Genetic Diversity of Olive (Olea europaea L.) Cultivars Assessed by Genotyping-by-Sequencing in Southern Peru
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Plant Material Collection
2.2. Morphological Characterization
- Tree traits (3): vigor, growth habit, canopy density.Tree vigor refers to the overall abundance of vegetative growth, encompassing canopy development in both height and volume [15]. Vigor was classified into three categories based on tree height and trunk diameter, measured at the time of phenotypic evaluation:
- -
- Low: trees with a height < 2.5 m and trunk diameter < 10 cm;
- -
- Medium: trees with a height between 2.5 and 3.5 m and diameter between 10 and 15 cm;
- -
- High: trees with a height > 3.5 m and trunk diameter > 15 cm.
The classification was performed during the same phenotypic evaluation period, and only trees at least 5 years old were considered to minimize the influence of developmental stage.Growth habit describes the natural orientation and structure of branches and shoots [15]. Growth habit was classified into two categories based on branch orientation and canopy architecture:- -
- Open: characterized by an initially orthogeotropic branching pattern, where primary branches develop outward or at oblique angles, resulting in a spreading canopy structure.
- -
- Upright: defined by branches that grow predominantly in a vertical direction, with strong apical dominance and a compact, vertical canopy form.
Canopy density refers to the overall abundance of canopy vegetation and was assessed visually based on the extent of foliage coverage and light penetration [15]. It was classified into three categories:- -
- Sparse: Associated with fast-growing cultivars with long internodes. Gaps in the canopy allow visible light to pass through from multiple angles.
- -
- Middle: Typical of the species; while vegetation is abundant, internode length allows some light diffusion and partial shading inside the canopy.
- -
- Dense: Typical of cultivars with short internodes and abundant branching. The canopy appears compact, with inner parts deeply shaded due to foliage density.
- Leaf traits (4): shape, length, width, longitudinal curvature.
- Fruit traits (10): weight, shape, symmetry (position A), apex, base, nipple presence, lenticel presence and size, ripening color, maximum transverse diameter (position B).
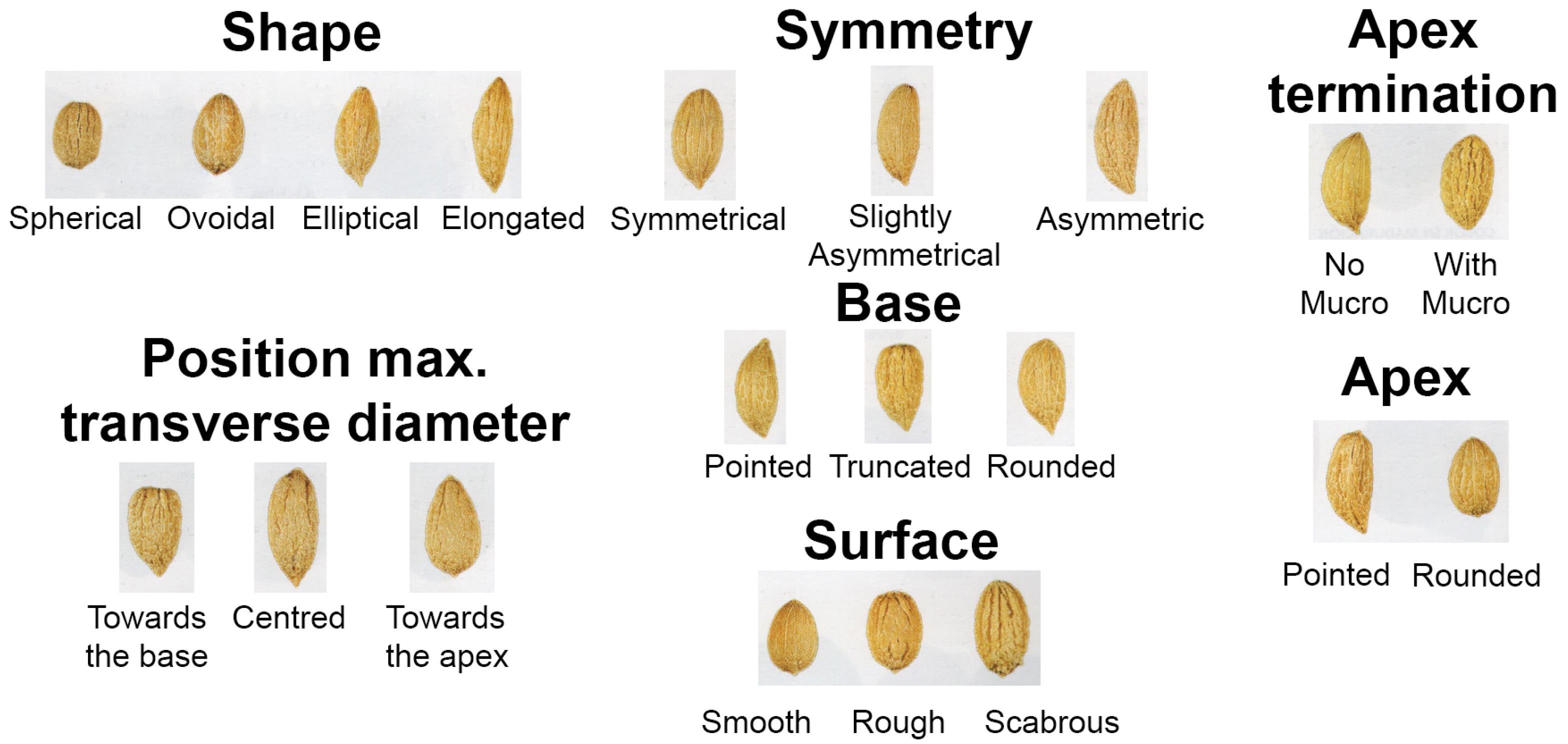
- Endocarp traits (8): shape, symmetry (positions A and B), apex, base, surface texture, number of fibrovascular grooves, apex termination.
