Abstract
Rice tillering is an important trait that is genetically and environmentally co-regulated. Nitorgen is one of the key nutrients affecting tillering, and straw return further affects tiller development by altering soil heterogeneity. In order to analyze the genetic regulation mechanism of rice tillering and its interactions with the environment, 124 recombinant inbred line (RIL) populations derived from two superior Peijiu lines, 9311 and PA64s, were used as materials in this study, and the dynamic tillering phenotypes were measured under three treatments (no nitrogen application, nitrogen application, and nitrogen + straw return) for two consecutive years. Using an existing genetic map, we conducted single-environment, multi-environment, and meta-QTL analyses to systematically identify tiller-related genetic loci and their environmental interactions. The main findings were as follows: (1) A total of 57 QTLs were identified in the single-environment QTL analysis, of which 44 were unreported new QTLs. Four QTLs showed temporal pleiotropy, ten QTLs contributed more than 10% to the phenotypes under the no-N treatment, and five QTLs contributed more than 10% under the straw return treatment. Among them, the phenotypic contribution of mks1-355 (qD1tn1-3) and mks1-352 (qD2TN1-2) both exceeded 40%. (2) Multi-environmental QTL analysis detected 15 QTLs. Of these, qmD1TN1 (mks1-356) showed no environmental interaction effect, while qmD1TN12 (mks12-267), qmD2TN1 (mks1-334), qmD2TN3-1 (mks3-105), and qmD5TN6 (mks6-71) exhibited antagonistic pleiotropy, suggesting that these QTL need to be considered for environmental specificity in breeding. (3) Meta-QTL analysis localized 52 MQTLs, of which MQTL3.1 and MQTL6.8 contained 82 and 59 candidate genes, respectively, and no reported tiller-related genes were found. (4) mks1-355 (qD1tn1-3), mks1-352 (qD2TN1-2), and mks1-356 (qmD1TN1) may be located in the same genetic locus, and their phenotypic contributions were more than 40%. These QTLs were detected stably for two consecutive years, and they may be the main effector QTLs in tillering that are less affected by the environment. Further analysis revealed that these QTLs corresponded to MQTL1.6, which contains 56 candidate genes. Of these, the expression level of OsSPL2 gene in the parental line 9311 was significantly higher than that of PA64s, and there were polymorphic differences in the coding region. It was hypothesized that OsSPL2 was the main effector gene of this QTL. This study provides important genetic resources for mining candidate genes related to tillering and nitrogen efficiency in rice and lays a theoretical foundation for directional breeding and molecular marker development in specific environments.
1. Introduction
Tiller number in rice is a quantitative trait controlled by multiple genes, and its development is regulated by both genetic and environmental factors. Genetically, a series of genes have been reported to be directly involved in tillering regulation, such as MOC1 [1], FON1 [2], IPA1 [3], OsTB1 [4], and D3 [5]. In terms of environment, fertilizer application, planting density, cultivation conditions, and climatic factors significantly affected tiller development [6,7]. Among them, nitrogen is one of the key nutrients affecting tillering in rice, and moderate nitrogen application can significantly promote tillering [8,9]. However, there is a threshold effect on the uptake of nitrogen fertilizer in rice. Over-application of nitrogen not only makes it difficult to further increase the number of tillers, but also reduces nitrogen fertilizer utilization and leads to an increase in the number of ineffective tillers [10,11]. Therefore, optimizing nitrogen fertilizer management is of great significance to improve rice yield and resource utilization efficiency. As an important agricultural resource, crop straw can provide an organic nitrogen source, carbon source, and nutrients such as phosphorus and potassium after decomposition by soil microorganisms, partially replacing chemical nitrogen fertilizers [12,13,14]. Returning straw to the field not only improves the decline in soil fertility and the degradation of physicochemical properties caused by long-term cultivation, but it also reduces the environmental pollution caused by straw burning and promotes the nutrient cycling of the ecosystem in the field [12,13,14]. This measure not only helps to reduce the cost of nitrogen fertilizer application, but also responds to the strategic needs of national rural revitalization and sustainable development.
Quantitative trait loci (QTL) localization is an important tool for mining yield-related genes in rice. Currently, more than 200 QTLs related to tiller number in rice have been reported [15]. However, studies on dynamic tillering QTLs and their interactions with the environment are still relatively limited. Previously, QTL analyses of dynamic tillering in rice have been conducted mainly using single segment substitution lines (SSSL) and double haploid (DH) populations [16,17,18], but the dynamic characteristics of tillering QTLs and their environmental interactions (QEIs) have not yet been systematically analyzed under straw-returned and nitrogen-stressed conditions. Xing et al. [19] pointed out that the interactions between genotypes and the environment have a significant effect on developmental and agronomic traits, and this interactions may lead to a significant increase in the number of tillers in rice. In addition, such interactions may lead to genetic variation in the same QTLs in different environments [20]. Therefore, it is important to analyze the environmental response mechanism of tillering QTLs for rice breeding.
Molecular marker-assisted selection (MAS) is an efficient and precise breeding strategy. In recent years, a series of nitrogen-efficient genes (e.g., OsNR2 [21], NRT1.1B [22], OsTCP19 [23], and OsLHT1 [24]) have been cloned, and related molecular markers have been developed for application in breeding practice [25]. Among them, OsTCP19 was shown to be a key gene for efficient response to tiller nitrogen [23]. In addition, meta-QTL analysis can significantly improve the accuracy of QTL localization and provide important clues for candidate gene mining by integrating QTL information from multiple studies [26]. Therefore, systematic analysis of the dynamic characteristics of tillering QTLs and their environmental interaction effects under straw return and N stress conditions not only helps to mine N-efficient related genes, but also provides a theoretical basis for molecular design breeding.
In this study, the phenotypic identification of dynamic tiller number in rice was systematically carried out using the parental lines 9311 and PA64s and 124 recombinant inbred lines (RIL) populations derived from them, and three treatments, namely, straw return + normal nitrogen (GNSB), normal nitrogen (GN), and nitrogen stress (NN), were set in 2021 and 2022. Combined with single-environment QTL analysis, multi-environment QTL analysis, and meta-QTL integration analysis, we aimed to explore tillering-related genetic loci and their environmental interaction effects and to provide theoretical support for rice N-efficient breeding and molecular marker development.
The specific objectives were as follows: 1. Identify dynamic tillering QTLs under three treatments (no N, N, N + straw return) across two years; 2. Detect QTL-by-environment interactions (QEI) and pleiotropic effects; 3. Validate stable QTLs using meta-analysis and identify candidate genes for nitrogen-efficient tillering. We hypothesized that nitrogen and straw return treatments would modulate the expression of tillering QTLs, with specific loci exhibiting stable effects across environments or environment-specific effects.
2. Materials and Methods
2.1. Test Materials and Field Trials
A population of 124 recombinant inbred lines derived from a cross between indica PA64s (female parent) and indica 9311 (male parent) was used as the material in this experiment. These materials were planted in Qinglian Town, Mianyang City, Sichuan Province. The seeds of the populations were arranged in sequence and transplanted in single rows of 10 plants each, with a plant spacing of 15 cm and a row spacing of 33 cm, and the plot area was designed to be 1.5 m2. The seeds were sown in the middle of April, transplanted in the second half of May, and subjected to conventional field management.
2.2. Investigation of Dynamic Tiller Number in Rice Under Three Treatments and Data Analysis
The 124 lines and their parental materials were grown under three treatments, namely, straw return + normal N, normal N, and no N application with two-year replications (2021 and 2022). The treatments under 2021 and 2022 are denoted by E1 (straw return + normal N in 2021), E2 (normal N in 2021), E3 (no N application in 2021), E4 (straw return + normal N in 2022), E5 (normal N in 2022), and E6 (no N application in 2022). The detailed data of soil treatment can be found in Table S4. From two weeks after transplanting, the number of tillers per line was counted randomly in five replicates for every 7 days until the highest number of tillers appeared in each plot. Tiller numbers were recorded continuously for six weeks (expressed as D1-D6). Individual tiller counts of plants in each plot at different measurement stages were used as raw data in the analyses. All tiller raw data were analyzed using ANOVA using SPSS 22.0 and QTL IciMapping v4.1
2.3. Genetic Mapping
Genetic linkage maps have been constructed in a previous study [24]. Having two parents (9311 and PA64s) that had been sequenced completely, 124 recombinant autogamous singletons (F18 generation) were then sequenced with low coverage and analyzed with recombinant bin. Only one genotype of 9311 or PA64s was present in each recombinant segment. Then, segments with the same genotype were combined into one recombinant bin, resulting in a total of 3524 bins for molecular markers. Finally, a high-density map was then obtained by constructing a genetic map in Mapmaker/EXP3.0 with a total length of 1381.9 cM and an average marker spacing of 0.392 cM. A high-density genotype map was successfully constructed by identifying SNPs and recombination sites using high-throughput resequencing (Figure 1).
