Candidate Pheromone Receptors of the Red-Belted Clearwing Moth Synanthedon myopaeformis Bind Pear Ester and Other Semiochemicals
Abstract
1. Introduction
2. Material and Methods
2.1. Insect Dissection and RNA Extraction
2.2. RNA Sequencing and Transcriptomics Analysis
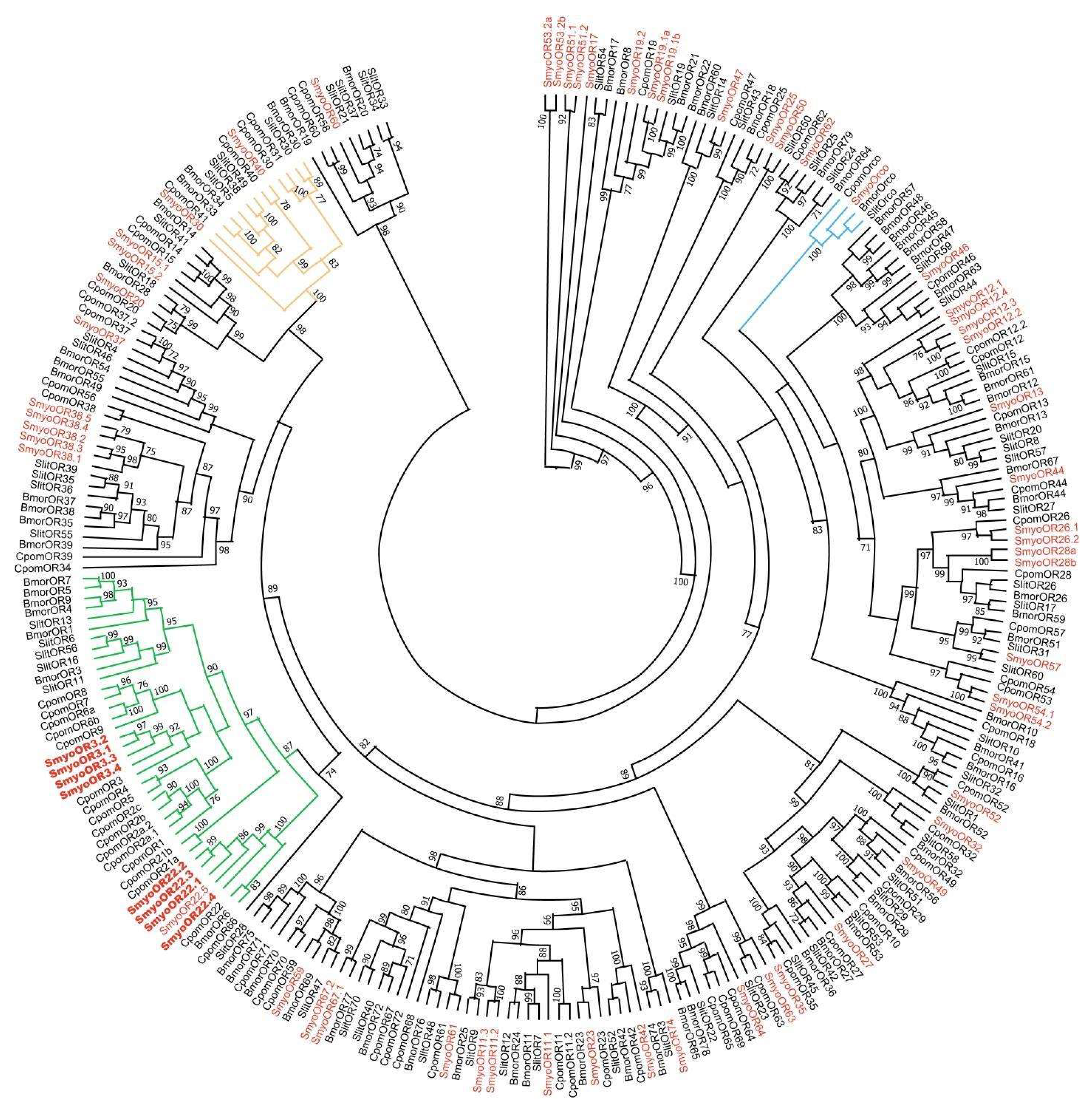
2.3. Phylogenetic Analysis of SmyoORs
2.4. Cloning of Olfactory Receptors for Expression in the Drosophila Empty Neuron System
2.5. Single Sensillum Recordings (SSRs)
2.6. Gas Chromatography Coupled with Single Sensillum Recordings (GC-SSR)
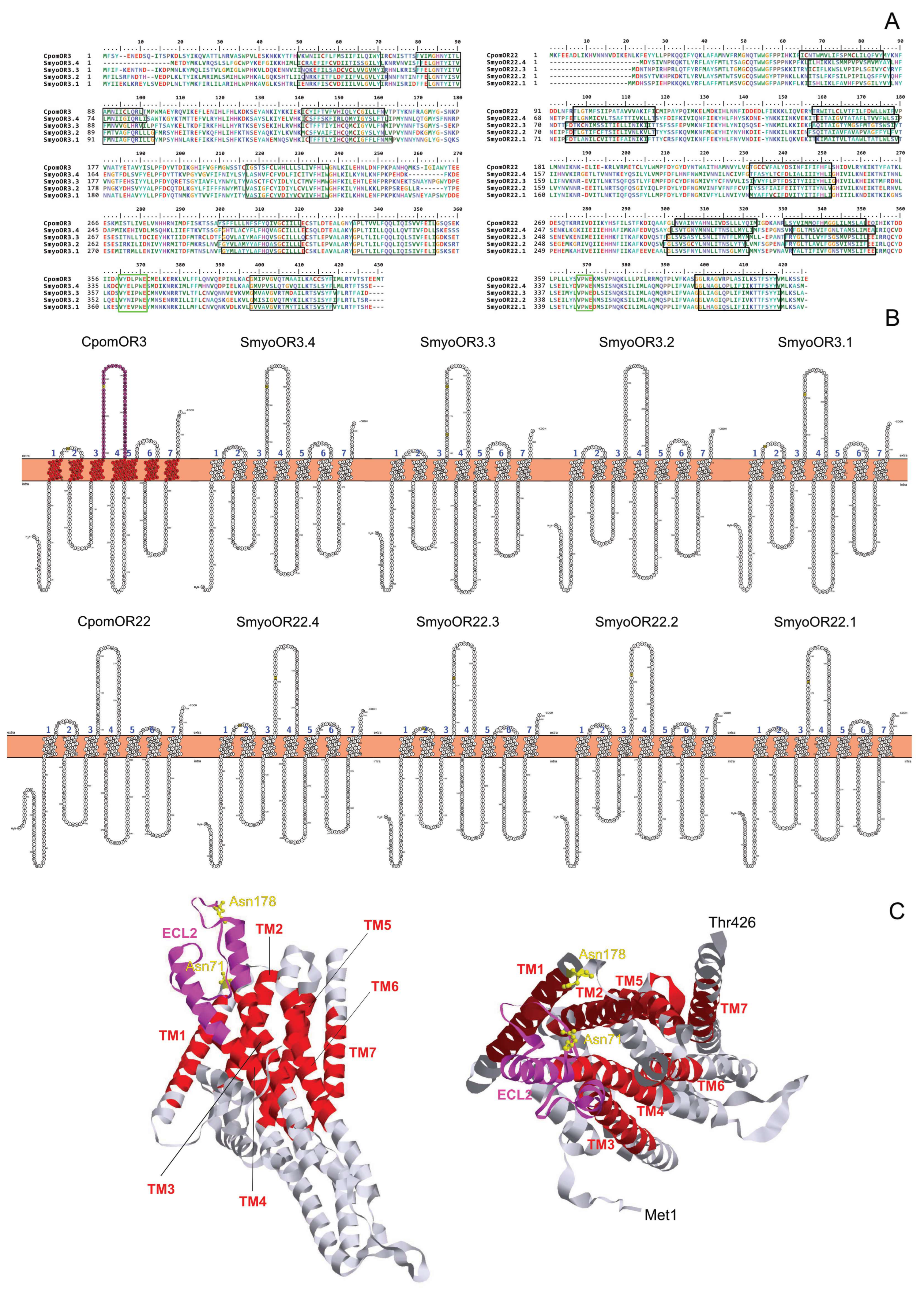
2.7. Sequence and Structural Analysis
3. Results
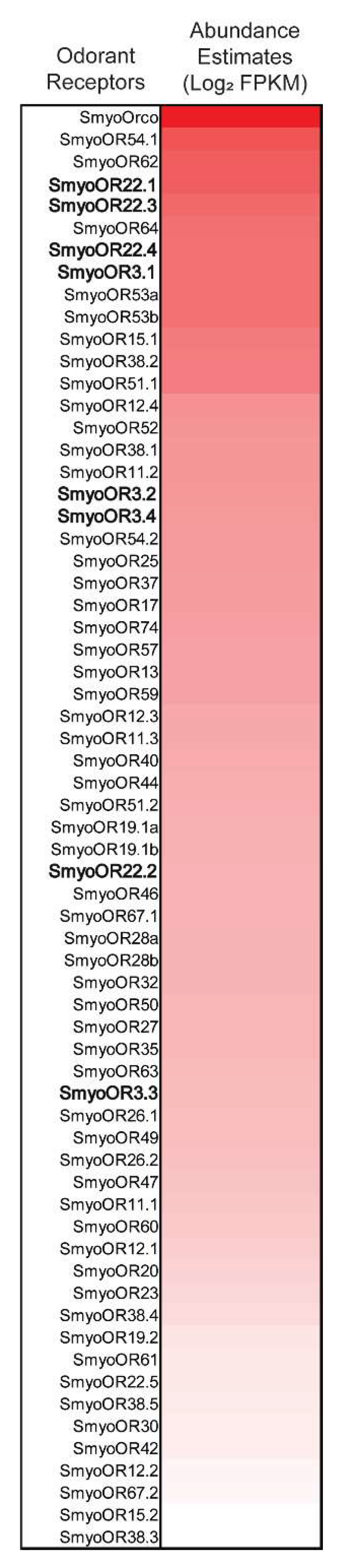
3.1. Phylogenetic Analysis and Relative Expression of SmyoORs
3.2. Functional Characterization and Activation of SmyoORs
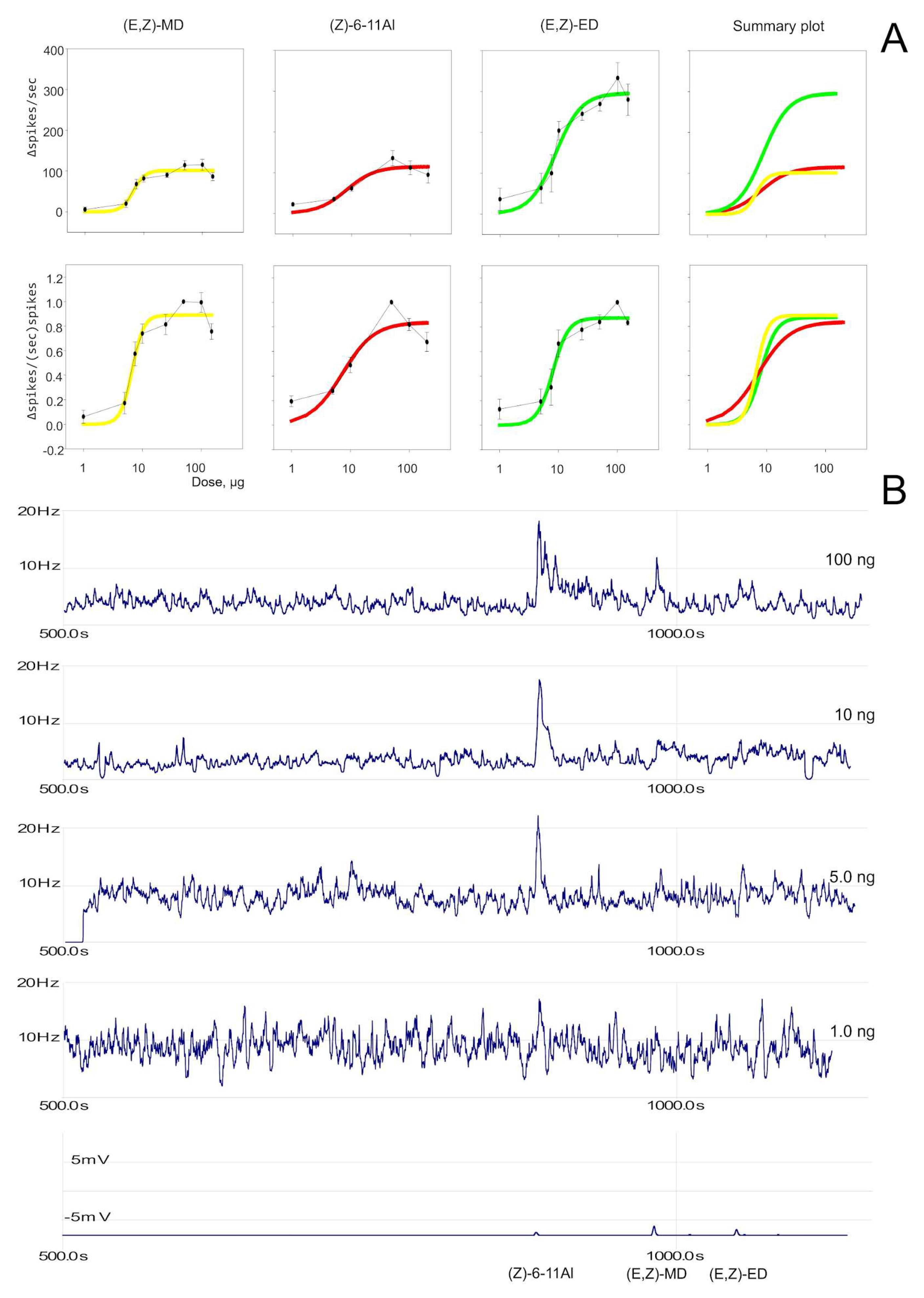
3.3. Dose–Response Experiments
3.4. GC-SSR Testing Headspace Collections
