Estimation of Genetic Parameters and Prediction for Body Weight of Angus Cattle
Abstract
1. Introduction
2. Materials and Methods
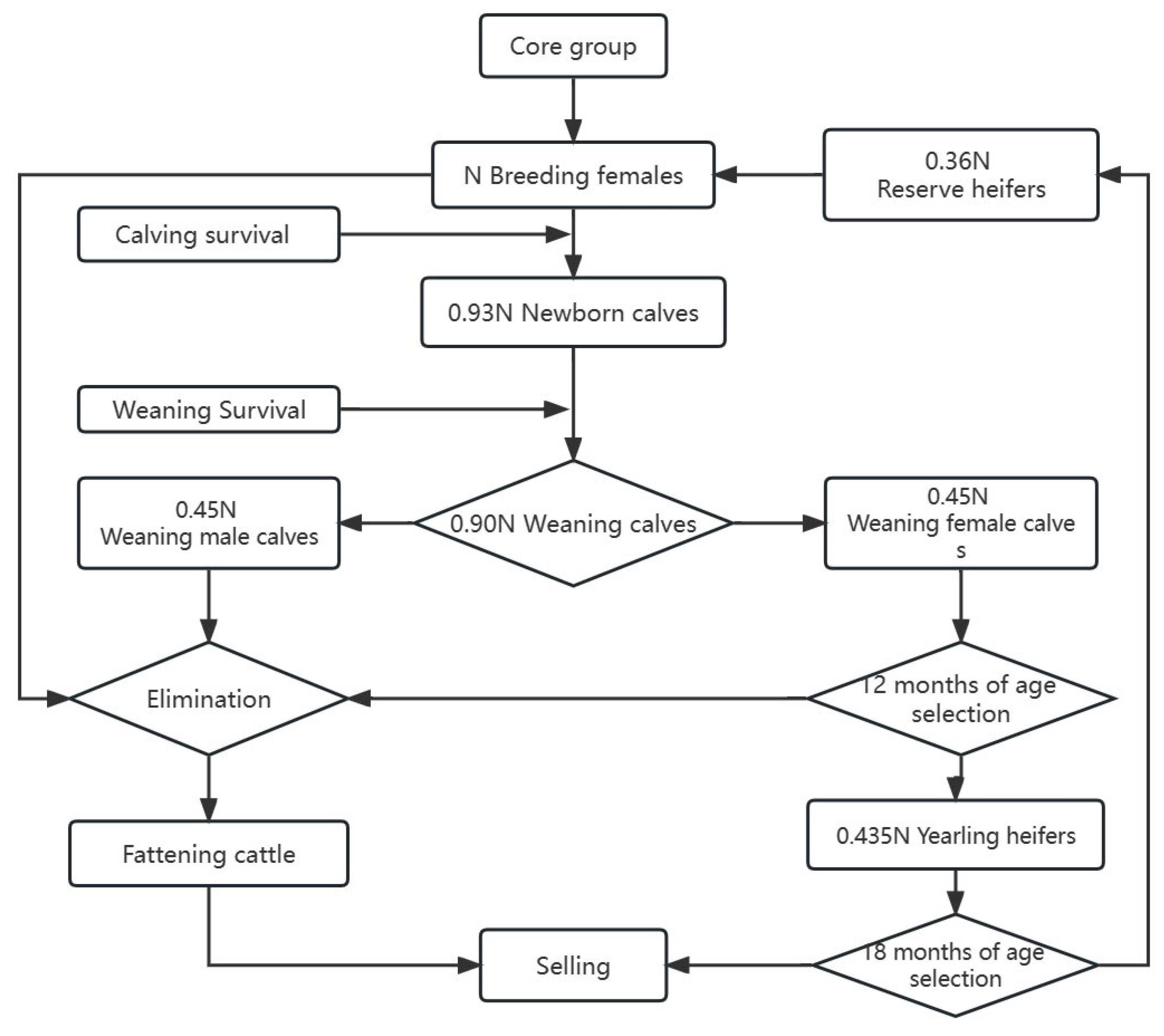
2.1. Data Sources and Animal Management
2.2. Data Collection and Statistical Analysis
2.3. Methods and Models for Estimating Genetic Parameters
3. Results and Discussion
3.1. Variance Components and Heritability
3.2. Genetic and Phenotypic Correlations
3.3. Regression Model for Early Growth Live Weight to Predict Late Growth Live Weight
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Norouzy, A.; Nassiry, M.R.; Shahrody, F.E.; Javadmanesh, A.; Abadi, M.R.M.; Sulimova, G.E. Identification of bovine leucocyte adhesion deficiency (BLAD) carriers in Holstein and Brown Swiss AI bulls in Iran. Russ. J. Genet. 2005, 41, 1409–1413. [Google Scholar] [CrossRef]
- Visscher, P.M.; Hill, W.G.; Wray, N.R. Heritability in the genomics era--concepts and misconceptions. Nat. Rev. Genet. 2008, 9, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; An, B.; Du, L.; Chang, T.; Liang, M.; Yang, B.G.; Xu, L.; Zhang, L.; Li, J.; E, G.; et al. Genome-Wide Association Analysis of Growth Curve Parameters in Chinese Simmental Beef Cattle. Animals 2021, 11, 192. [Google Scholar] [CrossRef]
- Dadi, H.; Schoeman, S.J.; Jordaan, G.F. Estimation of (Co)variance Components and Genetic Parameters of Growth Traits in Beef Cattle. J. Appl. Anim. Res. 2004, 26, 77–82. [Google Scholar] [CrossRef]
- Swalve, H.H. Estimation of direct and maternal (co)variance components for growth traits in Australian Simmental beef cattle. J. Anim. Breed. Genet. 1993, 110, 241–252. [Google Scholar] [CrossRef]
- Schoeman, S.J.; Jordaan, G. Multitrait estimation of direct and maternal (co)variances for growth and efficiency traits in a multibreed beef cattle herd. J. Natl. Cancer Inst. 1999, 29, 299–307. [Google Scholar] [CrossRef]
- Vostrý, L.; Přibyl, J.; Veselá, Z.; Jakubec, V. Selection of a suitable data set and model for the estimation of genetic parameters of the weaning weight in beef cattle. Arch. Anim. Breed. 2007, 50, 562–574. [Google Scholar] [CrossRef]
- Rumph, J.M.; Koch, R.M.; Gregory, K.E.; Cundiff, L.V.; Van Vleck, L.D. Comparison of models for estimation of genetic parameters for mature weight of Hereford cattle. J. Anim. Sci. 2002, 80, 583–590. [Google Scholar] [CrossRef]
- dos Santos Gonçalves Cyrillo, J.N.; de Alencar, M.M.; Razook, A.G.; Mercadante, M.E.Z.; de Figueiredo, L.A. Modelagem e estimação de parâmetros genéticos e fenotípicos para pesos do nascimento à seleção (378 dias) de machos Nelore. Rev. Bras. Zootec. 2004, 27, 575–583. [Google Scholar] [CrossRef]