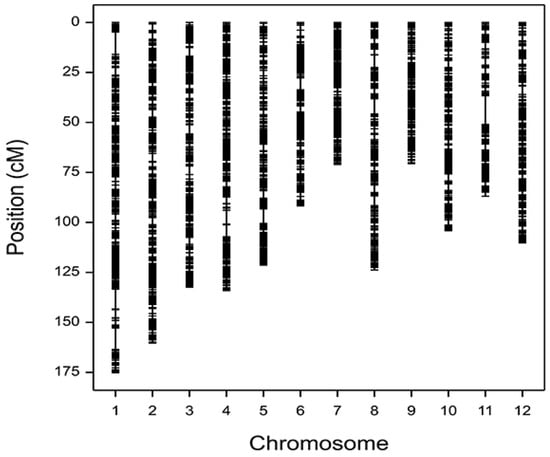
Figure 1.
Genetic mapping of recombinant inbred line (RIL) populations.
2.4. Single- and Multi-Environment QTL Analyses for Dynamic Tillering in Rice
Single-environment QTLs were detected using composite interval mapping (CIM) with a LOD threshold of 3.0 (corresponding to p < 0.01, validated via 1000 permutations). Multi-environment analysis used genome-wide significance (p < 0.05), and FDR correction (Benjamini–Hochberg, q < 0.10) was used to control the false discovery rate.
2.5. Tiller Number-Related QTL Collection and Meta-QTL Analysis
Thirty QTL articles related to tiller number from 2002 to 2022 were collected mainly through the Chinese National Knowledge Base, NBCI PubMed, and Web of Science. The collected QTL information included QTL name, population type, QTL chromosome, R2 value (phenotypic variation explained, PVE), LOD value, confidence interval, and map position. A QTL consensus map for tiller number trait in rice was constructed by integrating the collected markers and QTLs using BioMercator 4.2 software. The collected QTLs were then clustered and distributed using the mapping function of BioMercator 4.2 software. Finally, meta-analysis was performed on the entire dataset to determine the effect size and confidence level of each QTL. By comparing areas of overlap and effect sizes between studies, it was possible to determine which QTLs were reliable. The locations of reliable QTLs are compared to known genome sequences to identify potential candidate genes in the genome. Candidate genes are further subjected to functional annotation, expression analysis, and genetic validation to determine whether they are associated with the target complex trait.
2.6. Candidate Gene Analysis
Firstly, the genes were identified by https://archive.gramene.org/markers (accessed on 10 August 2022) to determine the physical location of the markers at both ends of the MQTL on the chromosome. All the genes with physical location within the two side markers were considered as candidate genes. The candidate genes within the interval were retrieved from the Rice Annotated Gene Database (RAPDB, http://RAPDB.dna.affrc.go.jp (accessed on 11 August 2022)), and the candidate genes within the interval were retrieved from the Chinese National Rice Gene Database (https://www.ricedata.cn/gene/ (accessed on 12 August 2022)) and https://funricegenes.github.io (accessed on 15 August 2022) to search for candidate genes that have been reported to be associated with tiller number.
3. Results
3.1. Phenotypic Variation of Dynamic Tillering in Rice Under Three Treatments
The number of tillers of parents 9311 and PA64s differed in different periods and treatments, except that the number of tillers of PA64s was slightly lower than that of 9311 in E1 and E3 treatments in D1 period, and the number of tillers of PA64s was significantly higher than that of 9311 in the rest of the periods (Table 1). The fastest tiller growth was observed from D1 to D3, and the number of tillers showed a decreasing trend from D5 to D6, which could be attributed to the production of ineffective tillers at the late tillering stage (Figure 2). The most serious stress on tiller number in the three periods of D2, D3, and D4 was caused by the no-N treatment, and the degree of stress gradually decreased. The number of tillers in this environment decreased by 31.5%, 17.2%, and 8% and 30%, 11.3%, and 9.9% compared with that in the same period of time under the straw-returned field and the normal N treatments, respectively, and there was no significant difference between the number of tillers under the straw-returned field and the normal N treatments in the respective periods (Table 1). Meanwhile, the violin plots of tiller numbers in the RIL population at different periods showed a normal distribution under all three treatments, and all showed superparental segregation (Figure 2), which is suitable for QTL analysis.

Table 1.
Statistical analysis of dynamic tiller number traits in the RIL population under three treatments.
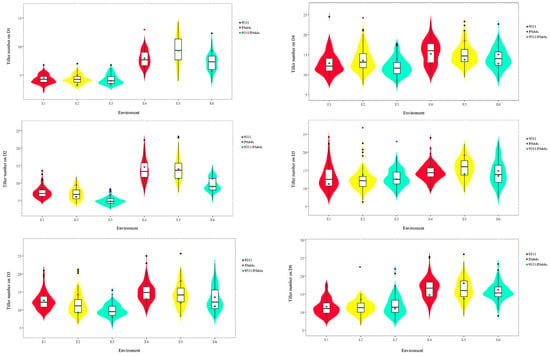
Figure 2.
Violin plots showing the distribution of tiller numbers in the RIL population and parents (9311: dark green; PA64s: purple) across six developmental stages (D1–D6) under three treatments: E1/E4 (straw + N), E2/E5 (N), and E3/E6 (no N). Superparental segregation indicates transgressive variation suitable for QTL analysis. Note: Dark green and purple colors indicate the parents 9311 and PA64s, respectively, and green colors indicate the same number of tillers. D1–D6 represent the tillering traits at each of the six periods.
3.2. QTL Correlation Analysis of Tiller Number Across Six Periods in Recombinant Inbred Lines of Rice
3.2.1. Single-Environment QTL Analysis of Tiller Number Traits in RIL Populations in Six Periods
Through the constructed genetic linkage map, a total of 57 QTLs were detected in tiller number across six periods of the population in mono-environment. The QTLs were distributed on chromosomes 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, and 12, of which 25 phenotypic incremental alleles were from PA64s and 32 phenotypic incremental alleles were from 9311 (Table 2). The LOD values of these 57 QTLs ranged from 3.05 to 6.01, and the phenotypic contribution ranged from 6.49% to 47.95%. Of the 57 QTLs, 44 were novel, including two major-effect QTL, namely, qD1tn1-3 (PVE = 41.61%, no N 2022) and qD2TN1-2 (PVE = 47.95%, N 2021), located 0.57 cM apart on chromosome 1, suggesting a shared genetic locus. (Table 2). Therefore, it was hypothesized that there was a primary QTL or primary gene near these two markers controlling tillering traits in this period and that this primary QTL or primary gene was stable and temporally multiplexed and could be detected in different environments and times. This QTL could be detected under normal nitrogen and no nitrogen treatments, suggesting that this QTL may be less affected by nitrogen and also have high contribution to population tillering under low nitrogen environments.

Table 2.
QTLs for number of tillers in the dynamic phase in a single environment detected in the RIL population of Pa64s × 9311.
Two QTLs, qD2tn1 and qD1tn1-2, were detected in the D1 and D2 periods, respectively. Their central markers were all located in mks1-305 and were detected under the E4 (straw return in 2022) treatment, and its LOD values were 3.19 and 3.23, with a phenotypic contribution of 9.05% and 7.38%. Two QTLs, qD2tn3 and qD5tn3, were detected in D2 and D5 periods. The LOD values of these two QTLs were 5.7 and 6.01, respectively, with phenotypic contributions of 13.93% and 18.71%, respectively, and their central markers were all located in mks3-165 detected under E5 (2022 normal nitrogen) treatment. qD3TN6 and qD5TN6, the central markers of these two QTLs, were detected in the D3 and D5 periods, respectively, and their central markers were all located in mks6-235, both detected under E1 (2021 straw return + normal N) treatment, with LODs of 3.22 and 3.31 and phenotypic contributions of 6.87% and 9.2%, respectively (Table 2). mks1-305 and mks3-165 both may have a QTL with temporal pleiotropy. qD3TN7 and qD6TN7 are two QTLs whose central markers are located in mks7-139. mks7-139 was detected under the E1 environment at the D3 period and the E3 environment at the D6 period with LOD values of 3.5 and 4.08, respectively, and contributions of 7.17% and 9.89%, respectively. This QTL may contain temporally and spatially pleiotropic genes controlling the phenotypes in these two periods. At the same time, this QTL may contain tiller nitrogen efficiency response genes. The QTL qD4tn9 was detected in E5 and E6, with LOD values of 5.64 and 3.77, respectively. The contributions to the phenotypes in the corresponding environments were 14.59% and 11.27%, respectively. Therefore, this QTL may be present in tiller nitrogen efficiency response-related genes.