3.5. Structural Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Dickler, E. Zur Biologie und Schadwirkung von Synanthedon myopaeformis Brkh. (Lepid., Aegeriidae), einem neuen Schädling in Apfeldichtpflanzungen. Z. Angew. Entomol. 1976, 82, 259–266. [Google Scholar] [CrossRef]
- Warner, J.; Hay, S. Observation, monitoring, and control of clearwing borers (Lepidoptera: Sesiidae) on apple in central Ontario. Can. Entomolol. 1985, 117, 1471–1478. [Google Scholar] [CrossRef]
- Bergh, J.C.; Leskey, T.C. Biology, ecology, and management of dogwood borer in eastern apple orchards. Can. Entomolol. 2003, 135, 615–635. [Google Scholar] [CrossRef]
- Ateyyat, M.A. Efficacy of some insecticides against the small red-belted clearwing borer, Synanthedon myopaeformis (Borkh.) (Lepidoptera: Sessiidae), in apple orchards in Jordan. Commun. Agric. Appl. Biol. Sci. 2005, 70, 759–765. [Google Scholar]
- Trematerra, P. On the possibility of mass-trapping Synanthedon myopaeformis Bkh. (Lep., Sesiidae). J. Appl. Entomol. 1993, 115, 476–483. [Google Scholar] [CrossRef]
