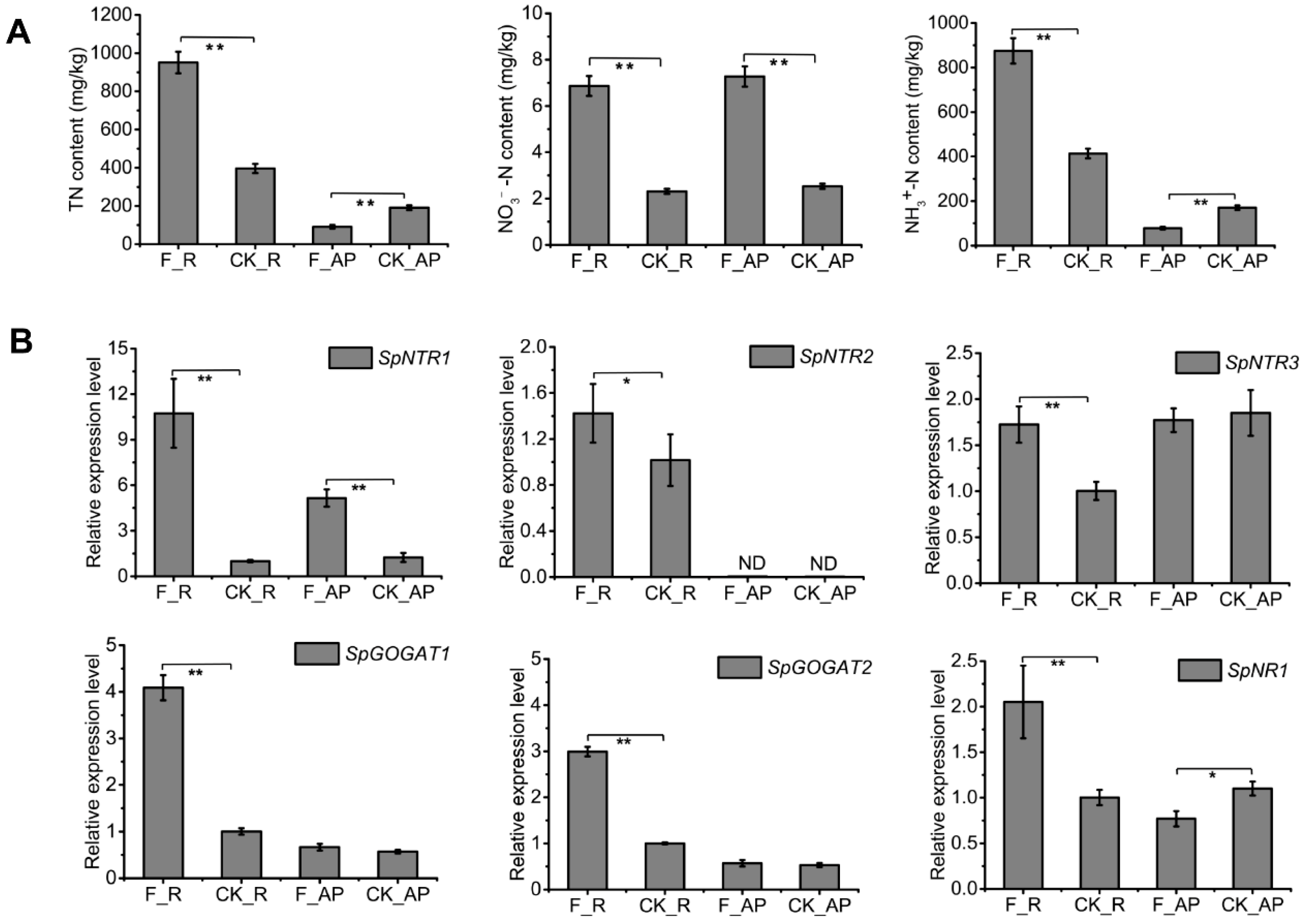
3.4. Cladosporium sp. ‘BF-F’ Inoculation Promotes Nitrogen Uptake in S. portulacastrum
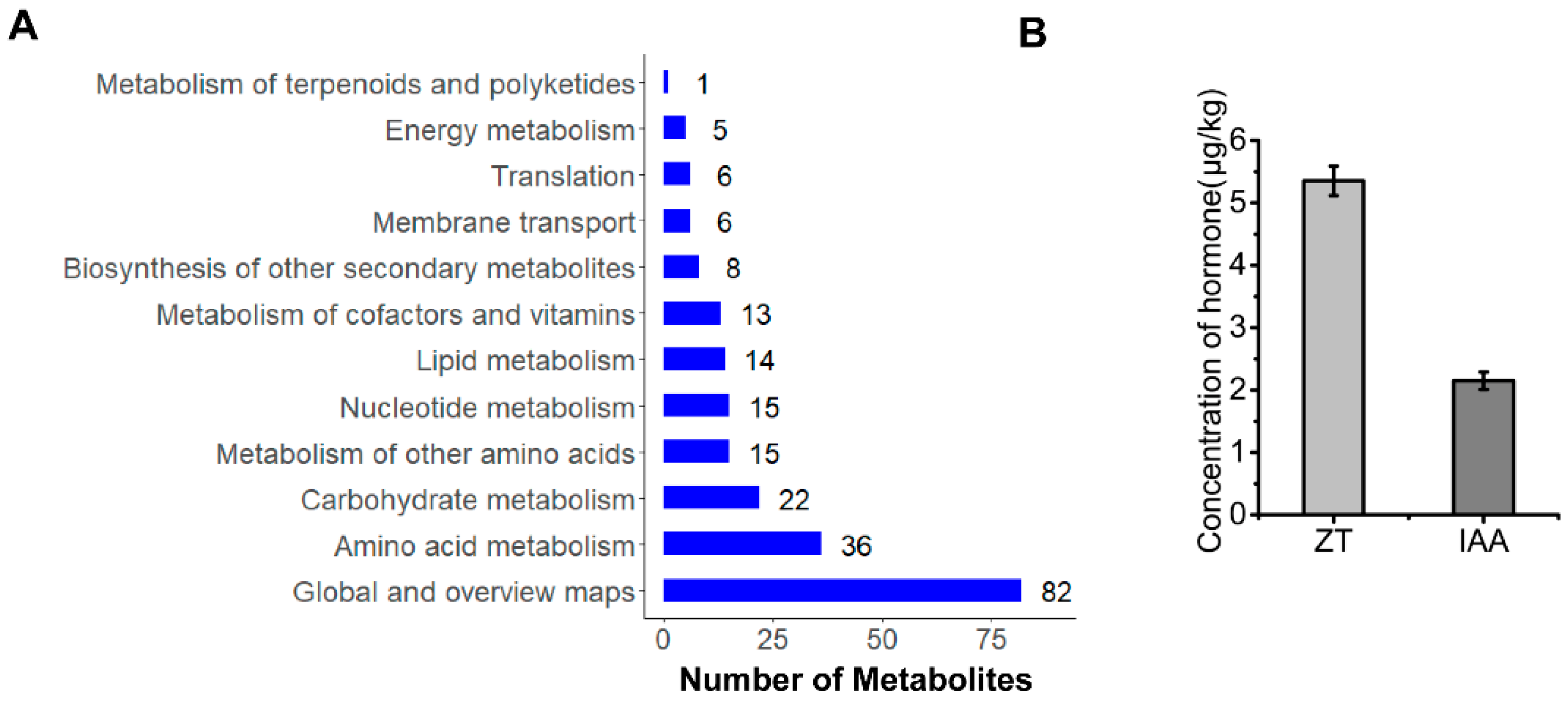
3.5. Cladosporium sp. ‘BF-F’ Promotes the Growth of S. portulacastrum by Synthesizing IAA and ZT
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jamil, A.; Riaz, S.; Ashraf, M.; Foolad, M.R. Gene expression profiling of plants under salt stress. Crit. Rev. Plant Sci. 2011, 30, 435–458. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cells 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Haro, R.; Benito, B. The role of soil fungi in K+ plant nutrition. Int. J. Mol. Sci. 2019, 20, 3169. [Google Scholar] [CrossRef]
- Zhou, L.S.; Tang, K.; Guo, S.X. The Plant Growth-Promoting Fungus (PGPF) Alternaria sp. A13 Markedly enhances Salvia miltiorrhiza root growth and active ingredient accumulation under greenhouse and field conditions. Int. J. Mol. Sci. 2018, 19, 270. [Google Scholar] [CrossRef] [PubMed]
- Ou, T.; Zhang, M.; Gao, H.; Wang, F.; Xu, W.; Liu, X.; Wang, L.; Wang, R.; Xie, J. Study on the potential for stimulating mulberry growth and drought tolerance of plant growth-promoting fungi. Int. J. Mol. Sci. 2023, 24, 4090. [Google Scholar] [CrossRef]
- Kazerooni, E.A.; Maharachchikumbura, S.S.N.; Al-Sadi, A.M.; Rashid, U.; Kang, S.M.; Lee, I.J. Actinomucor elegans and Podospora bulbillosa positively improves endurance to water deficit and salinity stresses in tomato plants. J. Fungi 2022, 8, 785. [Google Scholar] [CrossRef] [PubMed]
- El-Saadony, M.T.; Saad, A.M.; Soliman, S.M.; Salem, H.M.; Ahmed, A.I.; Mahmood, M.; El-Tahan, A.M.; Ebrahim, A.A.M.; El-Mageed, T.A.A.; Negm, S.H.; et al. Plant growth-promoting microorganisms as biocontrol agents of plant diseases: Mechanisms, challenges and future perspectives. Front. Plant Sci. 2022, 13, 923880. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, F.S.; Wu, X.Q.; Luan, F.G.; Zhang, L.P.; Fang, X.M.; Ye, J.R. Isolation and characterization of two phosphate-solubilizing fungi from rhizosphere soil of moso bamboo and their functional capacities when exposed to dierent phosphorus sources and pH environments. PLoS ONE 2018, 13, e0199625. [Google Scholar] [CrossRef]