3.2.2. Multi-Environmental QTL Analysis of Tiller Number Traits in RIL Populations Across Six Periods
To determine the genetic basis of tillering plasticity in rice under straw return and no N application treatments, the population was subjected to a multi-environmental QTL analysis, and a total of 15 QTLs were localized under the three treatments (Figure 3). There were 14 QTLs with Q × E interactions and only one QTL without Q × E interactions (Table 3). qmD1TN1 with its centromeric marker mks1-356 locus located at position 152.63 cM on chromosome 1 was detected at the D1 period. This QTL was separated from the qD2TN1-2 (mks1-352) and qD1tn1-3 (mks1-355) loci by 0.76 cM and 0.19 cM, respectively. This locus was detected with p-values less than 0.01 under all three treatments, and alleles with an additive effect value of 0.33 phenotypic increment were all from 9311. All of the phenotypic contributions exceeded 14% in 2021 and contributed slightly less to the locus in 2022. The absence of QTL–environment interaction effects for qmD1TN1 suggests that this QTL may not be affected by the environment and possesses a role in tillering regulation under no-N application treatment.
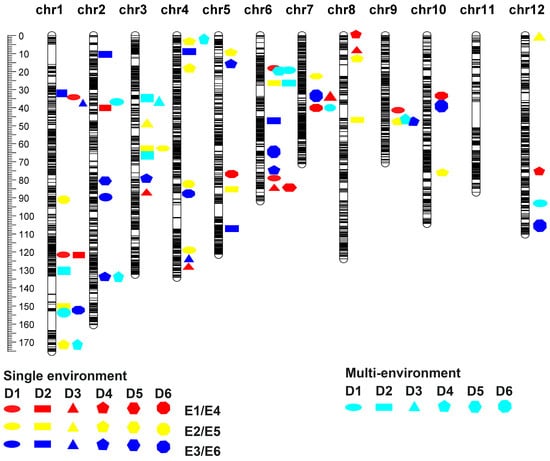
Figure 3.
Genetic map of QTLs associated with dynamic tiller number across six periods in the RIL population.

Table 3.
QTLs detected in the RIL population of A64s × 9311 for dynamic tillering in multiple environments.
The QTL qmD1TN12 (mks12-267) was detected under normal nitrogen and no nitrogen application treatments in the D1 period (p < 0.05). Three QTLs, qmD2TN1 (mks1-334), qmD2TN3-1 (mks3-105), and qmD5TN6 (mks6-71), were detected in the straw-returned field and normal nitrogen environment (p < 0.05), and antagonistic pleiotropy was present in all four QTLs (Table 3). qmD1TN12 (mks12-267) derived its phenotypic increment allele from PA64s under no nitrogen treatment and 9311 under normal nitrogen environments. qmD2TN1 (mks1-334) and qmD2TN3-1 (mks3-105) were detected in straw-returned environments. Their phenotypic increment alleles were from PA64s in the straw-returned environment and 9311 in the normal nitrogen environment. The qmD5TN6 (mks6-71) locus was detected in the E1 and E5 environments, and its phenotypic increment alleles were from 9311 in the straw-returned environment and PA64s in the normal nitrogen environment. qmD1TN2 (mks2-106) and qmD2TN6 (mks6-81) are two QTLs that were condition neutral. Both were detected under E5 (2022 normal nitrogen) treatment, suggesting that these two QTLs may only play a role in regulating tillering under normal nitrogen environments. The remaining QTLs in different environments all showed differential sensitivity, with qmD4TN4 (mks4-14) and qmD6TN7 (mks7-145) phenotypically potentiating alleles from PA64s in multiple environments. The QTL qmD4TN4 (mks4-14) was detected under the E2, E4, and E5 treatments, and the additive effect in the straw-returned treatment was higher than the effect value under the normal nitrogen treatment. The values were higher than those under normal nitrogen treatment, suggesting that this QTL may have a higher regulatory effect on tillering under straw return treatment than under normal nitrogen environment at the tillering bloom. qmD6TN7 (mks7-145) was detected under E3 and E4 treatments, with effect values of −0.79 and −0.84 under straw return and no nitrogen treatment, respectively, and a smaller difference in effect values was noted between the no-nitrogen treatment and normal nitrogen treatment. The difference between the effect values of −0.05 and −0.84 under the no-N treatment and the normal-N treatment was small, so this QTL may possess genes that act as potentiators for tillering under the no-N treatment. In addition, qmD1TN6 (mks6-71) was also detected in the D1 period, but showed conditionally neutral only under normal nitrogen environment. The phenotypic potentiation allele was from PA64s, and the LOD of this QTL reached 6.6 in multi-environmental analysis. The QTL was expressed under normal nitrogen treatment, and the results also indicated that there might be a gene that not only exhibited antagonistic pleiotropy but also was expressed under normal nitrogen treatment in the vicinity of the mks6-71 marker.
3.3. Meta-QTL Analysis of Tillering Traits in Rice
3.3.1. Collection of Information Related to QTLs for Tiller Number in Rice
A total of 222 QTLs related to tiller number were collected from 30 reported research populations between 2002 and 2021 (Table 4), and a pre-consensus map covering 12 chromosomes suitable for meta-QTL analyses was constructed by integrating the genetic maps of the studied tiller numbers (Figure 4). The map also had 1865 highly saturated markers with a full length of 2523.2 cM and an average genetic distance of 0.93 to 3.32 cM per marker (Table 5). The highest number of QTLs were localised on chromosomes 1 and 6, with 36 and 37, respectively, and the lowest number of QTLs were localised on chromosome 12, with eight.

Table 4.
QTL information reported in previous studies.
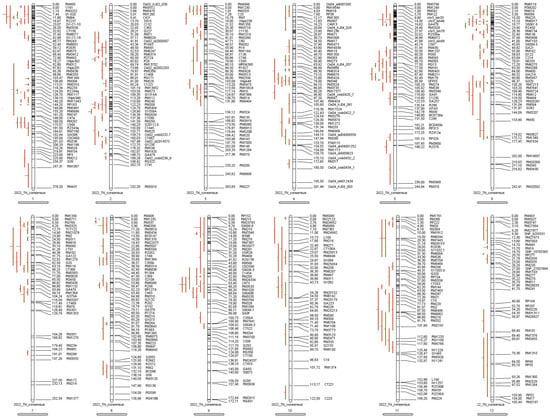
Figure 4.
Consensus mapping of tiller number traits.

Table 5.
Integration of information from mapping.
3.3.2. Meta-QTL Analysis of Tiller Number Traits in Rice
Meta-analysis of the 222 QTLs collected was performed using BioMercator V4.2 software. A total of 52 MQTLs with 95% confidence intervals associated with tiller number were calculated. The distribution of MQTLs on each staining ranged from 0 to 8. No MQTLs were detected on chromosomes 10 and 12, and the highest number of MQTLs detected on chromosome 6 was 8 (Table 6). The PVE values of the MQTL ranged from 4 to 43%, with 23 MQTLs with PVE values exceeded 20%. MQTL 11.2 had the highest PVE value of 43%.

Table 6.
Meta-QTL analysis of tiller number traits in rice.