2.3. DNA Extraction and Quality Assessment
2.4. Genotyping by Sequencing (GBS) and Bioinformatics Analysis
3. Results
3.1. Morphological Variation Among Olive Varieties
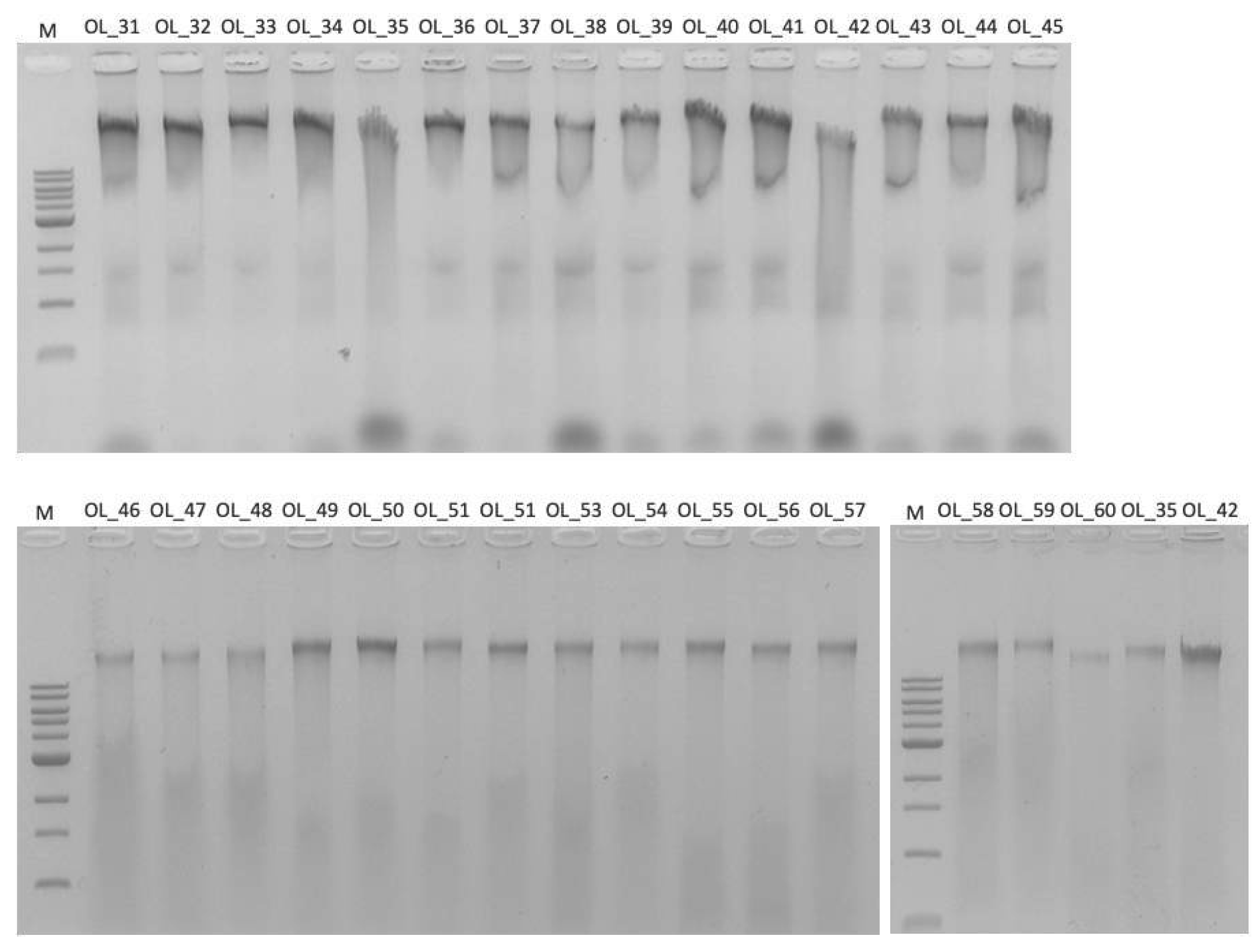
3.2. DNA Extraction and Protocol Optimization
3.3. Sequencing and Quality Control
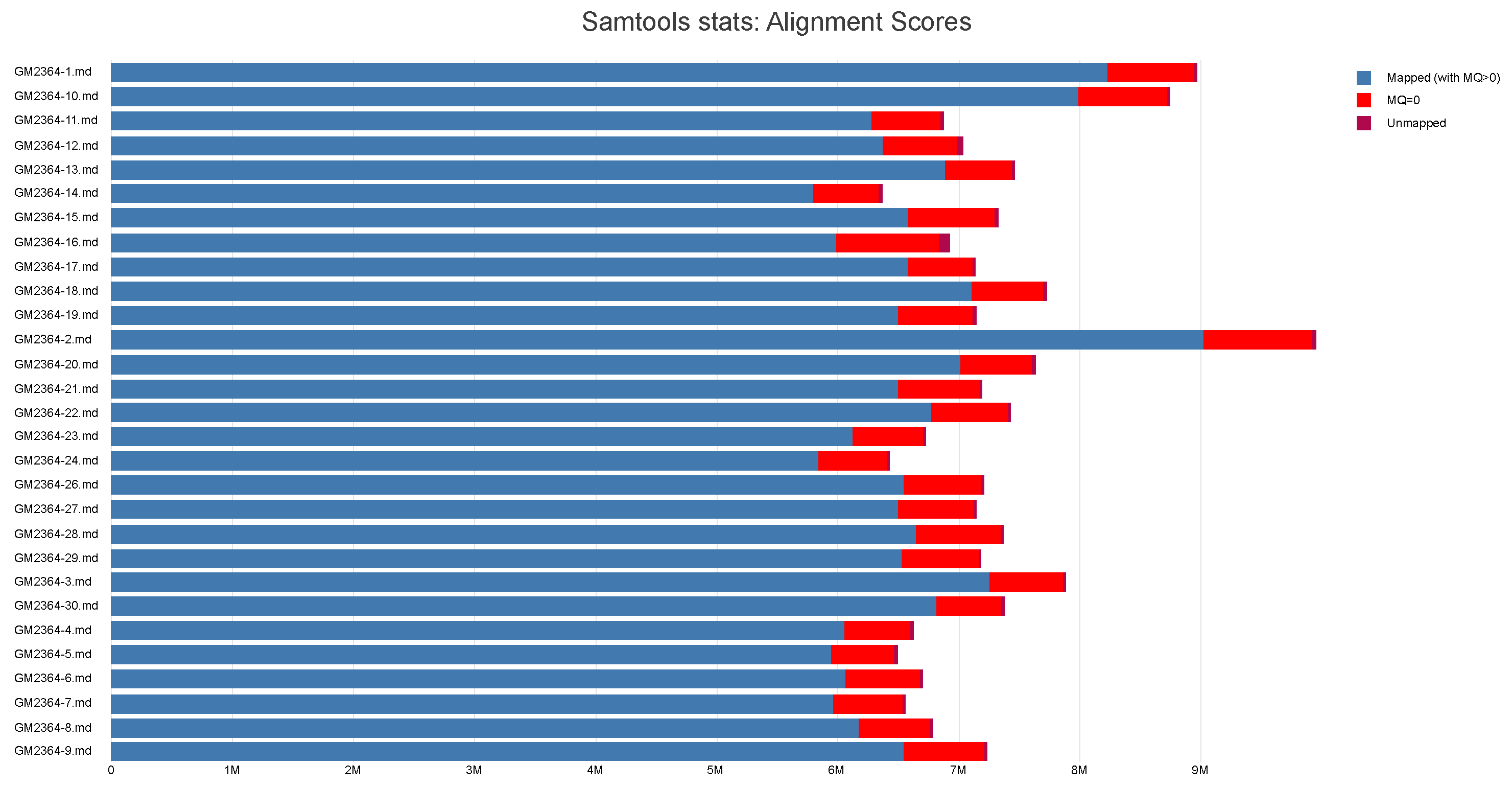