- Khan, A.L.; Waqas, M.; Lee, I.J. Resilience of Penicillium resedanum LK6 and exogenous gibberellin in improving Capsicum annuum growth under abiotic stresses. J. Plant Res. 2015, 128, 259–268. [Google Scholar] [CrossRef]
- Halifu, S.; Deng, X.; Song, X.; Song, R. Effects of two Trichoderma strains on plant growth, rhizosphere soil nutrients, and fungal community of Pinus sylvestris var. mongolica annual seedlings. Forests 2019, 10, 758. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhang, C.; Zhou, W.; Li, W.; Chu, L.; Yan, J.; Li, H. Diversity and plant growth-promoting ability of endophytic fungi from the five flower plant species collected from Yunnan, Southwest China. J. Plant Interact. 2014, 9, 585–591. [Google Scholar] [CrossRef]
- Zhang, F.; Meng, X.; Feng, C.; Ran, W.; Yu, G.; Zhang, Y.; Shen, Q. Hydrolytic amino acids employed as a novel organic nitrogen source for the preparation of PGPF-containing bio-organic fertilizer for plant growth promotion and characterization of substance transformation during BOF production. PLoS ONE 2016, 11, e0149447. [Google Scholar] [CrossRef]
- Kang, S.M.; Shahzad, R.; Bilal, S.; Khan, A.L.; Park, Y.G.; Lee, K.E.; Asaf, S.; Khan, M.A.; Lee, I.J. Indole-3-acetic-acid and ACC deaminase producing Leclercia adecarboxylata MO1 improves Solanum lycopersicum L. growth and salinity stress tolerance by endogenous secondary metabolites regulation. BMC Microbiol. 2019, 19, 80. [Google Scholar] [CrossRef]
- Tarroum, M.; Romdhane, W.B.; Al-Qurainy, F.; Ali, A.A.M.; Al-Doss, A.; Fki, L.; Hassairi, A. A novel PGPF Penicillium olsonii isolated from the rhizosphere of Aeluropus littoralis promotes plant growth, enhances salt stress tolerance, and reduces chemical fertilizers inputs in hydroponic system. Front. Microbiol. 2022, 13, 996054. [Google Scholar] [CrossRef] [PubMed]
- Palberg, D.; Kisiała, A.; Jorge, G.L.; Emery, R.J.N. A survey of Methylobacterium species and strains reveals widespread production and varying profiles of cytokinin phytohormones. BMC Microbiol. 2022, 22, 49. [Google Scholar] [CrossRef]
- Guo, D.J.; Singh, P.; Yang, B.; Singh, R.K.; Verma, K.K.; Sharma, A.; Khan, Q.; Qin, Y.; Chen, T.S.; Song, X.P.; et al. Complete genome analysis of sugarcane root associated endophytic diazotroph Pseudomonas aeruginosa DJ06 revealing versatile molecular mechanism involved in sugarcane development. Front. Microbiol. 2023, 14, 1096754. [Google Scholar] [CrossRef]