Of the 57 QTLs detected in the RIL population by crossing 9311 to PA64s, 13 QTLs were covered by MQTLs. Among them, on chromosome 1, two QTLs were covered by the same MQTL. Specifically, qD1tn1-3 and qD2tn1-2 were covered by MQTL1.6. On chromosome 2 two QTLs were covered, including qD2tn2 by MQTL2.1 and qD4TN2-1 by MQTL2.4. On chromosome 3, qD4TN3 was covered by MQTL3.3. On chromosome 4, two QTLs were covered by MQTLs, including qD2tn4-1 by MQTL4.1 and qD6TN4 by MQTL4.3. On chromosome 5, qD2tn5-2 was covered by MQTL5.6. On chromosome 6, three QTLs were covered by MQTL, including qD1TN6-2 covered by MQTL6.3, qD2tn6-1 covered by MQTL6.4, and qD4TN6 covered by MQTL6.6. On chromosome 7, qD5tn7 was covered by MQTL7.2. On chromosome 8, qD4TN8-1 was covered by MQTL8.1.
The raw QTLs on each MQTL ranged from 1 to 10, with a maximum of 10 raw QTLs for MQTL2.2 and a minimum of 1 for MQTL8.1. The 95% confidence intervals for the MQTLs ranged from 0.29 to 12.12 cM, and the physical lengths ranged from 0.27 to 6.0 Mb. There were only three MQTLs, MQTL6.6, MQTL7.2, and MQTL8.3, with physical lengths greater than 5 Mb, and the rest of the MQTLs were less than 5 Mb. The physical lengths of five MQTLs, MQTL1.6, MQTL2.6, MQTL5.5, MQTL6.8, and MQTL8.1, were all less than 0.5 Mb, and the physical lengths of MQTL6.8 and MQTL8.1 were the shortest at 0.27 Mb. These five MQTLs can be further studied and analyzed to mine tillering-related genes.
3.4. Analysis of Candidate Genes Related to Tillering in Rice
3.4.1. MQTL Candidate Gene Analysis
The obtained MQTLs were screened for candidate genes. The physical locations of the MQTLs were obtained using the Gramene database (https://archive.gramene.org/markers/ (accessed on 10 August 2022)), and the 52 MQTLs included a total of 19,274 candidate genes (Table S2). MQTL6.8 encompassed a minimum of 59 genes with a physical length of about 0.27 Mb. MQTL7.2 contains the most candidate genes with 936 genes and a physical length of about 6 Mb. Five MQTLs, MQTL1.6, MQTL2.6, MQTL3.1, MQTL6.8, and MQTL8.1, contain less than 100 candidate genes, which allows for the mining relevant genes related to tiller number. MQTL3.1, MQTL3.2, MQTL3.3, MQTL5.2, MQTL5.3, MQTL7.2, and MQTL8.2 do not have genes that have been reported to be related to tillering and can be further mined for tiller-related candidate genes. MQTL3.1 has a physical length of 0.37 Mb and a phenotypic contribution rate of 29%, and the interval is between 3,964,690 and 4,333,950. This MQTL can be screened for tillering-related candidate genes. MQTL1.6 (56 candidate genes) contains OsSPL2, showing higher expression in 9311, and a coding SNP (T441G) that is hypothesized to be the main effector gene for the stable QTL cluster (qmD1TN1, qD1tn1-3, qD2TN1-2).
Candidate gene screening of 52 MQTLs using the Rice Data Centre library (https://www.ricedata.cn/gene/ (accessed on 12 August 2022)) yielded 104 candidate genes related to tillering (Table 5). Some played important roles in rice tiller growth, e.g., MOC1 [17], FON1 [1], OsTB1 [3], and D3 [5]. Some genes regulate tiller response to nitrogen, e.g., OsTCP19 [20], OsNPF7.1 [25], OsNPF7.2 [26], and OsNR2 [18].
At a tiller number of 6 in 9311 and 11 in PA64s, RNA from parental tiller shoots was extracted (Figure 5A), and the relative expression of some genes related to positive or negative tiller regulation reported by Wang et al. [56] was determined (Table S1). HTD1 [57] and D3 [5] negatively regulated the growth of tiller shoots, and DLT [58] positively regulated the growth of tillers. The results determined in this study showed that the relative expression levels of D3 and HTD1 genes were significantly higher in the parental line 9311 than in PA64s. The relative expression levels of DLT genes were significantly higher in the parental line PA64s than in 9311, and the expression of genes related to the negative regulation of tillering was relatively higher in 9311 than in PA64s (Figure 5B).
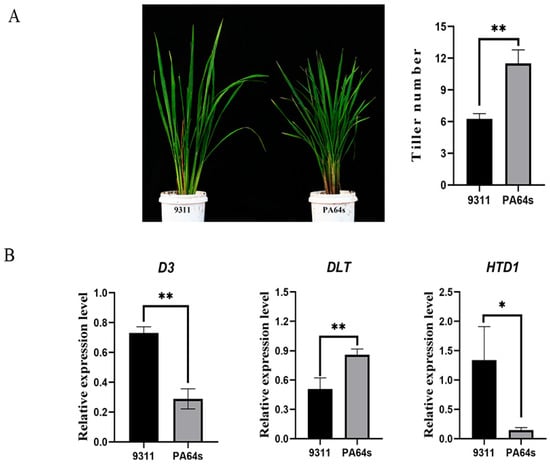
Figure 5.
Expression levels of genes related to regulating tiller number in rice. (A): Graph of parental monocots at the tillering stage; (B): number of parental tillers at the time of taking tiller buds; (B-D3): expression level of the D3 gene in parents; (B-DLT): expression level of the DLT gene in parents; (B-HTD1): expression level of the HTD1 gene in parents. *, p < 0.05; **, p < 0.01.
3.4.2. Analysis of MQTL1.6-Related Candidate Genes
By analyzing the QTLs detected in a single environment, it was found that the two QTLs, qD1tn1-3 and qD2TN1-2, both of which provided extremely high contributions to the tiller number phenotype in the pre-tiller stage, reached more than 40%, and the positional distance between the highest points of the two QTLs differed by 0.57 cM. One QTL (mks1-356) was detected in the pre-tiller stage in multi-environmental QTL analyses, and it was identified that mks1-356 did not exist in the pre-tiller stage. It was located 0.76 cM and 0.19 cM away from the qD2TN1-2 and qD1tn1-3 loci, and no environmental interactions were identified for mks1-356 using multi-environmental QTL analysis. MQTL1.6 covers this segment, which contains 56 candidate genes (Table S3), and the analysis of related genes that have been reported for tiller number by the National Rice Data Centre (https://www.ricedata.cn/gene/ (accessed on 12 August 2022)) revealed that this segment contains an OsSPL2 gene that has been reported as a Squamosa promoter-binding protein. This gene has not been studied in depth, and information is limited [59]. Quantitative gene analysis of OsSPL2 in 9311 and PA64s (Table S1) revealed that the relative expression level of the OsSPL2 gene in 9311 was significantly higher than that in PA64s during the pre-tillering stage. In addition, there was a single nucleotide mutation (SNP) (T → G) in the third exon at position 441, where the amino acid was mutated from phenylalanine to valine (Figure 6). Thus, we hypothesized that this locus caused the expression difference between the parents. The candidate gene for qD1tn1-3 and qD2TN1-2 is OsSPL2, and this gene may not be affected by the environment as a tiller nitrogen efficiency response gene.
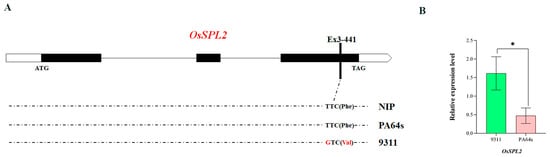
Figure 6.
SNPs and relative expression of the OsSPL2 gene in the parental strains. (A): Structure and comparison of the OsSPL2 gene in parental strains PA64s and 9311 and the reference Nipponbare; (B): indicates the expression level of the OsSPL2 gene in different parental strains. *, indicates the significance of the difference in expression levels between PA64s and 9311 at p < 0.05.
4. Discussion
4.1. Comparison of the QTLs Identified in This Study with Reported QTLs
Some of the QTLs detected in this study were located in the same or similar genomic regions as those reported by previous authors. For example, qD2TN1-2, qD1tn1-3, qmD1TN1, qD4TN8-1, and qmD6TN7 were located in similar positions as qNPT1-2, qNTT8-1, and qNTT7-2, respectively, as reported by B. Rajurkar [29]. qD2tn2 and qD6TN4 were located in similar positions as qTN-2 and qTN-4 as reported by Liang [55]. qD4TN2-1 and qmD4TN2 are close to qTNL2-1 as reported by Kwon [30]. In addition, qD4TN3 and qD5tn7 are similar in positions as qTN3 and qTN7 as reported by Liang [42]. qD2tn4-1 is similar in position to qPTN4.1 as reported by Beerelli [32]. qD2tn5-2 is similar in position to qMTN5-2 as reported by Sun [46]. qD1TN6-2, qD2tn6-1, qmD2TN3-1, qmD3TN3, qmD2TN6, and qmD4TN4 were similar in positions to qCTN6-5, qCTN6-6, qCTN3-1, and qCTN4-3 as reported by Jiang [16], and qD4TN6 was similar in position to qTN-6 as reported by Wang [27]. These results further verified the reliability of the QTL localization in this study and also indicated that some of the QTLs were conserved under different genetic backgrounds and environmental conditions.