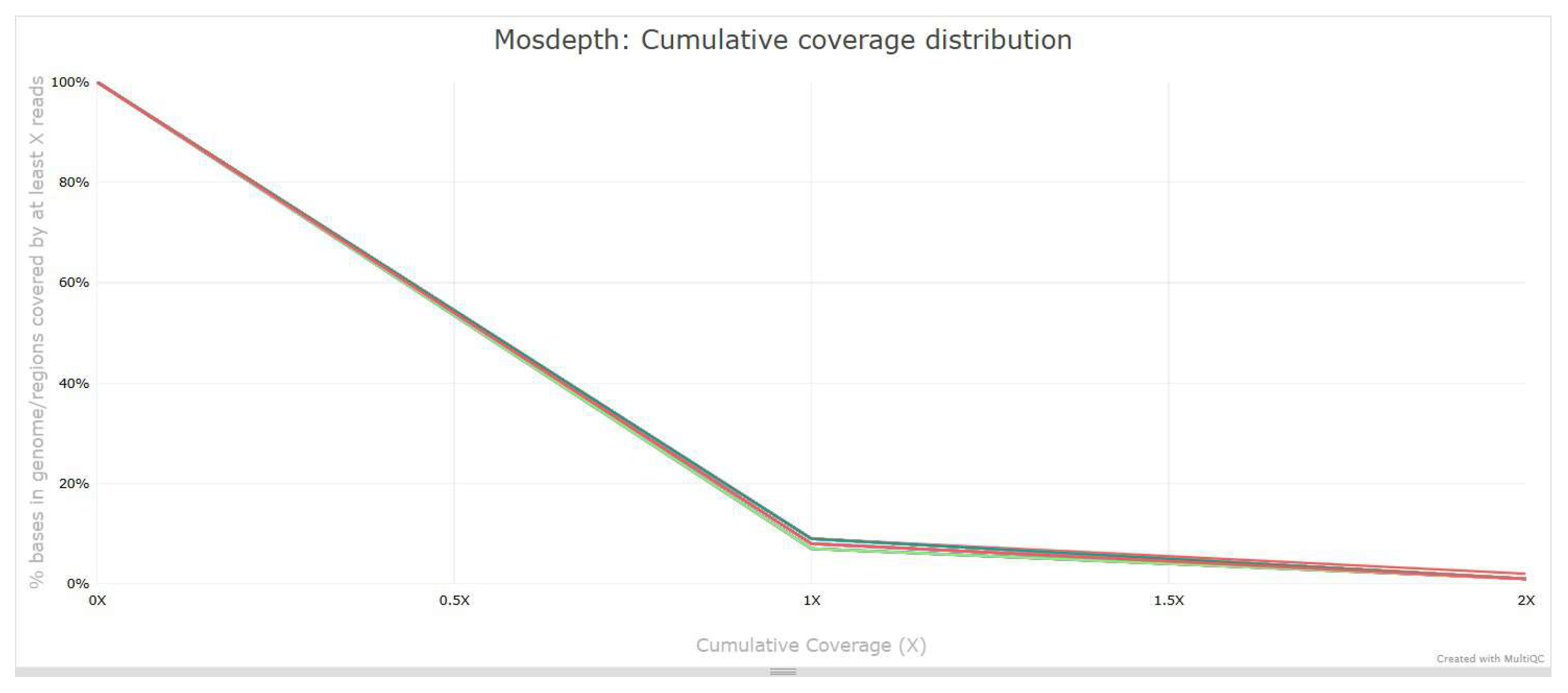
3.4. Alignment and Coverage Analysis
3.5. Duplicate Read Analysis
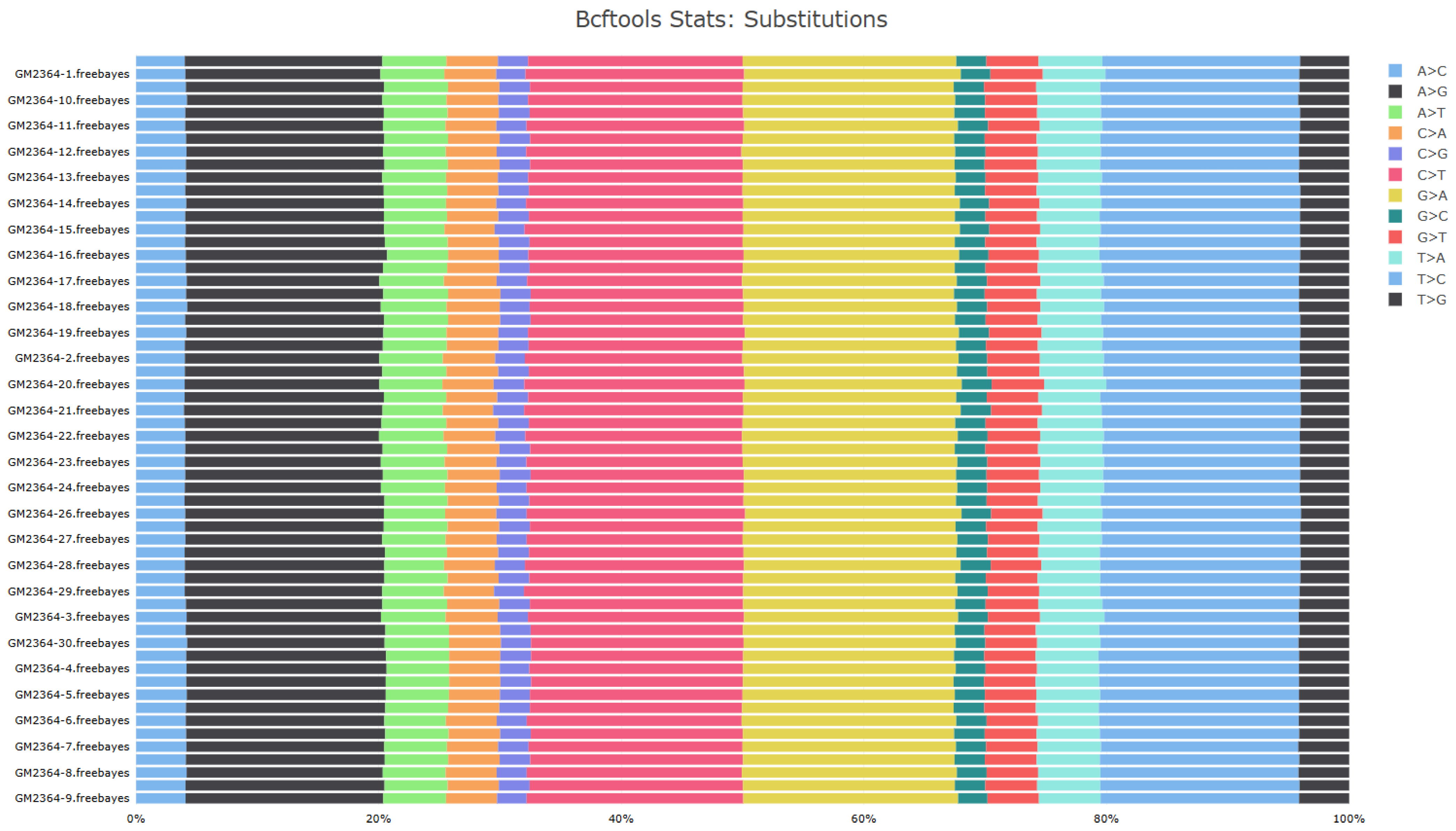
3.6. Genetic Variant Identification and Diversity Analysis
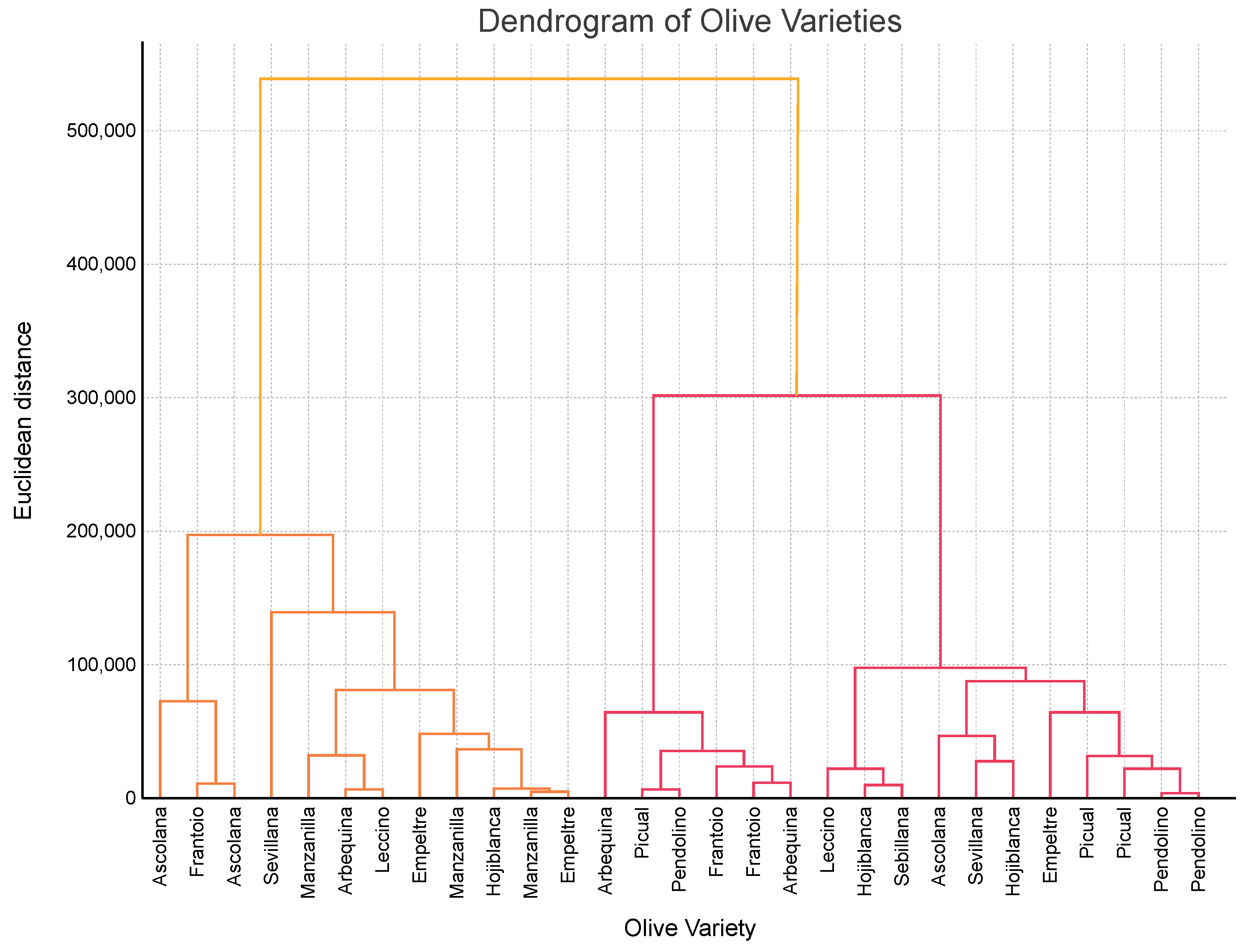
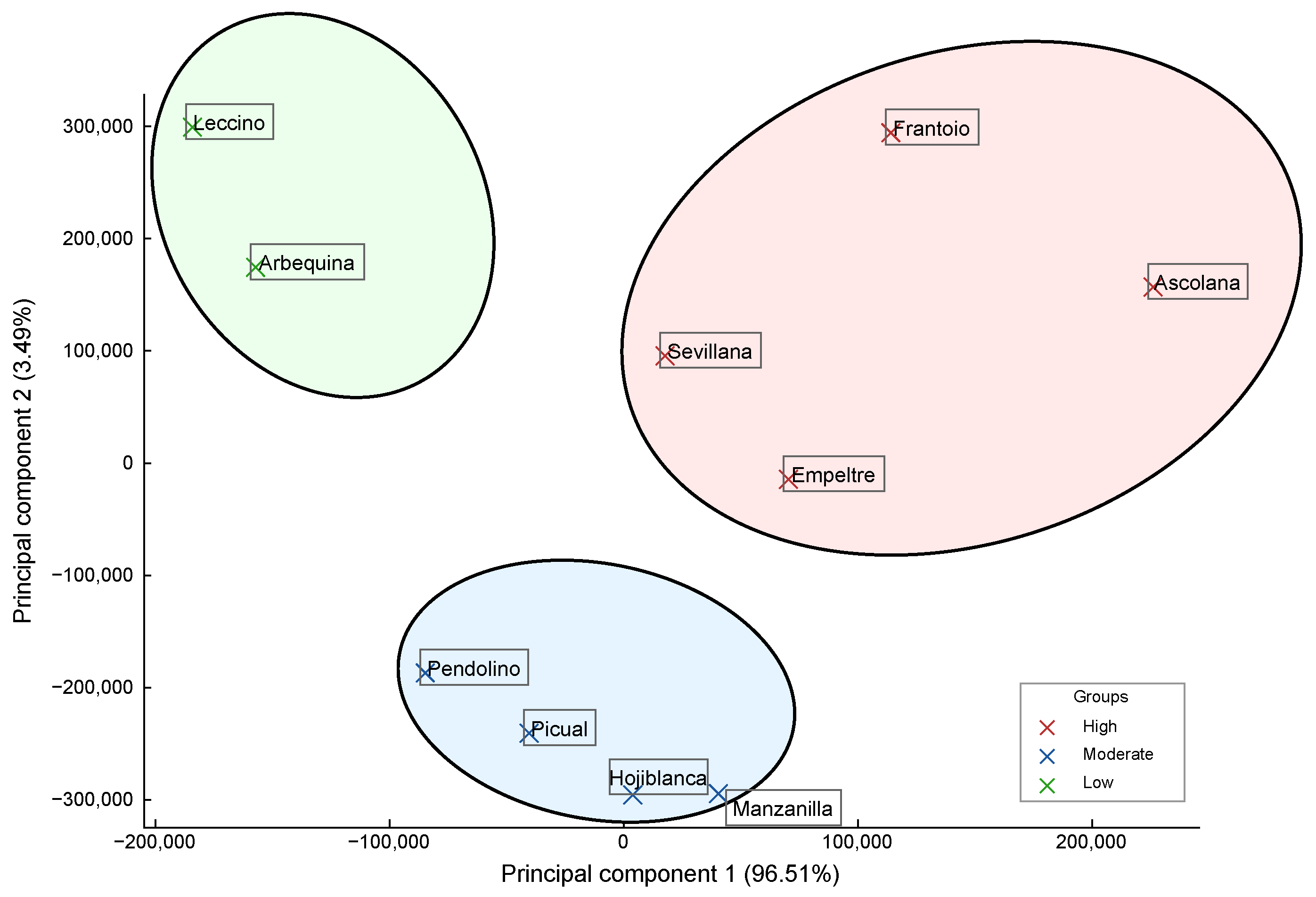
3.7. Clustering of Genetic Diversity
- Group 1—High Genetic Variability: Ascolana, Frantoio, Empeltre, and Sevillana, characterized by high SNPs, haplotypes, and indels.
- Group 2—Moderate Genetic Variability: Manzanilla, Hojiblanca, Picual, and Pendolino, with intermediate genetic parameters.
- Group 3—Low Genetic Variability: Arbequina and Leccino, showing lower SNP and indel counts.