- Willham, R.L. The role of maternal effects in animal breeding. 3. Biometrical aspects of maternal effects in animals. J. Anim. Sci. 1972, 35, 1288–1293. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Wang, M.; Chen, B.; Sun, Y.; Chen, K.; Du, Q.; Pi, X.; Wang, Y. Adsorption of methylene blue from aqueous solution using gelatin-based carboxylic acid-functionalized carbon nanotubes@metal-organic framework composite beads. Nanomaterials 2022, 12, 2533. [Google Scholar] [CrossRef] [PubMed]
- Molina, A.; Demyda-Peyrás, S.; Sánchez, M.; Serradilla, J.M.; Menéndez-Buxadera, A. Genetic analysis of the effects of heat stress before and after lambing on pre-weaning live weight in Spanish Merino lambs. Vet. Med. Sci. 2022, 8, 1721–1734. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, V.R.; Vega Murillo, V.; Costa, R.G.; Parraguirre Lezama, C.; Valencia de Ita, M.L.A.; Romero-Arenas, O. Evaluation of genetic parameters of growth of pelibuey and blackbelly sheep through pedigree in Mexico. Animals 2022, 12, 691. [Google Scholar] [CrossRef]
- Vargas Jurado, N.; Notter, D.R.; Taylor, J.B.; Brown, D.J.; Mousel, M.R.; Lewis, R.M. Model definition for genetic evaluation of purebred and crossbred lambs including heterosis. J. Anim. Sci. 2022, 100, skac188. [Google Scholar] [CrossRef]
- Goshu, H.A. Estimation of nonadditive and additive genetic effects on growth traits of Horro cattle. Trop. Anim. Health Prod. 2022, 54, 349. [Google Scholar] [CrossRef]
- Costa, R.B.; Misztal, I.; Elzo, M.A.; Bertrand, J.K.; Silva, L.O.; Łukaszewicz, M. Estimation of genetic parameters for mature weight in Angus cattle. J. Anim. Sci. 2011, 89, 2680–2686. [Google Scholar] [CrossRef]
- Estrada-León, R.J.; Magaña-Monforte, J.G.; Segura-Correa, J.C. Estimation of genetic parameters for preweaning growth traits of Brahman cattle in Southeastern Mexico. Trop. Anim. Health Prod. 2014, 46, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Jonker, A.; Hickey, S.M.; Rowe, S.J.; Janssen, P.H.; Shackell, G.H.; Elmes, S.; Bain, W.E.; Wing, J.; Greer, G.J.; Bryson, B.; et al. Genetic parameters of methane emissions determined using portable accumulation chambers in lambs and ewes grazing pasture and genetic correlations with emissions determined in respiration chambers. J. Anim. Sci. 2018, 96, 3031–3042. [Google Scholar] [CrossRef]
- Yadav, N.; Illa, S.K.; Mukherjee, S.; Gujar, G.; Mukherjee, A. Bayesian estimates for genetic and phenotypic parameters of growth traits in Sahiwal cattle. Trop. Anim. Health Prod. 2022, 55, 30. [Google Scholar] [CrossRef]