4.2. Adaptive QTLs Associated with Dynamic Tillering in Rice RIL Populations Under Three Treatments
Phenotypic plasticity is the ability of the same genotype to exhibit different phenotypes in different environments [60,61]. Studies have shown that the plasticity of tillering traits to soil heterogeneity is universal, and this plasticity helps rice adapt to stress related to nitrogen deficits [61]. In this study, single-environment QTL analysis identified 10 QTLs (e.g., qD2tn2, qD5TN2-1, qD1tn2, qD4TN3, etc.) that contributed more than 10% under no-N treatment, and these QTLs may contain the main effector genes for the efficient response of tillering to nitrogen. Among them, the efficiency alleles of qD1tn2, qD1tn4, qD2tn2, and qD4TN3 were derived from PA64s, suggesting that these QTLs positively regulate tiller number under low nitrogen conditions. In addition, qD1TN6-2, qD3TN8-1, qD4TN12, and qD6tn5 contributed more than 10% under straw return treatment, suggesting that these QTLs may be involved in regulating rice adaptation to soil heterogeneity.
QTL–environment interaction (QEI) analyses reveal differences in phenotypic plasticity of different genotypes in different environments [62,63]. In this study, qmD1TN1 (mks1-356) did not have environmental interactions effects, and its contribution was higher in the normal nitrogen environment than under the no-nitrogen treatment condition, suggesting that this QTL may attenuate the repressive effect of 9311-related tiller genes under low nitrogen conditions. In addition, four QTLs with antagonistic pleiotropy (e.g., qmD1TN12, qmD2TN1, qmD2TN3-1, and qmD5TN6) were identified in this study, and these QTLs showed opposite effect values under different environments. For example, qmD1TN12 (mks12-267) increased the number of tillers under no-N treatment, whereas it decreased the number of tillers under the normal N environment. qmD2TN1 (mks1-334) and qmD2TN3-1 (mks3-105) increased the number of tillers under straw return condition, whereas they showed opposite effects under the normal N environment. These results suggest that the environmental specificity of QTLs needs to be considered in breeding for optimal trait selection in specific environments [64].
4.3. Dynamic Tillering in Rice RIL Populations Under Three Treatments Associated with Pleiotropic QTLs and Primary QTLs
QTL pleiotropy refers to the phenomenon that a single QTL locus can regulate multiple phenotypic traits. QTLs related to rice quality and grain traits have been shown to be pleiotropic [65,66]. Liu et al. [17] reported dynamic QTLs for tiller number over time, revealing differences in tiller QTLs at different developmental stages [16,67]. In this study, four QTLs with temporal pleiotropy were identified, of which mks3-165 (qD2tn3, qD5tn3) and mks6-235 (qD3TN6, qD5TN6) were detected in the rapid tiller growth and later stages of tiller development, respectively, suggesting that these QTLs may regulate rapid tiller growth and the formation of the highest tiller number. In addition, mks1-305 (qD1tn1-2, qD2tn1) was detected in the middle and early stages of tillering and might be related to the formation of tiller buds. In addition, mks7-139 (qD6TN7, qD3TN7) was detected in the rapid growth and late stages of tillering and showed significant effects under different treatments. These results indicate that this QTL is spatio-temporally multiplexed and might regulate the final effective spike formation.
In addition, two main effector QTLs (qD1tn1-3 and qD2TN1-2) with more than 40% phenotypic contribution were identified in this study, and both of their potentiated alleles were derived from 9311. In the multi-environmental analysis, qmD1TN1 (mks1-356) did not have environmental reciprocal effects and was located in close proximity to qD1tn1-3 and qD2TN1-2, suggesting that these three QTLs may be located at the same genetic locus. This main effect QTL may play a key role in tiller shoot formation and rapid growth period and is not affected by the environment, suggesting that it may be a main effect gene for efficient response to tiller nitrogen.
4.4. Meta-QTLs and Candidate Genes Associated with Tiller Traits in Rice
Meta-QTL analysis can significantly improve QTL localization accuracy and provide important clues for candidate gene mining by integrating QTL information from multiple studies [22,68]. In this study, 52 MQTLs were identified, of which MQTL3.1 and MQTL6.8 contained 82 and 59 candidate genes, respectively, and no reported tillering-related genes were found, suggesting that these intervals may contain new tillering-regulated genes. In addition, this study covered several nitrogen-efficient related genes (e.g., OsTCP19 [23], NPF7.1 [69], OsNPF7.2 [70], and OsNR2 [21]). Of these, OsTCP19 was confirmed to be a key gene in the nitrogen-efficient response of tillering, and a 29 bp deletion in its promoter region was associated with a nitrogen-efficient phenotype. This gene is located within the qD2tn6-1 interval and could be a priority target for the development of molecular markers for nitrogen efficiency in tillers.
The primary QTLs identified in this study, qD1tn1-3 and qD2TN1-2, were covered by MQTL1.6, an interval containing 56 candidate genes. Of these, OsSPL2 is the only reported tillering-related gene, and its function has not been analyzed in depth. OsSPL2 belongs to the Squamosa promoter binding protein-like (SPL) family of transcription factors, members of which play key regulatory roles in plant growth and development and stress response [59,71,72]. In this study, we found that the expression level of OsSPL2 in 9311 was significantly higher than that of PA64s, and its coding region was polymorphically different among the parents (site 441 of the third exon was mutated from T to G). This was hypothesized to be a possible cause of the functional difference. Therefore, OsSPL2 may be a key gene for this primary QTL, and its independence from environmental influences has the potential to be developed as a molecular marker. In addition, other candidate genes within the MQTL1.6 interval (e.g., LOC_Os01g69980 and LOC_Os01g70050) may also be involved in tillering regulation, which is worthy of further investigation.
4.5. Breeding Implications
The stable QTL qmD1TN1 (mks1-356) and its candidate gene OsSPL2, unaffected by environmental interactions, represent ideal targets for MAS to enhance tiller establishment under low N or straw return. Molecular markers linked to mks1-355/mks1-352 could efficiently introgress high-tillering alleles from 9311 into breeding lines. Pyramiding environment-specific QTLs with stable major-effect QTLs may optimize tiller architecture for sustainable systems. Candidate lines carrying the MQTL1.6 haplotype from 9311, combined with PA64s-derived alleles for qD2tn2 (low N tolerance), could balance tiller number and nitrogen efficiency.
4.6. Limitations of Year-to-Year Reproducibility and Statistical Stringency
In this study, although 57 tiller-related QTLs were identified, limitations still exist in their year-to-year reproducibility and statistical rigor. Inter-annual environmental variations (such as soil nitrogen and temperature) led to unstable detection of some key QTLs. Genotype–environment interaction (G × E) analysis indicates that their expression is regulated by specific environmental conditions, suggesting that QTL effects are context-dependent rather than universally stable. In terms of statistical methods, although the FDR correction (q < 0.10) was used to control false positives, the population of 124 recombinant inbred lines (RILs) had insufficient power to detect small-effect QTLs (PVE < 10%), and some minor-effect loci might have been missed. Future research can improve the reliability and universality of QTL detection by expanding the population size, conducting controlled-environment experiments, or integrating multi-model analysis, providing more robust genetic targets for precision breeding.