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Breton, C.; Guerin, J.; Ducatillion, C.; Médail, F.; Kull, C.A.; Bervillé, A. Taming the Wild and ‘Wilding’ the Tame: Tree Breeding and Dispersal in Australia and the Mediterranean. Plant Sci. 2008, 175, 197–205. [Google Scholar] [CrossRef]
- Laval, H.; Leveau, P.; Marinval, P.; Medus, J. L’olivier et sa culture en Provence. Donnees historiques, palynologiques et carpologiques [archéobotanique]. Ecol. Mediterr. 1990, 16, 427–435. [Google Scholar]
- Leveau, P.; Heinz, C.; Laval, H.; Marinval, P.; Medus, J. Les origines de l’oléiculture en Gaule du Sud. Revue d’Archéométrie 1991, 15, 83–94. [Google Scholar] [CrossRef]
- Diez, C.M.; Trujillo, I.; Martinez-Urdiroz, N.; Barranco, D.; Rallo, L.; Marfil, P.; Gaut, B.S. Olive Domestication and Diversification in the Mediterranean Basin. New Phytol. 2015, 206, 436–447. [Google Scholar] [CrossRef]
- Besnard, G.; Khadari, B.; Navascués, M.; Fernández-Mazuecos, M.; El Bakkali, A.; Arrigo, N.; Baali-Cherif, D.; Brunini-Bronzini de Caraffa, V.; Santoni, S.; Vargas, P.; et al. The Complex History of the Olive Tree: From Late Quaternary Diversification of Mediterranean Lineages to Primary Domestication in the Northern Levant. Proc. R. Soc. B 2013, 280, 20122833. [Google Scholar] [CrossRef]
- Turcato, A.; Mattar, S. Olive oils from South America. In Olive Oil Sensory Science; Willett, W., Ed.; Wiley: Hoboken, NJ, USA, 2014; pp. 337–357. [Google Scholar]
- León, L.; Casanova, D.; Palma, J.; González, J. Caracterización agromorfológica de plantas madre del banco de germoplasma de “olivo” Olea europaea (Oleaceae) en la región Tacna. Arnaldoa 2021, 28, 593–612. [Google Scholar]
- Morgil, H.; Gercek, Y.C.; Tulum, I. Single Nucleotide Polymorphisms (SNPs) in Plant Genetics and Breeding. In The Recent Topics in Genetic Polymorphisms; InTech: London, UK, 2020; p. 53. [Google Scholar]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A Robust, Simple Genotyping-by-Sequencing (GBS) Approach for High Diversity Species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Cruz, F.; Julca, I.; Gómez-Garrido, J.; Loska, D.; Marcet-Houben, M.; Cano, E.; Galán, B.; Frias, L.; Ribeca, P.; Derdak, S.; et al. Genome Sequence of the Olive Tree, Olea europaea. Gigascience 2016, 5, 29. [Google Scholar] [CrossRef]
- Gómez-Gálvez, F.J.; Ninot, A.; Rodríguez, J.C.; Compañ, S.P.; Andreva, J.U.; Rubio, J.A.G.; Aragón, I.P.; Viñuales-Andreu, J.; Casanova-Gascón, J.; Šatović, Z.; et al. New Insights in the Spanish Gene Pool of Olive (Olea europaea L.) Preserved Ex Situ and In Situ Based on High-Throughput Molecular Markers. Front. Plant Sci. 2023, 14, 1267601. [Google Scholar] [CrossRef]
- Unver, T.; Wu, Z.; Sterck, L.; Turktas, M.; Lohaus, R.; Li, Z.; Yang, M.; He, L.; Deng, T.; Escalante, F.J.; et al. Genome of Wild Olive and the Evolution of Oil Biosynthesis. Proc. Natl. Acad. Sci. USA 2017, 114, E9413–E9422. [Google Scholar] [CrossRef]
- Besnard, G.; Hernández, P.; Khadari, B.; Dorado, G.; Savolainen, V. Genomic Profiling of Plastid DNA Variation in the Mediterranean Olive Tree. BMC Plant Biol. 2011, 11, 80. [Google Scholar] [CrossRef] [PubMed]
- Ministerio de Agricultura. Aceituna: Un Campo fértil Para Sus Inversiones y el Desarrollo de Sus Exportaciones; Gobierno del Perú: Lima, Peru, 2020. Available online: https://www.midagri.gob.pe/portal/download/pdf/herramientas/cendoc/manuales-boletines/aceituna/aceituna_feb11.pdf (accessed on 10 January 2025).
- International Union for the Protection of New Varieties of Plants. Olive UPOV Code: OLEAA_EUR Olea europaea L. Guidelines for the Conduct of Tests for Distinctness, Uniformity and Stability. 2011. Available online: https://www.upov.int/edocs/tgdocs/en/tg099.pdf (accessed on 23 September 2024).
- Nolan, K.A.; Callahan, J.E. Beachcomber Biology: The Shannon-Weiner Species Diversity Index. Proc. Workshop ABLE 2006, 27, 334–338. [Google Scholar]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.scienceopen.com/book?vid=45ad53d9-9259-4c9a-b773-54a1bd5f0287 (accessed on 12 September 2024).