- Lopes, F.B.; Ferreira, J.L.; Lobo, R.B.; Rosa, G.J.M. Bayesian analyses of genetic parameters for growth traits in Nellore cattle raised on pasture. Genet. Mol. Res. 2017, 16, gmr16039606. [Google Scholar] [CrossRef]
- Boligon, A.A.; Silva, J.A.; Sesana, R.C.; Sesana, J.C.; Junqueira, J.B.; Albuquerque, L.G. Estimation of genetic parameters for body weights, scrotal circumference, and testicular volume measured at different ages in Nellore cattle. J. Anim. Sci. 2010, 88, 1215–1219. [Google Scholar] [CrossRef] [PubMed]
- Vargas, G.; Buzanskas, M.E.; Guidolin, D.G.; Grossi Ddo, A.; Bonifácio Ada, S.; Lôbo, R.B.; da Fonseca, R.; Oliveira, J.A.; Munari, D.P. Genetic parameter estimation for pre- and post-weaning traits in Brahman cattle in Brazil. Trop. Anim. Health Prod. 2014, 46, 1271–1278. [Google Scholar] [CrossRef]
- Eler, J.P.; Van Vleck, L.D.; Ferraz, J.B.; Lôbo, R.B. Estimation of variances due to direct and maternal effects for growth traits of Nelore cattle. J. Anim. Sci. 1995, 73, 3253–3258. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Herrera-Ojeda, J.B.; Parra-Bracamonte, G.M.; Lopez-Villalobos, N.; Martínez-González, J.C.; Magaña-Monforte, J.G.; Morris, S.T.; López-Bustamante, L.A. Genetic variances and covariances of live weight traits in Charolais cattle by multi-trait analysis. J. Appl. Genet. 2019, 60, 385–391. [Google Scholar] [CrossRef]
- Fitzmaurice, S.; Conington, J.; Fetherstone, N.; Pabiou, T.; McDermott, K.; Wall, E.; Banos, G.; McHugh, N. Genetic analyses of live weight and carcass composition traits in purebred Texel, Suffolk and Charollais lambs. Animal 2020, 14, 899–909. [Google Scholar] [CrossRef]
- Kamprasert, N.; Duijvesteijn, N.; Van der Werf, J.H.J. Estimation of genetic parameters for BW and body measurements in Brahman cattle. Animal 2019, 13, 1576–1582. [Google Scholar] [CrossRef] [PubMed]
- Bertipaglia, T.S.; Carreño, L.O.D.; Aspilcueta-Borquis, R.R.; Boligon, A.A.; Farah, M.M.; Gomes, F.J.; Machado, C.H.C.; Rey, F.S.B.; da Fonseca, R. Estimates of genetic parameters for growth traits in Brahman cattle using random regression and multitrait models. J. Anim. Sci. 2015, 93, 3814–3819. [Google Scholar] [CrossRef]
- Cucco, D.C.; Ferraz, J.B.; Pinto, L.F.; Eler, J.P.; Balieiro, J.C.; Mattos, E.C. Genetic parameters for pre-weaning traits in Braunvieh cattle. Genet. Mol. Res. 2009, 8, 291–298. [Google Scholar] [CrossRef]
| Traits | Number of Records | Mean | Standard Deviation | Coefficient of Variation | Number of Sires | Number of Dams | Number of Dams with Records |