5. Conclusions
This study systematically analyzed the molecular basis of genetic regulation of tillering and environmental interactions in rice and revealed the dynamic characteristics of tillering-related QTLs and key candidate genes in multiple environments. Using single-environment, multi-environment, and meta-QTL integration analyses, 44 new unreported QTLs were identified, among which three main effective QTLs (mks1-355, mks1-352, and qmD1TN1) contributed more than 40% in two consecutive years of experiments and were unaffected by environmental interactions. These QTLs were further locked by the MQTL1.6 interval and might be the same genetic locus. It is worth noting that there are significant expression differences and polymorphisms of the candidate gene OsSPL2 among the parents, suggesting that it may be the main effector gene of tillering, and OsSPL2 may be a priority target for molecular marker development. In addition, four QTLs with antagonistic pleiotropy (e.g., qmD1TN12) were identified, which provides a theoretical basis for the targeted selection of favorable traits under specific environments. These QTLs exhibit stage- and environment-specific stability, with consistent effects during early tillering under defined nitrogen/straw treatments, rather than universal stability across all conditions. In the future, independent validation in a larger RIL population (n > 200) and transgenic analysis of OsSPL2 will solidify the functional relevance of the MQTL1.6 cluster. These steps are crucial for translating stage-specific QTLs into widely applicable breeding tools.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agriculture15111115/s1, Table S1: Primers for gene wxpression analysis and sequencing; Table S2: Analysis of Rice Tiller Nunber Rleated MQTL candidate Genes; Table S3: 56.MQTL 1.6 candidate genes; Table S4: Soil treatment information form.
Author Contributions
Conceptualization, Y.S., F.G. and Y.P.; methodology, Y.S., F.G. and P.Q.; software, Y.S., F.G. and P.Q.; validation, Y.S., F.G., P.Q. and S.Y.; formal analysis, Y.S., F.G., P.Q. and Y.P.; investigation, Y.S., F.G. and P.Q.; resources, Y.P.; data curation, F.G. and Y.S.; writing—review and editing, Y.S. and F.G.; visualization, Y.S.; supervision, W.Y., Y.H. and S.Y.; project administration, W.Y., Y.H. and Y.P.; funding acquisition, W.Y., Y.H. and S.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Central Guidance for Local Science and Technology Development Fund Projects, grant number 2024ZYD0339; Sichuan University-Zigong City Campus Local Cooperation Project, grant number 2022CDZG-22; the Key Research and Development Program of Sichuan, grant number 2021YFYZ0016; and the Major Science and Technology Projects in Sichuan Province, grant number 2022ZDZX0012.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The data reported in this study are contained within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Li, X.; Qian, Q.; Fu, Z.; Wang, Y.; Xiong, G.; Zeng, D.; Wang, X.; Liu, X.; Teng, S.; Hiroshi, F.; et al. Control of tillering in rice. Nature 2003, 422, 618–621. [Google Scholar] [CrossRef] [PubMed]
- Shao, G.; Lu, Z.; Xiong, J.; Wang, B.; Jing, Y.; Meng, X.; Liu, G.; Ma, H.; Liang, Y.; Chen, F.; et al. Tiller Bud Formation Regulators MOC1 and MOC3 Cooperatively Promote Tiller Bud Outgrowth by Activating FON1 Expression in Rice. Mol. Plant 2019, 12, 1090–1102. [Google Scholar] [CrossRef]
- Song, X.; Lu, Z.; Yu, H.; Shao, G.; Xiong, J.; Meng, X.; Jing, Y.; Liu, G.; Xiong, G.; Duan, J.; et al. IPA1 functions as a downstream transcription factor repressed by D53 in strigolactone signaling in rice. Cell Res. 2017, 27, 1128–1141. [Google Scholar] [CrossRef]
- Takeda, T.; Suwa, Y.; Suzuki, M.; Kitano, H.; Ueguchi-Tanaka, M.; Ashikari, M.; Matsuoka, M.; Ueguchi, C. The OsTB1 gene negatively regulates lateral branching in rice. Plant J. Cell Mol. Biol. 2003, 33, 513–520. [Google Scholar] [CrossRef]
- Ishikawa, S.; Maekawa, M.; Arite, T.; Onishi, K.; Takamure, I.; Kyozuka, J. Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol. 2005, 46, 79–86. [Google Scholar] [CrossRef]
- Zhang, M.; Hu, D.; Zhang, D. Advances in molecular mechanism of rice tillering. Biot. Resour. 2022, 44, 26–35. [Google Scholar] [CrossRef]
- Zhou, W.; Yan, F.; Chen, Y.; Ren, W. Optimized nitrogen application increases rice yield by improving the quality of tillers. Plant Prod. Sci. 2022, 25, 311–319. [Google Scholar] [CrossRef]
- Xu, J.; Zha, M.; Li, Y.; Ding, Y.; Chen, L.; Ding, C.; Wang, S. The interaction between nitrogen availability and auxin, cytokinin, and strigolactone in the control of shoot branching in rice (Oryza sativa L.). Plant Cell Rep. 2015, 34, 1647–1662. [Google Scholar] [CrossRef]
- Zhong, X.; Peng, S.; Sheehy, J.E.; Visperas, R.M.; Liu, H. Relationship between tillering and leaf area index: Quantifying critical leaf area index for tillering in rice. J. Agric. Sci. 2002, 138, 269–279. [Google Scholar] [CrossRef]
- McAllister, C.H.; Beatty, P.H.; Good, A.G. Engineering nitrogen use efficient crop plants: The current status. Plant Biotechnol. J. 2012, 10, 1011–1025. [Google Scholar] [CrossRef]
- Sang, S.; Cao, M.; Wang, Y.; Wang, J.; Sun, X.; Zhang, W.; Ji, S. Research progress of nitrogen efficiency related genes in rice. Sci. Agric. Sin. 2022, 55, 1479–1491. [Google Scholar]
- Kong, D. Effect on Nitrogen and Carbon Content and Microbial Community Structure of Wheatsoybean Rotation Soil under Straw Return and Fertilizer Application Treatments. Ph.D. Thesis, North West Agriculture and Forestry University, Xianyang, China, 2020. [Google Scholar]
- Wang, X.; Zhao, S.; Zheng, X.; Wang, Z.; He, G. Effects of Straw Returning and Nitrogen Application Rate on Grain Yield and Nitrogen Utilization of Winter Wheat. Sci. Agric. Sin. 2021, 54, 5043–5053. [Google Scholar]
- Yin, H.; Zhao, W.; Li, T.; Cheng, X.; Liu, Q. Balancing straw returning and chemical fertilizers in China: Role of straw nutrient resources. Renew. Sustain. Energy Rev. 2018, 81, 2695–2702. [Google Scholar] [CrossRef]
- Lei, L.; Zheng, H.L.; Wang, J.G.; Liu, H.L.; Sun, J.; Zhao, H.W.; Yang, L.M.; Zou, D.T. Genetic dissection of rice (Oryza sativa L.) tiller, plant height, and grain yield based on QTL mapping and metaanalysis. Euphytica 2018, 214, 109. [Google Scholar] [CrossRef]
- Jiang, H.; Jiang, L.; Guo, L.; Gao, Z.; Zeng, D.; Zhu, L.; Liang, G.; Qian, Q. Conditional and unconditional mapping of quantitative trait loci underlying plant height and tiller number in rice (Oryza sativa L.) grown at two nitrogen levels. Prog. Nat. Sci.-Mater. Int. 2008, 18, 1539–1547. [Google Scholar] [CrossRef]