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An Ultra-fast All-in-one FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows–Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce Framework for Analyzing Next-Generation DNA Sequencing Data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Garrison, E.; Marth, G. Haplotype-based Variant Detection from Short-read Sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- Pedersen, B.S.; Quinlan, A.R. Mosdepth: Quick Coverage Calculation for Genomes and Exomes. Bioinformatics 2018, 34, 867–868. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The Variant Call Format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Bartolini, G.; Prevost, G.; Messeri, C.; Carignani, G. Olive Germplasm: Cultivars and World-Wide Collections; FAO Library: Rome, Italy, 1998; 459p. [Google Scholar]
- Trujillo, I.; Ojeda, M.A.; Urdiroz, N.M.; Potter, D.; Barranco, D.; Rallo, L.; Diez, C.M. Identification of the Worldwide Olive Germplasm Bank of Córdoba (Spain) Using SSR and Morphological Markers. Tree Genet. Genomes 2014, 10, 141–155. [Google Scholar] [CrossRef]
- Belaj, A.; Muñoz-Díez, C.; Baldoni, L.; Satovic, Z.; Barranco, D. Genetic Diversity and Relationships of Wild and Cultivated Olives at Regional Level in Spain. Sci. Hortic. 2010, 124, 323–330. [Google Scholar] [CrossRef]
- Baldoni, L.; Cultrera, N.G.; Mariotti, R.; Ricciolini, C.; Arcioni, S.; Vendramin, G.G.; Buonamici, A.; Porceddu, A.; Sarri, V.; Ojeda, M.A.; et al. A Consensus List of Microsatellite Markers for Olive Genotyping. Mol. Breed. 2009, 24, 213–231. [Google Scholar] [CrossRef]
- Besnard, G.; Terral, J.-F.; Cornille, A. On the Origins and Domestication of the Olive: A Review and Perspectives. Ann. Bot. 2018, 121, 587–603. [Google Scholar] [CrossRef]
- Zhu, S.; Niu, E.; Shi, A.; Mou, B. Genetic Diversity Analysis of Olive Germplasm (Olea europaea L.) with Genotyping-by-Sequencing Technology. Front. Genet. 2019, 10, 755. [Google Scholar] [CrossRef]
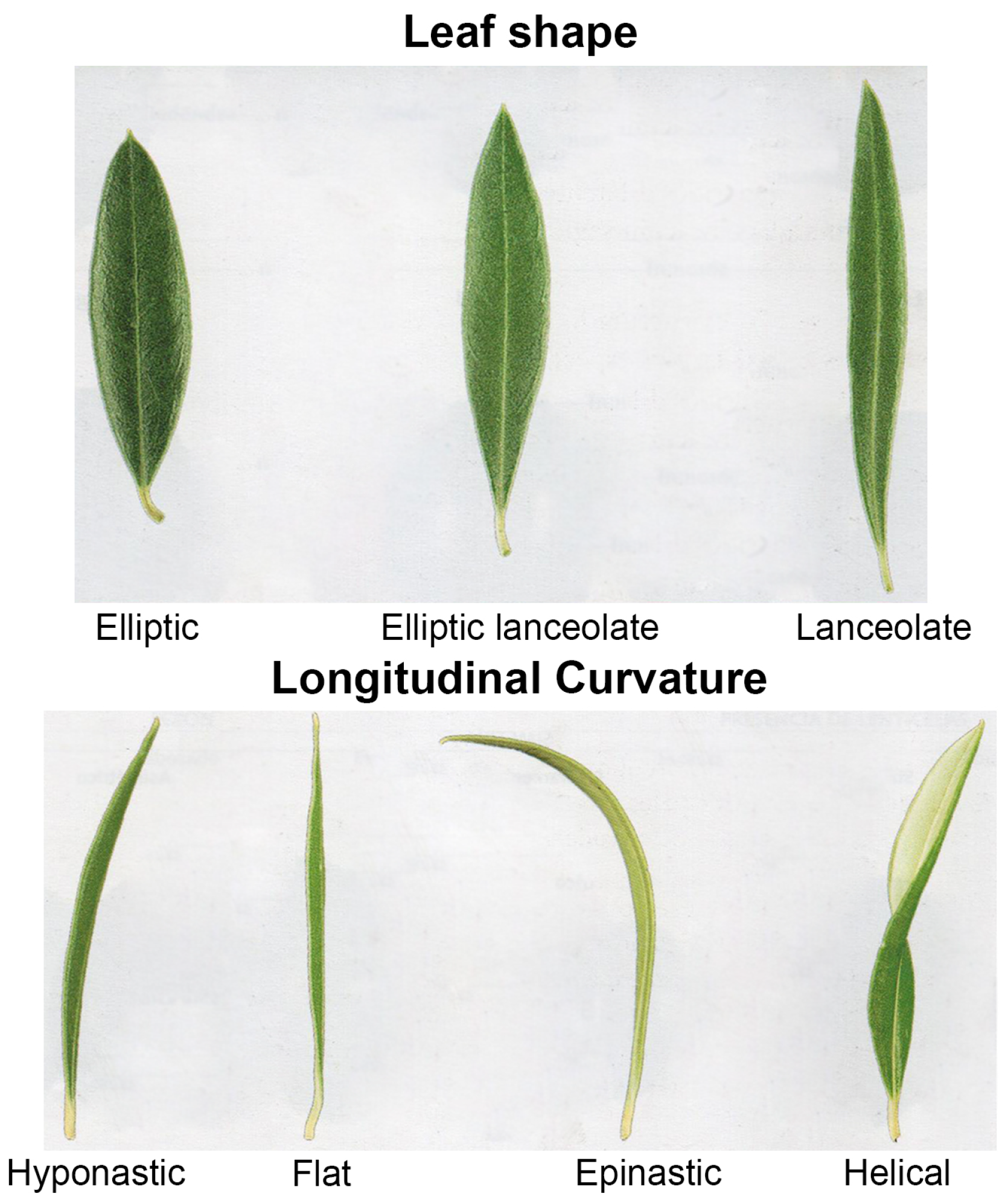


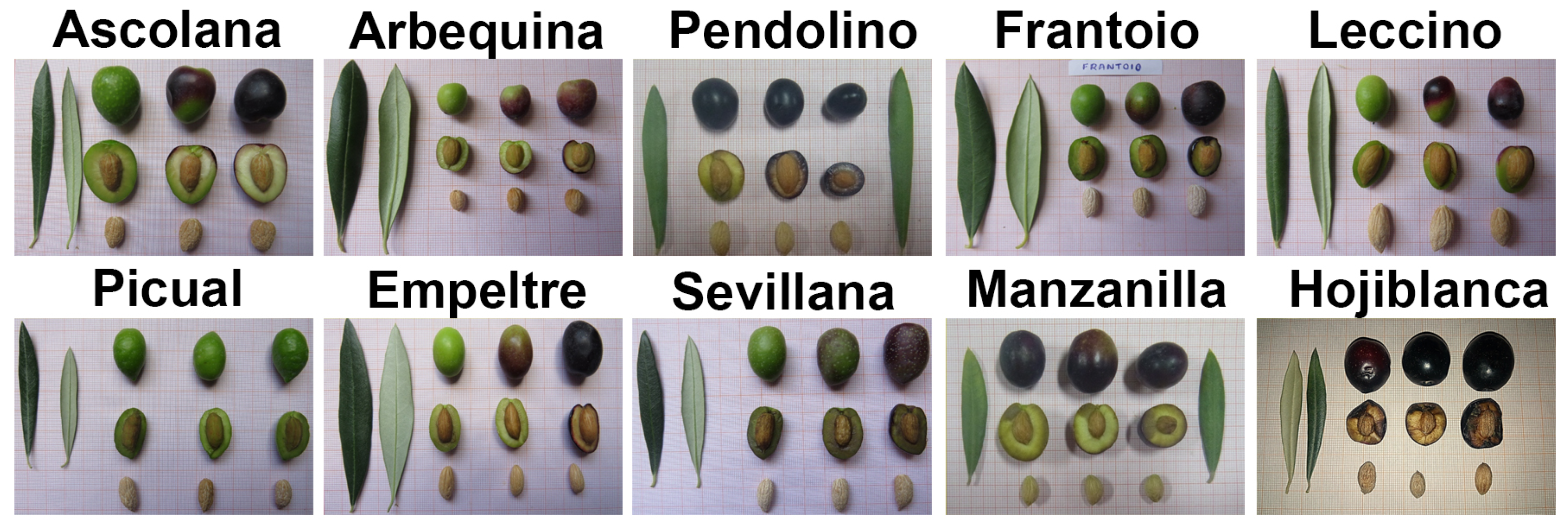
| Nº | Variety | Vigor | Growth Habit | Canopy Density | Leaf Shape | Length | Width | Longitudinal Curvature of the Leaf | Weight | Fruit Shape | Degree of Symmetry (Position A) | Diameter Transverse Maximum (Position B) | Apex | Base | Nipple | Presence of Lenticels | Size of Lenticels | Maturing Color |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Ascolana tenera | low | open | middle | lanceolate | large | middle | epinastic | very elevated | ovoid | slightly asymm. | centered | pointed | truncated | absent | abundant | large | violet |
| 2 | Arbequina | low | open | middle | elliptic | short | middle | epinastic | low | spherical | symmetric | base | redounded | truncated | absent | sparse | small | black |
| 3 | Pendolino | low | open | middle | elliptic lanceolate | middle | narrow | flat | middle | ovoid | slightly asymm. | centered | redounded | truncated | absent | abundant | small | black |
| 4 | Frantoio | high | open | middle | elliptic lanceolate | middle | middle | epinastic | middle | ovoid | slightly asymm. | apex | redounded | redounded | absent | abundant | small | violet |
| 5 | Leccino | high | open | middle | elliptic lanceolate | middle | middle | flat | middle | ovoid | slightly asymm. | centered | redounded | truncated | absent | abundant | small | black |
| 6 | Picual | middle | open | dense | elliptic lanceolate | middle | middle | hyponastic | middle | ovoid | asymm. | centered | rounded | truncated | absent | abundant | small | black |
| 7 | Empeltre | middle | upright | dense | elliptic lanceolate | middle | middle | flat | middle | elongated | slightly asymm. | centered | rounded | truncated | absent | abundant | small | black |
| 8 | Sevillana | high | open | dense | lanceolate | large | middle | flat | very high | ovoid | asymm. | centered | pointed | truncated | absent | abundant | small | black |
| 9 | Manzanilla | middle | open | middle | elliptic | middle | middle | flat | raised | spherical | symmetric | centered | rounded | truncated | absent | abundant | small | black |
| 10 | Hojiblanca | high | upright | middle | lanceolate | large | middle | flat | raised | ovoid | symm. | centered | rounded | truncated | absent | abundant | small | violet |
| Commercial Use | Variety | Shape | Symmetry (A and B) | Position Max. Transverse Diameter | Apex | Base | Surface | Fibrovascular Grooves | Apex Termination |
|---|---|---|---|---|---|---|---|---|---|
| Table | Ascolana tenera | elliptical | slightly asymmetrical | centered | pointed | pointed | scabrous | high | with mucro |