- Shahzad, R.; Khan, A.L.; Bilal, S.; Waqas, M.; Kang, S.; Lee, I. Inoculation of abscisic acid-producing endophytic bacteria enhances salinity stress tolerance in Oryza sativa. Environ. Exp. Bot. 2017, 136, 68–77. [Google Scholar] [CrossRef]
- Singh, V.; Singh, P.N.; Yadav, R.L.; Awasthi, S.K.; Joshi, B.B.; Singh, R.K.; Lal, R.J.; Duttamajumder, S.K. Increasing the efficacy of Trichoderma harzianum for nutrient uptake and control of red rot in sugarcane. J. Hortic. For. 2010, 2, 66–71. [Google Scholar]
- Li, Z.; Bai, T.; Dai, L.; Wang, F.; Tao, J.; Meng, S.; Hu, Y.; Wang, S.; Hu, S. A study of organic acid production in contrasts between two phosphate solubilizing fungi: Penicillium oxalicum and Aspergillus niger. Sci. Rep. 2016, 6, 25313. [Google Scholar] [CrossRef]
- Murali, M.; Naziya, B.; Ansari, M.A.; Alomary, M.N.; AlYahya, S.; Almatroudim, A.; Thriveni, M.C.; Gowtham, H.G.; Singh, S.B.; Aiyaz, M.; et al. Bioprospecting of rhizosphere-resident fungi: Their role and importance in sustainable agriculture. J. Fungi 2021, 7, 314. [Google Scholar] [CrossRef]
- Yadav, R.L.; Shukla, S.K.; Suman, A.; Singh, P.N. Trichoderma inoculation and trash management effects on soil microbial biomass, soil respiration, nutrient uptake and yield of ratoon sugarcane under subtropical conditions. Biol. Fertil. Soils 2009, 45, 461–468. [Google Scholar] [CrossRef]
- Fiorentino, N.; Ventorino, V.; Woo, S.L.; Pepe, O.; De Rosa, A.; Gioia, L.; Romano, I.; Lombardi, N.; Napolitano, M.; Colla, G.; et al. Trichoderma-based biostimulants modulate rhizosphere microbial populations and improve N uptake efficiency, yield, and nutritional quality of leafy vegetables. Front. Plant Sci. 2018, 9, 743. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, D.; Cao, D.; Chen, J.; Wei, X. Exploring the potentials of Sesuvium portulacastrum L. for edibility and bioremediation of saline soils. Front. Plant Sci. 2024, 15, 1387102. [Google Scholar] [CrossRef] [PubMed]
- Ayyappan, D.; Sathiyaraj, G.; Ravindran, K.C. Phytoextraction of heavy metals by Sesuvium portulacastrum L. a salt marsh halophyte from tannery effluent. Int. J. Phytoremediat. 2016, 18, 453–459. [Google Scholar] [CrossRef]