- Liu, G.; Zhu, H.; Liu, S.; Zeng, R.; Zhang, Z.; Li, W.; Ding, X.; Zhao, F.; Zhang, G. Unconditional and conditional QTL mapping for the developmental behavior of tiller number in rice (Oryza sativa L.). Genetica 2010, 138, 885–893. [Google Scholar] [CrossRef]
- Liu, G.; Zhu, H.; Zhang, G.; Li, L.; Ye, G. Dynamic analysis of QTLs on tiller number in rice (Oryza sativa L.) with single segment substitution lines. Theor. Appl. Genet. 2012, 125, 143–153. [Google Scholar] [CrossRef]
- Xing, Z.; Tan, F.; Hua, P.; Sun, L.; Xu, G.; Zhang, Q. Characterization of the main effects, epistatic effects and their environmental interactions of QTLs on the genetic basis of yield traits in rice. Theor. Appl. Genet. 2002, 105, 248–257. [Google Scholar] [CrossRef]
- Gong, J.; Zheng, X.; Du, B.; Qian, Q.; Chen, S.; Zhu, L.; He, P. Comparative study of QTLs for agronomic traits of rice (Oriza sativa L.) between salt stress and nonstress environment. Sci. China Ser. C Life Sci. 2001, 44, 73–82. [Google Scholar] [CrossRef]
- Gao, Z.; Wang, Y.; Chen, G.; Zhang, A.; Yang, S.; Shang, L.; Wang, D.; Ruan, B.; Liu, C.; Jiang, H.; et al. The indica nitrate reductase gene OsNR2 allele enhances rice yield potential and nitrogen use efficiency. Nat. Commun. 2019, 10, 5207. [Google Scholar] [CrossRef]
- Hu, B.; Wang, W.; Ou, S.; Tang, J.; Li, H.; Che, R.; Zhang, Z.; Chai, X.; Wang, H.; Wang, Y.; et al. Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 2015, 47, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, H.; Jiang, Z.; Wang, W.; Xu, R.; Wang, Q.; Zhang, Z.; Li, A.; Liang, Y.; Ou, S.; et al. Genomic basis of geographical adaptation to soil nitrogen in rice. Nature 2021, 590, 600–605. [Google Scholar] [CrossRef] [PubMed]
- Guo, N.; Hu, J.; Yan, M.; Qu, H.; Luo, L.; Tegeder, M.; Xu, G. Oryza sativa Lysine-Histidine-type Transporter 1 functions in root uptake and root-to-shoot allocation of amino acids in rice. Plant J. Cell Mol. Biol. 2020, 103, 395–411. [Google Scholar] [CrossRef]
- Tao, Y.; Zhu, J.; Wang, J.; Fan, F.; Xu, Y.; Li, W.; Wang, F.; Chen, Z.; Jiang, Y.; Zhu, J.; et al. Development of functional markers and genotype screening for nitrogen use efficiency genes in rice. Acta Agron. Sin. 2022, 48, 3045–3056. [Google Scholar]
- Li, L.; Peng, Y.; Tang, S.; Yu, D.; Tian, M.; Guo, F.; Chen, Y.; Yang, G.; Wang, X.; Hu, Y. Mapping QTL for leaf pigment content at dynamic development stage and analyzing Meta-QTL in rice. Euphytica 2021, 217, 90. [Google Scholar] [CrossRef]
- Wang, X.; Li, W.; OUyang, L.; Xu, J.; Chen, X.; Bian, J.; Hu, L.; Peng, X.; He, X.; Fu, J.; et al. QTL mapping for plant architecture in rice based on chromosome segment substitution lines. Acta Agron. Sin. 2022, 48, 1141–1151. [Google Scholar] [CrossRef]
- Luo, L.; Lei, L.; Liu, J.; Zhang, R.; Jin, G.; Cui, D.; Li, M.; Ma, X.; Zhao, Z.; Han, L. Mapping QTLs for yield-related traits using chromosome segment substitution lines of Dongxiang common wild rice (Oryza rufipogon Griff.) and Nipponbare (Oryza sativa L.). Acta Agron. Sin. 2021, 47, 1391–1401. [Google Scholar]
- Rajurkar, A.B.; Muthukumar, C.; Ayyenar, B.; Thomas, H.B.; Babu, R.C. Mapping consistent additive and epistatic QTLs for plant production traits under drought in target populations of environment using locally adapted landrace in rice (Oryza sativa L.). Plant Prod. Sci. 2021, 24, 388–403. [Google Scholar] [CrossRef]
- Kwon, Y.-H.; Kabange, N.-R.; Lee, J.-Y.; Lee, S.-M.; Cha, J.-K.; Shin, D.-J.; Cho, J.-H.; Kang, J.-W.; Ko, J.-M.; Lee, J.-H. Novel QTL Associated with Shoot Branching Identified in Doubled Haploid Rice (Oryza sativa L.) under Low Nitrogen Cultivation. Genes 2021, 12, 745. [Google Scholar] [CrossRef]
- Suman, K.; Neeraja, C.N.; Madhubabu, P.; Rathod, S.; Bej, S.; Jadhav, K.P.; Kumar, J.A.; Chaitanya, U.; Pawar, S.C.; Rani, S.H.; et al. Identification of Promising RILs for High Grain Zinc Through Genotype x Environment Analysis and Stable Grain Zinc QTL Using SSRs and SNPs in Rice (Oryza sativa L.). Front. Plant Sci. 2021, 12, 587482. [Google Scholar] [CrossRef]
- Beerelli, K.; Balakrishnan, D.; Addanki, K.R.; Surapaneni, M.; Rao Yadavalli, V.; Neelamraju, S. Mapping of QTLs for Yield Traits Using F2:3:4 Populations Derived From Two Alien Introgression Lines Reveals qTGW8.1 as a Consistent QTL for Grain Weight From Oryza nivara. Front. Plant Sci. 2022, 13, 790221. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Anis, G.B.; Wang, R.; Wu, W.; Yu, N.; Shen, X.; Zhan, X.; Cheng, S.; Cao, L. Genetic dissection of QTL against phosphate deficiency in the hybrid rice ‘Xieyou9308’. Plant Growth Regul. 2018, 84, 123–133. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, H.; Hu, J.; Wang, X.; Huang, M.; Wang, H. Further QTL mapping for yield component traits using introgression lines in rice (Oryza sativa L.) under drought field environments. Euphytica 2018, 214, 33. [Google Scholar] [CrossRef]
- Li, Z.; Hua, Z.; Dong, L.; Zhu, W.; He, G.; Qu, L.; Qi, N.; Xu, Z.; Wang, F. Quantitative Trait Locus Mapping for Yield-Associated Agronomic Traits in a BC2F6 Population of Japonica Hybrid Rice Liaoyou 5218. J. Plant Growth Regul. 2020, 39, 60–71. [Google Scholar] [CrossRef]
- Zhou, X.; Lin, J.; Yao, W.; Zhai, Y.; Li, B.; Ding, X.; Liu, X.; Xiao, Y.; Liu, J. Localisation of QTL for tiller number traits using recombinant inbred line populations in rice. Mol. Plant Breed. 2019, 17, 7770–7777. [Google Scholar] [CrossRef]
- Emami, M.P.; Mohebalipour, N.; Ebadi, A.A.; Nourafcan, H.; Ajali, J. Qtl Mapping of Some Morphological Traits of Rice in a Rils Population Derived from Hashemi and Modified Nemat Varieties Hybrid. Genetika 2020, 52, 1087–1106. [Google Scholar] [CrossRef]
- Liang, Y.; Zhang, Q.; Wang, S.; Deng, Q.; Li, P.; Li, Y.; Yan, S. Construction of SSR genetic linkage maps of sequenced rice varieties and their gene locus analysis for agronomic traits. Chin. J. China Biotechnol. 2007, 27, 28–34. [Google Scholar] [CrossRef]
- Ye, S.; Zhang, Q.; Li, J.; Zhao, B.; Li, P. QTL localisation for six agronomic traits in rice using PA64S/Nippobare F2 population. China J. Rice Sci. 2007, 21, 39–43. [Google Scholar] [CrossRef]
- Venu, R.C.; Ma, J.; Jia, Y.; Liu, G.; Jia, M.H.; Nobuta, K.; Sreerekha, M.V.; Moldenhauer, K.; McClung, A.M.; Meyers, B.C.; et al. Identification of Candidate Genes Associated with Positive and Negative Heterosis in Rice. PLoS ONE 2014, 9, e95178. [Google Scholar] [CrossRef]
- Xu, Y.; This, D.; Pausch, R.C.; Vonhof, W.M.; Coburn, J.R.; Comstock, J.P.; McCouch, S.R. Leaf-level water use efficiency determined by carbon isotope discrimination in rice seedlings: Genetic variation associated with population structure and QTL mapping. Theor. Appl. Genet. 2009, 118, 1065–1081. [Google Scholar] [CrossRef]