|---|---|---|---|---|---|---|---|
| BW0 | 3493 | 32.63 | 4.84 | 14.74 | 78 | 2356 | 538 |
| BW3 | 3002 | 119.50 | 24.19 | 19.50 | 63 | 2192 | 386 |
| BW6 | 2931 | 188.53 | 33.61 | 17.83 | 53 | 2236 | 496 |
| BW12 | 2323 | 235.48 | 63.68 | 19.56 | 45 | 1721 | 785 |
| BW18 | 1858 | 561.93 | 68.91 | 15.84 | 36 | 1206 | 162 |
| Model | ram | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BW0 | ||||||||||||
| 1 | 11.001 | 7.885 | 18.886 | 0.582 ± 0.017 | 0.418 ± 0.017 | 0.582 ± 0.012 | ||||||
| 2 | 8.134 | 3.322 | 6.988 | 18.444 | 0.441 ± 0.014 | 0.180 ± 0.013 | 0.379 ± 0.015 | 0.441 ± 0.012 | ||||
| 3 | 8.123 | 3.284 | 7.064 | 18.472 | 0.440 ± 0.018 | 0.178 ± 0.009 | 0.382 ± 0.016 | 0.529 ± 0.016 | ||||
| 4 | 12.709 | 8.241 | −8.524 | 6.373 | 18.798 | 0.676 ± 0.014 | 0.438 ± 0.015 | 0.339 ± 0.011 | 0.215 ± 0.007 | −0.969 | ||
| 5 | 8.071 | 0.945 | 2.427 | 6.995 | 18.438 | 0.438 ± 0.011 | 0.051 ± 0.019 | 0.132 ± 0.015 | 0.379 ± 0.015 | 0.463 ± 0.015 | ||
| 6 | 12.709 | 8.241 | −8.524 | 0.000 | 6.373 | 18.798 | 0.676 ± 0.013 | 0.438 ± 0.015 | 0.000 ± 0.000 | 0.339 ± 0.010 | 0.215 ± 0.007 | −0.969 |
| BW3 | ||||||||||||
| 1 | 123.588 | 164.726 | 288.314 | 0.429 ± 0.019 | 0.571 ± 0.022 | 0.429 ± 0.030 | ||||||
| 2 | 125.259 | 0.000 | 162.658 | 287.917 | 0.435 ± 0.021 | 0.000 ± 0.000 | 0.565 ± 0.030 | 0.435 ± 0.028 | ||||
| 3 | 121.886 | 4.959 | 160.649 | 287.495 | 0.424 ± 0.021 | 0.017 ± 0.017 | 0.559 ± 0.025 | 0.433 ± 0.024 | ||||
| 4 | 307.850 | 108.600 | −167.986 | 175.822 | 424.286 | 0.726 ± 0.027 | 0.256 ± 0.022 | 0.414 ± 0.026 | 0.340 ± 0.021 | −0.919 | ||
| 5 | 121.886 | 4.959 | 0.000 | 160.649 | 287.495 | 0.424 ± 0.024 | 0.017 ± 0.019 | 0.000 ± 0.000 | 0.559 ± 0.026 | 0.433 ± 0.024 | ||
| 6 | 307.850 | 108.600 | −167.986 | 0.000 | 175.822 | 424.286 | 0.726 ± 0.027 | 0.256 ± 0.025 | 0.000 ± 0.000 | 0.414 ± 0.023 | 0.340 ± 0.020 | −0.919 |
| BW6 | ||||||||||||
| 1 | 91.406 | 261.656 | 353.062 | 0.259 ± 0.042 | 0.741 ± 0.056 | 0.259 ± 0.0 | ||||||
| 2 | 95.452 | 0.000 | 256.893 | 352.345 | 0.271 ± 0.045 | 0.000 ± 0.000 | 0.729 ± 0.051 | 0.271 ± 0.0 | ||||
| 3 | 95.452 | 0.000 | 256.893 | 352.345 | 0.271 ± 0.044 | 0.000 ± 0.000 | 0.729 ± 0.063 | 0.271 ± 0.0 | ||||
| 4 | 247.984 | 71.223 | −128.721 | 187.660 | 378.146 | 0.656 ± 0.051 | 0.188 ± 0.028 | 0.496 ± 0.043 | 0.239 ± 0.035 | −0.917 | ||
| 5 | 95.452 | 0.000 | 0.000 | 256.893 | 352.345 | 0.271 ± 0.047 | 0.000 ± 0.000 | 0.000 ± 0.000 | 0.729 ± 0.050 | 0.271 ± 0.048 | ||
| 6 | 247.984 | 71.223 | −128.721 | 0.000 | 187.660 | 378.146 | 0.656 ± 0.051 | 0.188 ± 0.028 | 0.000 ± 0.000 | 0.496 ± 0.043 | 0.239 ± 0.057 | −0.917 |