| Oil | Arbequina | ovoidal | symmetrical | centered | rounded | rounded | rough | medium | with mucro |
| Pendolino | elliptical | symmetrical | towards the apex | rounded | rounded | smooth | medium | with mucro | |
| Frantoio | elliptical | symmetrical | centered | rounded | rounded | smooth | low | with mucro | |
| Leccino | elliptical | asymmetric | centered | rounded | rounded | rough | high | with mucro | |
| Picual | elliptical | asymmetric | centered | pointed | rounded | scabrous | medium | with mucro | |
| Double aptitude (table and oil) | Empeltre | elongated | asymmetric | towards the apex | pointed | pointed | rough | high | with mucro |
| Sevillana | elliptical | asymmetric | towards the base | pointed | truncated | rough | high | with mucro | |
| Manzanilla | ovoidal | slightly asymmetrical | towards the apex | rounded | pointed | rough | medium | with mucro | |
| Hojiblanca | elliptical | slightly asymmetrical | centered | rounded | rounded | rough | medium | with mucro |
| Commercial Destination | Varieties | Weight (g) | Length (mm) | Diameter (mm) | Number of Fibrovascular Grooves |
|---|---|---|---|---|---|
| Table | Ascolana tenera | 3.01 | 18.16 | 8.51 | 14 |
| Oil | Arbequina | 0.36 | 10.92 | 7.25 | 10 |
| Pendolino | 0.48 | 14.59 | 7.51 | 10 | |
| Frantoio | 0.46 | 14.42 | 7.31 | 6 | |
| Leccino | 0.79 | 18.10 | 7.22 | 11 | |
| Picual | 2.98 | 18.01 | 7.23 | 10 | |
| Double option (table and oil) | Empeltre | 0.84 | 20.92 | 8.71 | 12 |
| Sevillana | 0.95 | 18.93 | 8.31 | 14 | |
| Manzanilla | 0.55 | 13.85 | 7.55 | 10 | |
| Hojiblanca | 0.66 | 15.32 | 7.36 | 9 |
| Morphological Characteristics | Characteristic to Be Evaluated | Number of Parameters | Shannon Index |
|---|---|---|---|
| Endocarp (Stone) | Weight | 4 | 2.13 |
| Shape | 4 | 2.30 | |
| Degree of symmetry | 3 | 2.23 | |
| Transverse diameter | 3 | 2.28 | |
| Apex | 2 | 2.27 | |
| Base | 3 | 2.29 | |
| Surface | 3 | 2.26 | |
| Furrows | 3 | 2.28 | |
| Furrow distribution | 2 | 2.27 | |
| Apex termination | 2 | 2.27 | |
| Fruit | Weight | 4 | 2.18 |
| Shape | 3 | 2.30 | |
| Degree of symmetry | 3 | 2.23 | |
| Maximum transverse diameter | 3 | 2.28 | |
| Apex | 2 | 2.27 | |
| Base | 2 | 2.27 | |
| Nipple | 3 | 2.30 | |
| Color | 3 | 2.15 | |
| Leaf | Length | 3 | 2.28 |
| Width | 3 | 2.29 | |
| Shape | 3 | 2.29 | |
| Body curvature | 4 | 2.26 | |
| Tree | Vigor | 3 | 2.22 |
| Growth habit | 2 | 2.25 | |
| Canopy density | 3 | 2.28 |
| Number of Mother Plants | Variety Studied | Pi | Pi×log2Pi |
|---|---|---|---|
| 12 | Ascolana Ternera | 0.1304 | −0.2657 |
| 08 | Arbequina | 0.0870 | −0.2124 |
| 10 | Pendolino | 0.1087 | −0.2412 |
| 08 | Frantoio | 0.0870 | −0.2124 |
| 05 | Leccino | 0.0543 | −0.1583 |
| 06 | Picual | 0.0652 | −0.1780 |
| 13 | Empeltre | 0.1413 | −0.2765 |
| 15 | Sevillana | 0.1630 | −0.2957 |
| 10 | Manzanilla | 0.1087 | −0.2412 |
| 05 | Hojiblanca | 0.0543 | −0.1583 |
| Shannon index | 2.239705132 | ||
| Morphological Traits | Characteristic to Evaluate | Number of Parameters | Discriminating Power |
|---|---|---|---|
| Endocarp (Stone) | Weight | 4 | 0.08 |
| Shape | 4 | 0.01 | |
| Degree of Symmetry | 3 | 0.06 | |
| Transverse Diameter | 3 | 0.08 | |
| Apex | 2 | 0.09 | |
| Base | 3 | 0.05 | |
| Surface | 3 | 0.08 | |
| Grooves | 3 | 0.06 | |
| Groove Distribution | 2 | 0.03 | |
| Apex Termination | 2 | 0.02 | |
| Fruit | Weight | 4 | 0.07 |
| Shape | 4 | 0.09 | |
| Degree of Symmetry | 3 | 0.33 | |
| Maximum Transverse Diameter | 3 | 0.35 | |
| Apex | 2 | 0.06 | |
| Base | 2 | 0.16 | |
| Nipple | 3 | 0.09 | |
| Color | 3 | 0.78 | |
| Leaf | Length | 3 | 0.06 |
| Width | 3 | 0.05 | |
| Shape | 3 | 0.09 | |
| Limb Curvature | 4 | 0.13 | |
| Tree | Vigor | 3 | 0.21 |
| Growth Habit | 2 | 0.38 | |
| Canopy Density | 3 | 0.24 |
| Plant Part | Commercial Purpose | Evaluation Method | Test Statistic | p-Value |
|---|---|---|---|---|
| Endocarp | Oil | Multiple Comparisons | – | 0.793 |
| Levene | 0.06 | 0.831 | ||
| Dual Purpose | Multiple Comparisons | – | 0.912 | |
| Levene | 0.11 | 0.941 | ||
| Table | Multiple Comparisons | – | 0.995 | |
| Levene | 0.08 | 1.000 | ||
| Fruit | Oil | Multiple Comparisons | – | 0.891 |
| Levene | 0.31 | 0.733 | ||
| Dual Purpose | Multiple Comparisons | – | 1.000 | |
| Levene | 0.06 | 1.000 | ||
| Table | Multiple Comparisons | – | 0.996 | |
| Levene | 0.14 | 1.000 | ||
| Leaf | Oil | Multiple Comparisons | – | 0.869 |