- Feng, J.; Lin, Y.; Yang, Y.; Shen, Q.; Huang, J.; Wang, S.; Zhu, X.; Li, Z. Tolerance and bioaccumulation of Cd and Cu in Sesuvium portulacastrum. Ecotoxicol. Environ. Saf. 2018, 147, 306–312. [Google Scholar] [CrossRef]
- Uddin, M.M.; Chen, Z.F.; Huang, L.F. Cadmium accumulation, subcellular distribution and chemical fractionation in hydroponically grown Sesuvium portulacastrum [Aizoaceae]. PLoS ONE. 2020, 15, e0244085. [Google Scholar] [CrossRef]
- Liu, X.H.; Pu, X.M.; Luo, D.L.; Lu, J.; Liu, Z.L. Model assessment of nutrient removal via planting Sesuvium portulacastrum in floating beds in eutrophic marine waters: The case of aquaculture areas of Dongshan Bay. Acta Oceanol. Sin. 2019, 38, 91–100. [Google Scholar] [CrossRef]
- Senff, P.; Blanc, P.P.; Slater, M.; Kunzmann, A. Low-technology recirculating aquaculture system integrating milkfish Chanos chanos, sea cucumber Holothuria scabra and sea purslane Sesuvium portulacastrum. Aquac. Environ. Int. 2020, 12, 471–484. [Google Scholar] [CrossRef]
- Yang, N.; Zhang, W.; Wang, D.; Cao, D.; Cao, Y.; He, W.; Lin, Z.; Chen, X.; Ye, G.; Chen, Z.; et al. A novel endophytic fungus strain of Cladosporium: Its identification, genomic analysis, and effects on plant growth. Front. Microbiol. 2023, 14, 1287582. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Dewey, C.N.; Li, B. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011, 12, 323. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wang, D.; Yang, N.; Zhang, C.; He, W.; Ye, G.; Chen, J.; Wei, X. Transcriptome analysis reveals molecular mechanisms underlying salt tolerance in halophyte Sesuvium portulacastrum. Front. Plant Sci. 2022, 13, 973419. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Akhter, W.; Bhuiyan, M.K.A.; Sultana, F.; Hossain, M.M. Integrated effect of microbial antagonist, organic amendment and fungicide in controlling seedling mortality (Rhizoctonia solani) and improving yield in pea (Pisum sativum L.). Comptes Rendus Biol. 2015, 338, 21–28. [Google Scholar] [CrossRef]
- Vejan, P.; Abdullah, R.; Khadiran, T.; Ismail, S.; Nasrulhaq, B.A. Role of plant growth promoting rhizobacteria in agricultural sustainability-A review. Molecules 2016, 21, 573. [Google Scholar] [CrossRef] [PubMed]