| BW12 | ||||||||||||
| 1 | 785.542 | 1029.232 | 1814.774 | 0.433 ± 0.066 | 0.567 ± 0.055 | 0.433 ± 0.039 | ||||||
| 2 | 785.542 | 0.000 | 1029.231 | 1814.774 | 0.433 ± 0.054 | 0.000 ± 0.000 | 0.567 ± 0.050 | 0.433 ± 0.061 | ||||
| 3 | 766.657 | 41.628 | 1005.516 | 1813.801 | 0.423 ± 0.039 | 0.023 ± 0.019 | 0.554 ± 0.043 | 0.434 ± 0.072 | ||||
| 4 | 1293.711 | 248.986 | −491.143 | 831.320 | 1882.873 | 0.687 ± 0.064 | 0.105 ± 0.025 | 0.350 ± 0.062 | 0.362 ± 0.044 | −0.865 | ||
| 5 | 766.657 | 41.628 | 0.000 | 1005.516 | 1813.801 | 0.423 ± 0.078 | 0.023 ± 0.044 | 0.000 ± 0.000 | 0.554 ± 0.082 | 0.434 ± 0.058 | ||
| 6 | 1293.711 | 248.986 | −491.143 | 0.000 | 831.320 | 1882.873 | 0.687 ± 0.059 | 0.132 ± 0.052 | 0.000 ± 0.000 | 0.442 ± 0.049 | 0.362 ± 0.046 | −0.865 |
| BW18 | ||||||||||||
| 1 | 1371.163 | 2481.217 | 3852.380 | 0.356 ± 0.059 | 0.644 ± 0.044 | 0.356 ± 0.067 | ||||||
| 2 | 1324.442 | 1.273 | 2473.359 | 3799.075 | 0.349 ± 0.061 | 0.000 ± 0.000 | 0.651 ± 0.058 | 0.349 ± 0.082 | ||||
| 3 | 1364.411 | 0.000 | 2481.941 | 3846.352 | 0.355 ± 0.056 | 0.000 ± 0.000 | 0.645 ± 0.043 | 0.355 ± 0.075 | ||||
| 4 | 2069.838 | 526.834 | −957.769 | 2341.229 | 3980.132 | 0.419 ± 0.042 | 0.107 ± 0.017 | 0.474 ± 0.066 | 0.225 ± 0.048 | −0.833 | ||
| 5 | 1324.442 | 0.000 | 1.273 | 2473.359 | 3799.075 | 0.349 ± 0.034 | 0.000 ± 0.000 | 0.000 ± 0.000 | 0.651 ± 0.044 | 0.349 ± 0.055 | ||
| 6 | 2007.014 | 479.849 | −899.738 | 1.989 | 2336.090 | 3925.204 | 0.511 ± 0.091 | 0.122 ± 0.025 | 0.001 ± 0.000 | 0.595 ± 0.037 | 0.229 ± 0.063 | −0.833 |
| Model | Number of Parameters | BW0 | BW3 | BW6 | BW12 | BW18 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| −2lnL | AIC | −2lnL | AIC | −2lnL | AIC | −2lnL | AIC | −2lnL | AIC | ||
| 1 | 2 | 12,369.300 | 12,373.300 | 12,369.300 | 12,373.300 | 17,353.990 | 17,357.990 | 17,353.990 | 17,357.990 | 20,078.055 | 20,082.055 |
| 2 | 3 | 12,369.300 | 12,375.300 | 12,369.300 | 12,375.300 | 17,154.936 | 17,160.936 | 17,154.936 | 17,160.936 | 19,628.659 | 19,634.659 |
| 3 | 3 | 12,365.441 | 12,371.441 | 12,365.441 | 12,371.441 | 17,154.936 | 17,160.936 | 17,154.936 | 17,160.936 | 19,628.738 | 19,634.738 |
| 4 | 4 | 12,336.133 | 12,344.133 | 12,336.133 | 12,344.133 | 17,109.369 | 17,117.369 | 17,109.369 | 17,117.369 | 19,610.121 | 19,618.121 |
| 5 | 4 | 12,365.441 | 12,373.441 | 12,365.441 | 12,373.441 | 17,154.936 | 17,162.936 | 17,154.936 | 17,162.936 | 19,628.659 | 19,636.659 |
| 6 | 5 | 12,336.133 | 12,346.133 | 12,336.133 | 12,346.133 | 17,109.369 | 17,119.369 | 17,109.369 | 17,119.369 | 19,609.962 | 19,619.962 |