| Levene | 0.07 | 0.992 | ||
| Dual Purpose | Multiple Comparisons | – | 1.000 | |
| Levene | 0.03 | 1.000 | ||
| Table | Multiple Comparisons | – | 0.998 | |
| Levene | 0.01 | 1.000 |
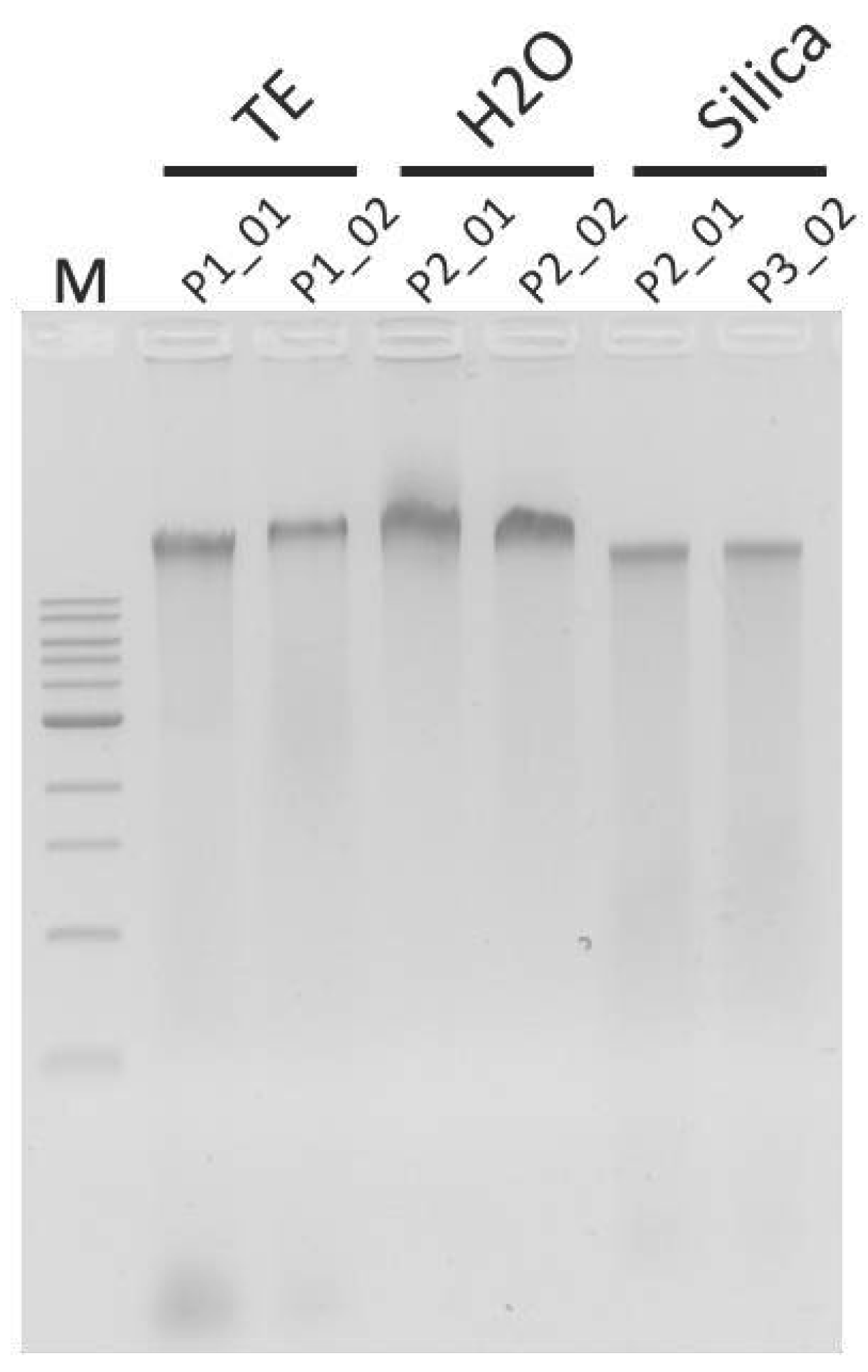
| Sample | DNA Concentration (ng/µL) |
|---|---|
| P1_01 | 21.4 |
| P1_02 | 38.2 |
| P2_01 | 16.8 |
| P2_02 | 2.24 |
| P3_01 | 4.14 |
| P3_02 | 1.72 |
| Sample Code | Olive Variety | Sample Weight (mg) |
|---|---|---|
| OL31 | Frantoio_P1 | 103 |
| OL32 | Frantoio_P2 | 120 |
| OL33 | Frantoio_P3 | 126 |
| OL34 | Ascolana_P1 | 114 |
| OL62 | Ascolana_P2 | 89 |
| OL36 | Ascolana_P3 | 124 |
| OL37 | Manzanilla_P1 | 118 |
| OL38 | Manzanilla_P2 | 122 |
| OL39 | Manzanilla_P3 | 123 |
| OL40 | Hoji Blanca_P1 | 123 |
| OL41 | Hoji Blanca_P2 | 123 |
| OL61 | Hoji Blanca_P3 | 95 |
| OL43 | Sevillana_P1 | 119 |
| OL44 | Sevillana_P2 | 118 |
| OL45 | Sevillana_P3 | 112 |
| OL46 | Picual_P1 | 100 |
| OL47 | Picual_P2 | 90 |
| OL48 | Picual_P3 | 92 |
| OL49 | Empeltre_P1 | 92 |
| OL50 | Empeltre_P2 | 92 |
| OL51 | Empeltre_P3 | 100 |
| OL52 | Arbequina_P1 | 91 |
| OL53 | Arbequina_P2 | 93 |
| OL54 | Arbequina_P3 | 94 |
| OL55 | Leccino_P1 | 96 |
| OL56 | Leccino_P2 | 96 |
| OL57 | Leccino_P3 | 91 |
| OL58 | Pendolino_P1 | 95 |
| OL59 | Pendolino_P2 | 92 |
| OL60 | Pendolino_P3 | 98 |
| Source | Variety | Sample | Absorbance A260 | DNA Concentration (ng/µL) |
|---|---|---|---|---|
| Los Palos | Frantoio | OL31 | 25.551 | 1277.5 |
| Los Palos | Frantoio | OL32 | 9.758 | 487.9 |
| Los Palos | Frantoio | OL33 | 9.973 | 498.6 |
| Magollo | Ascolana tenera | OL34 | 25.431 | 1271.6 |
| Magollo | Ascolana tenera | OL62 | 13.735 | 686.7 |
| Magollo | Ascolana tenera | OL36 | 13.981 | 699.1 |
| La Yarada | Manzanilla | OL37 | 11.725 | 586.3 |
| La Yarada | Manzanilla | OL38 | 28.789 | 1439.5 |
| La Yarada | Manzanilla | OL39 | 17.207 | 860.4 |
| La Yarada | Hojiblanca | OL40 | 19.003 | 950.2 |
| La Yarada | Hojiblanca | OL41 | 21.734 | 1086.7 |
| La Yarada | Hojiblanca | OL61 | 9.77 | 488.5 |
| Sama Las Yaras | Sevillana | OL43 | 25.576 | 1278.8 |
| Sama Las Yaras | Sevillana | OL44 | 17.626 | 881.3 |
| Sama Las Yaras | Sevillana | OL45 | 23.345 | 1167.3 |
| UNJBG-TACNA | Picual | OL46 | 5.202 | 260.1 |
| UNJBG-TACNA | Picual | OL47 | 5.015 | 250.8 |
| UNJBG-TACNA | Picual | OL48 | 1.838 | 91.9 |
| Para Grande | Empeltre | OL49 | 6.501 | 325.1 |
| Para Grande | Empeltre | OL50 | 5.171 | 258.5 |
| Para Grande | Empeltre | OL51 | 5.742 | 287.1 |
| Los Palos | Arbequina | OL52 | 5.241 | 262.1 |
| Los Palos | Arbequina | OL53 | 6.798 | 339.9 |
| Los Palos | Arbequina | OL54 | 4.708 | 235.4 |
| Los Palos | Leccino | OL55 | 6.777 | 338.9 |
| Los Palos | Leccino | OL56 | 6.51 | 325.5 |
| Los Palos | Leccino | OL57 | 6.705 | 335.3 |
| Inclán | Pendolino | OL58 | 7.602 | 380.1 |
| Inclán | Pendolino | OL59 | 4.433 | 221.7 |
| Inclán | Pendolino | OL60 | 8.96 | 448 |
| Magollo | Ascolana | OL35 | 46.662 | 2333.1 |
| La Yarada | Hojiblanca | OL42 | 15.654 | 782.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Casilla García, M.E.; Becerra, R.A.; Cotrado Cotrado, J.; Casilla Rondán, J.I.; Huatuco Coaquira, J.L.; Bedoya Justo, E.V. Genetic Diversity of Olive (Olea europaea L.) Cultivars Assessed by Genotyping-by-Sequencing in Southern Peru. Agriculture 2025, 15, 1237. https://doi.org/10.3390/agriculture15121237
Casilla García ME, Becerra RA, Cotrado Cotrado J, Casilla Rondán JI, Huatuco Coaquira JL, Bedoya Justo EV. Genetic Diversity of Olive (Olea europaea L.) Cultivars Assessed by Genotyping-by-Sequencing in Southern Peru. Agriculture. 2025; 15(12):1237. https://doi.org/10.3390/agriculture15121237
Chicago/Turabian StyleCasilla García, Martín Eloy, Rina Alvarez Becerra, José Cotrado Cotrado, Juan Iván Casilla Rondán, Janet Libertad Huatuco Coaquira, and Edgar Virgilio Bedoya Justo. 2025. "Genetic Diversity of Olive (Olea europaea L.) Cultivars Assessed by Genotyping-by-Sequencing in Southern Peru" Agriculture 15, no. 12: 1237. https://doi.org/10.3390/agriculture15121237
APA StyleCasilla García, M. E., Becerra, R. A., Cotrado Cotrado, J., Casilla Rondán, J. I., Huatuco Coaquira, J. L., & Bedoya Justo, E. V. (2025). Genetic Diversity of Olive (Olea europaea L.) Cultivars Assessed by Genotyping-by-Sequencing in Southern Peru. Agriculture, 15(12), 1237. https://doi.org/10.3390/agriculture15121237