- Sahu, P.K.; Singh, D.P.; Prabha, R.; Meena, K.K.; Abhilash, P.C. Connecting microbial capabilities with the soil and plant health: Options for agricultural sustainability. Ecol. Indic. 2018, 105, 601–612. [Google Scholar] [CrossRef]
- Zhang, C.; Yu, Z.; Zhang, M.; Li, X.; Wang, M.; Li, L.; Li, X.; Ding, Z.; Tian, H. Serratia marcescens PLR enhances lateral root formation through supplying PLR-derived auxin and enhancing auxin biosynthesis in Arabidopsis. J. Exp. Bot. 2022, 73, 3711–3725. [Google Scholar] [CrossRef]
- Lacombe, B.; Achard, P. Long-distance transport of phytohormones through the plant vascular system. Curr. Opin. Plant Biol. 2016, 34, 1–8. [Google Scholar] [CrossRef]
- Rehan, M.; Al-Turki, A.; Abdelmageed, A.H.A.; Abdelhameid, N.M.; Omar, A.F. Performance of Plant-Growth-Promoting Rhizobacteria (PGPR) isolated from sandy soil on growth of tomato (Solanum lycopersicum L.). Plants 2023, 12, 1588. [Google Scholar] [CrossRef]
- Paul, S.; Rakshit, A. Evaluating lignification, antioxidative defense, and physiochemical changes in soybean through biopriming under graded soil fertilization. J. Soil Sci. Plant Nutr. 2022, 22, 2295–2306. [Google Scholar] [CrossRef]
- Xu, G.; Fan, X.; Miller, A.J. Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 2012, 63, 153–182. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cao, H.; Li, S.; Dong, X.; Zhao, Z.; Jia, Z.; Yuan, L. Genetic and molecular mechanisms underlying nitrogen use efficiency in maize. J. Genet. Genom. 2025, 52, 276–286. [Google Scholar] [CrossRef] [PubMed]
| Gene ID | Log2FoldChange | Description |
|---|---|---|
| DEGs expressed in roots | ||
| Cluster-1879.21594 | 3.61 | High-affinity nitrate transporter 2.5 OS = Arabidopsis thaliana |
| Cluster-1879.22814 | 2.62 | High-affinity nitrate transporter 2.5 OS = Arabidopsis thaliana |
| Cluster-1879.23037 | 1.59 | Probable peptide/nitrate transporter At3g43790 OS = Arabidopsis thaliana |
| Cluster-1879.39351 | 1.15 | Probable peptide/nitrate transporter At3g43790 OS = Arabidopsis thaliana |
| Cluster-1879.56503 | −1.84 | High-affinity nitrate transporter 2.6 OS = Arabidopsis thaliana |
| Cluster-1879.56504 | −1.64 | High-affinity nitrate transporter 2.4 OS = Arabidopsis thaliana |