- Liang, Y.; Zhan, X.; Gao, Z.; Lin, Z.; Yang, Z.; Zhang, Y.; Shen, X.; Cao, L.; Cheng, S. Mapping of QTLs associated with important agronomic traits using three populations derived from a super hybrid rice Xieyou9308. Euphytica 2012, 184, 1–13. [Google Scholar] [CrossRef]
- Zhou, S.; Zhu, M.; Wang, F.; Huang, J.; Wang, G. Mapping of Qtls for Yield and Its Components in a Rice Recombinant Inbred Line Population. Pak. J. Bot. 2013, 45, 183–189. [Google Scholar]
- Sun, P.; Cai, H.; Wei, X. QTL analysis of maximum tiller number and effective tiller number in rice. Henan Agric. Sci. 2014, 43, 12–15. [Google Scholar] [CrossRef]
- Lim, J.-H.; Yang, H.-J.; Jung, K.-H.; Yoo, S.-C.; Paek, N.-C. Quantitative Trait Locus Mapping and Candidate Gene Analysis for Plant Architecture Traits Using Whole Genome Re-Sequencing in Rice. Mol. Cells 2014, 37, 149–160. [Google Scholar] [CrossRef]
- Sun, J.; Wang, J.; Liu, H.; Xie, D.; Zheng, H.; Zhao, H.; Zou, D.; Luan, F. Dynamic QTL Analysis of Rice Seedling Height and Tiller Number Under Salt Stress. J. Nucl. Agric. Sci. 2015, 29, 235–243. [Google Scholar]
- Xu, F.; Huang, Y.; Bao, J. Identification of QTLs for agronomic traits in indica rice using an RIL population. Genes Genom. 2015, 37, 809–817. [Google Scholar] [CrossRef]
- Surapaneni, M.; Balakrishnan, D.; Mesapogu, S.; Addanki, K.R.; Yadavalli, V.R.; Venkata, T.V.G.N.; Neelamraju, S. Identification of Major Effect QTLs for Agronomic Traits and CSSLs in Rice from Swarna/Oryza nivara Derived Backcross Inbred Lines. Front. Plant Sci. 2017, 8, 1027. [Google Scholar] [CrossRef]
- Jiang, H.; Jiang, L.; Sun, H.; Zeng, L.; Xue, D.; Liang, G. Developmental QTL analysis of stem tiller extinction in two rice DH populations. J. Yangzhou Univ. Agric. Life Sci. Ed. 2008, 29, 53–60. [Google Scholar] [CrossRef]
- Jiang, H. Analysis and Mutant Gene Mapping of Tillering and Leaf-related Trait in Rice (Oryza sativa L.). Ph.D. Thesis, Yangzhou University, Yangzhou, China, 2008. [Google Scholar]
- Kennard, C.; Phillips, L.; Porter, A. Genetic dissection of seed shattering, agronomic, and color traits in American wildrice (Zizania palustris var. interior L.) with a comparative map. Theor. Appl. Genet. 2002, 105, 1075–1086. [Google Scholar] [CrossRef]
- Kobayashi, S.; Fukuta, Y.; Sato, T.; Osaki, M.; Khush, G.S. Molecular marker dissection of rice (Oryza sativa L.) plant architecture under temperate and tropical climates. Theor. Appl. Genet. 2003, 107, 1350–1356. [Google Scholar] [CrossRef]
- Ren, X.; Weng, Q.; Zhu, L.; He, G. Mapping Quantitative Trait Loci for Tillering Ability in Rice. Wuhan Univ. J. Nat. Sci. Ed. 2003, 49, 533–537. [Google Scholar]
- Miyamoto, N.; Goto, Y.; Matsui, M.; Ukai, Y.; Morita, M.; Nemoto, K. Quantitative trait loci for phyllochron and tillering in rice. Theor. Appl. Genet. 2004, 109, 700–706. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Zhang, Q.; Ye, S.; Liu, M.; Yin, D.; Li, P. QTL mapping and analysis for tiller and its relafed traits in rice. Chin. Agric. Sci. Bull. 2005, 21, 47–52. [Google Scholar]
- Wang, Y.; Li, J. Branching in rice. Curr. Opin. Plant Biol. 2011, 14, 94–99. [Google Scholar] [CrossRef]
- Zheng, X.; Yang, X.; Chen, Z.; Xie, W.; Yue, X.; Zhu, H.; Chen, S.; Sun, X. Arabidopsis SMAX1 overaccumulation suppresses rosette shoot branching and promotes leaf and petiole elongation. Biochem. Biophys. Res. Commun. 2021, 553, 44–50. [Google Scholar] [CrossRef]
- Oikawa, T.; Kyozuka, J. Two-Step Regulation of LAX PANICLE1 Protein Accumulation in Axillary Meristem Formation in Rice. Plant Cell 2009, 21, 1095–1108. [Google Scholar] [CrossRef]
- Zhang, Z.; Sun, X.; Ma, X.; Xu, B.; Zhao, Y.; Ma, Z.; Li, G.; Khan, N.U.; Pan, Y.; Liang, Y.; et al. GNP6, a novel allele of MOC1, regulates panicle and tiller development in rice. Crop J. 2021, 9, 57–67. [Google Scholar] [CrossRef]
- Lu, Z.; Yu, H.; Xiong, G.; Wang, J.; Jiao, Y.; Liu, G.; Jing, Y.; Meng, X.; Hu, X.; Qian, Q.; et al. Genome-Wide Binding Analysis of the Transcription Activator Ideal Plant Architecture1 Reveals a Complex Network Regulating Rice Plant Architecture. Plant Cell 2013, 25, 3743–3759. [Google Scholar] [CrossRef]
- Zheng, J.; Hong, K.; Zeng, L.; Wang, L.; Kang, S.; Qu, M.; Dai, J.; Zou, L.; Zhu, L.; Tang, Z.; et al. Karrikin Signaling Acts Parallel to and Additively with Strigolactone Signaling to Regulate Rice Mesocotyl Elongation in Darkness. Plant Cell 2020, 32, 2780–2805. [Google Scholar] [CrossRef]
- Kulkarni, K.P.; Vishwakarma, C.; Sahoo, S.P.; Lima, J.M.; Nath, M.; Dokku, P.; Gacche, R.N.; Mohapatra, T.; Robin, S.; Sarla, N.; et al. A substitution mutation in OsCCD7 cosegregates with dwarf and increased tillering phenotype in rice. J. Genet. 2014, 93, 389–401. [Google Scholar] [CrossRef]
- Zou, J.; Zhang, S.; Zhang, W.; Li, G.; Chen, Z.; Zhai, W.; Zhao, X.; Pan, X.; Xie, Q.; Zhu, L. The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J. Cell Mol. Biol. 2006, 48, 687–698. [Google Scholar] [CrossRef]
- Miura, K.; Ikeda, M.; Matsubara, A.; Song, X.-J.; Ito, M.; Asano, K.; Matsuoka, M.; Kitano, H.; Ashikari, M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 2010, 42, 545–549. [Google Scholar] [CrossRef]
- Dai, Z.; Wang, J.; Yang, X.; Lu, H.; Miao, X.; Shi, Z. Modulation of plant architecture by the miR156f-OsSPL7-OsGH3.8 pathway in rice. J. Exp. Bot. 2018, 69, 5117–5130. [Google Scholar] [CrossRef]
- Leran, S.; Varala, K.; Boyer, J.-C.; Chiurazzi, M.; Crawford, N.; Daniel-Vedele, F.; David, L.; Dickstein, R.; Fernandez, E.; Forde, B.; et al. A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci. 2014, 19, 5–9. [Google Scholar] [CrossRef]
- Huang, W.; Nie, H.; Feng, F.; Wang, J.; Lu, K.; Fang, Z. Altered expression of OsNPF7.1 and OsNPF7.4 differentially regulates tillering and grain yield in rice. Plant Sci. 2019, 283, 23–31. [Google Scholar] [CrossRef] [PubMed]
- McSteen, P. Hormonal Regulation of Branching in Grasses. Plant Physiol. 2009, 149, 46–55. [Google Scholar] [CrossRef]
- Wu, J.; Guo, X.; Lu, J.; Wang, X.; Xu, Z.; Zhang, X. Effects of continuous straw mulching on soil physical and chemical properties and crop yields in paddy-upland rotation sys-tem. Plant Nutr. Fertil. Sci. 2012, 18, 587–594. [Google Scholar]
- Liu, Z.; Tao, L.; Liu, T.; Zhang, X.; Wang, W.; Song, J.; Yu, C.; Peng, X. Nitrogen application after low-temperature exposure alleviates tiller decrease in rice. Environ. Exp. Bot. 2019, 158, 205–214. [Google Scholar] [CrossRef]
- Kiba, T.; Takei, K.; Kojima, M.; Sakakibara, H. Side-Chain Modification of Cytokinins Controls Shoot Growth in Arabidopsis. Dev. Cell 2013, 27, 452–461. [Google Scholar] [CrossRef]
- Schaller, G.E.; Street, I.H.; Kieber, J.J. Cytokinin and the cell cycle. Curr. Opin. Plant Biol. 2014, 21, 7–15. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).