| Traits | BW0 | BW3 | BW6 | BW12 | BW18 |
|---|---|---|---|---|---|
| BW0 | 0.110 ± 0.062 | 0.061 ± 0.049 | 0.113 ± 0.030 | 0.160 ± 0.043 | |
| BW3 | 0.165 ± 0.057 | 0.417 ± 0.038 | 0.688 ± 0.041 | 0.704 ± 0.029 | |
| BW6 | 0.029 ± 0.028 | 0.337 ± 0.048 | 0.504 ± 0.026 | 0.828 ± 0.033 | |
| BW12 | 0.074 ± 0.035 | 0.396 ± 0.034 | 0.598 ± 0.062 | 0.785 ± 0.025 | |
| BW18 | 0.136 ± 0.017 | 0.499 ± 0.022 | 0.552 ± 0.020 | 0.755 ± 0.032 |
| Predicted Traits | Model | Regression Equation | Determination Coefficient |
|---|---|---|---|
| BW18 | 1: BW0 | Y = −256.495 + 25.092BW0 | 0.630 |
| 2: BW3 | Y = 72.509 + 3.631BW3 | 0.596 | |
| 3: BW6 | Y = −117.452 + 3.348BW6 | 0.797 | |
| 4: BW12 | Y = −1.415 + 1.585BW12 | 0.746 | |
| 5: BW0 + BW3 | Y = −251.826 + 16.293BW0 + 2.167BW3 | 0.765 | |
| 6: BW0 + BW3 + BW6 | Y = −174.789 + 4.960BW0 + 0.894BW3 + 2.263BW6 | 0.810 | |
| 7: BW0 + BW3 + BW6 + BW12 | Y = −187.472 + 4.448BW0 + 1.008BW3 + 2.981BW6−0.371BW12 | 0.811 | |
| 8: BW3 + BW6 + BW12 | Y = −141.586 + 0.865BW3 + 3.730BW6−0.472BW12 | 0.807 | |
| 9: BW3 + BW6 | Y = −118.011 + 0.693BW3 + 2.902BW6 | 0.804 | |
| 10: BW6 + BW12 | Y = −125.403 + 3.667BW6−0.160BW12 | 0.797 | |
| BW12 | 1: BW0 | Y = −102.988 + 13.966BW0 | 0.657 |
| 2: BW3 | Y = 65.309 + 2.141BW3 | 0.697 | |
| 3: BW6 | Y = −49.663 + 1.989BW6 | 0.947 | |
| 5: BW0 + BW3 | Y = −99.984 + 8.303BW0 + 1.394BW3 | 0.844 | |
| 7: BW0 + BW3 + BW6 | Y = −34.163−1.379BW0 + 0.307BW3 + 1.933BW6 | 0.955 | |
| 9: BW3 + BW6 | Y = −34.045 + 5.008BW3 + 0.562BW6 | 0.876 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, X.; Wang, Y.; Zhao, J.; Jiang, Q.; Chen, Y.; Gu, Y.; Guo, P.; Zheng, J. Estimation of Genetic Parameters and Prediction for Body Weight of Angus Cattle. Agriculture 2025, 15, 1216. https://doi.org/10.3390/agriculture15111216
Feng X, Wang Y, Zhao J, Jiang Q, Chen Y, Gu Y, Guo P, Zheng J. Estimation of Genetic Parameters and Prediction for Body Weight of Angus Cattle. Agriculture. 2025; 15(11):1216. https://doi.org/10.3390/agriculture15111216
Chicago/Turabian StyleFeng, Xiaofang, Yu Wang, Jie Zhao, Qiufei Jiang, Yafei Chen, Yaling Gu, Penghui Guo, and Juanshan Zheng. 2025. "Estimation of Genetic Parameters and Prediction for Body Weight of Angus Cattle" Agriculture 15, no. 11: 1216. https://doi.org/10.3390/agriculture15111216
APA StyleFeng, X., Wang, Y., Zhao, J., Jiang, Q., Chen, Y., Gu, Y., Guo, P., & Zheng, J. (2025). Estimation of Genetic Parameters and Prediction for Body Weight of Angus Cattle. Agriculture, 15(11), 1216. https://doi.org/10.3390/agriculture15111216