| Cluster-1879.26502 | 1.49 | Ammonium Transporter Family/Translocation protein Sec62 |
| Cluster-1879.62558 | 1.48 | Glutamine synthetase type III N terminal |
| Cluster-1879.41276 | 1.79 | Glutamate synthase 1 [NADH], chloroplastic-like isoform X3 [Chenopodium quinoa] |
| Cluster-1879.28631 | 5.68 | Glutamate synthase 1 [NADH], chloroplastic [Vitis vinifera] |
| Cluster-1879.38837 | 3.81 | PREDICTED: Chenopodium quinoa glutamate synthase 1 [NADH], chloroplastic-like (LOC110728845), transcript variant X1, mRNA |
| Cluster-1879.43022 | 2.64 | PREDICTED: Beta vulgaris subsp. vulgaris ferredoxin-dependent glutamate synthase, chloroplastic (LOC104889481), mRNA |
| Cluster-1879.35615 | 2.20 | Nitrate reductase [NADH] OS = Spinacia oleracea OX = 3562 GN = NIA PE = 2 SV = 1 |
| DEGs expressed in the aboveground part | ||
| Cluster-1879.41276 | 1.79 | Glutamate synthase 1 [NADH], chloroplastic OS = Arabidopsis thaliana |
| Cluster-1879.38837 | 3.81 | Glutamate synthase 1 [NADH], chloroplastic OS = Arabidopsis thaliana |
| Cluster-1879.28631 | 5.68 | Glutamate synthase 2 [NADH], chloroplastic OS = Oryza sativa subsp. japonica |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.; Zhang, W.; Cao, D.; Wei, X. Transcriptome Analysis Elucidates the Mechanism of an Endophytic Fungus Cladosporium sp. ‘BF-F’ in Enhancing the Growth of Sesuvium portulacastrum. Agriculture 2025, 15, 1522. https://doi.org/10.3390/agriculture15141522
Wang D, Zhang W, Cao D, Wei X. Transcriptome Analysis Elucidates the Mechanism of an Endophytic Fungus Cladosporium sp. ‘BF-F’ in Enhancing the Growth of Sesuvium portulacastrum. Agriculture. 2025; 15(14):1522. https://doi.org/10.3390/agriculture15141522
Chicago/Turabian StyleWang, Dan, Wenbin Zhang, Dinging Cao, and Xiangying Wei. 2025. "Transcriptome Analysis Elucidates the Mechanism of an Endophytic Fungus Cladosporium sp. ‘BF-F’ in Enhancing the Growth of Sesuvium portulacastrum" Agriculture 15, no. 14: 1522. https://doi.org/10.3390/agriculture15141522
APA StyleWang, D., Zhang, W., Cao, D., & Wei, X. (2025). Transcriptome Analysis Elucidates the Mechanism of an Endophytic Fungus Cladosporium sp. ‘BF-F’ in Enhancing the Growth of Sesuvium portulacastrum. Agriculture, 15(14), 1522. https://doi.org/10.3390/agriculture15141522