- Balázs, K.; Bujáki, G.; Farkas, K. Incorporation of apple clearwing (Synanthedon myopaeformis Bork.) control into the IPM system of apple. Acta Hortic. 1996, 422, 134–139. [Google Scholar] [CrossRef]
- Judd, G.J.R.; Bedford, K.; Cossentine, J. Control of Apple Clearwing Moth, Synanthedon myopaeformis, with Tree-trunk Applications of Reduced-risk Insecticides, Nematodes and Barriers. J. Entomol. Soc. Br. Columbia 2015, 112, 69–83. [Google Scholar] [CrossRef]
- El-Ashry, R.M.; El-Sheikh, M.F.M.; Azazi, A.M.; Olphat, E.A. Field Control of Synanthedon myopaeformis Borkh and Zeuzera pyrina L. Using Entomopathogenic Nematodes, Insecticides and Microbial Agents. Egypt. J. Agronematol. 2018, 17, 121–131. [Google Scholar] [CrossRef]
- Erler, F. Efficacy of tree trunk coating materials in the control of the apple clearwing, Synanthedon myopaeformis. J. Insect Sci. 2010, 10, 63. [Google Scholar] [CrossRef]
- Ateyyat, M.A. Effect of three apple rootstocks on the population of the small red-belted clearwing borer, Synanthedon myopaeformis. J. Insect Sci. 2006, 6, 40. [Google Scholar] [CrossRef]
- Witzgall, P.; Stelinski, L.; Gut, L.; Thomson, D. Codling moth management and chemical ecology. Ann. Rev. Entomol. 2008, 53, 503–522. [Google Scholar] [CrossRef] [PubMed]
- Tumlinson, J.H.; Yonce, C.E.; Doolittle, R.E.; Heath, R.R.; Gentry, C.R.; Mitchell, E.R. Sex pheromones and reproductive isolation of the lesser peachtree borer and the peachtree borer. Science 1974, 185, 614–616. [Google Scholar] [CrossRef]
- Voerman, S.; Minks, A.K.; Van Wetswinker, G.; Tumlinson, J.H. Activity of 3,13-octadecadien-1-ol acetates to the male clearwing moth Synanthedon myopaeformis (Borkhausen) (Lepidoptera, Sesiidae). Entomol. Exp. Applicata 1978, 23, 301–304. [Google Scholar] [CrossRef]
- Szöcs, G.; Tóth, M.; Sziráki, G.Y.; Schwarz, M. 2,13- and 3,13-octadecadienyl compounds composing sex attractants for tineid and sesiid moths (Lepidoptera). Biochem. Syst. Ecol. 1989, 17, 417–422. [Google Scholar] [CrossRef]
- El-Sayed, A.M. The Pherobase: Database of Insect Pheromones and Semiochemicals [Online]. 2009. Available online: http://www.pherobase.com (accessed on 28 March 2024).
- Vogt, R.G. Molecular Basis of Pheromone Detection in Insects. In Comprehensive Insect Physiology, Biochemistry, Pharmacology and Molecular Biology; Gilbert, L.I., Iatro, K., Gill, S., Eds.; Vol. 3 Endocrinology; Elsevier: London, UK, 2005; pp. 753–804. [Google Scholar] [CrossRef]
- Fleischer, J.; Krieger, J. Insect pheromone receptors—Key elements in sensing intraspecific chemical signals. Front. Cell. Neurosci. 2018, 12, 425. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Pellegrino, M.; Nakagawa, T.; Nakagawa, T.; Vosshall, L.B.; Touhara, K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 2008, 452, 1002–1006. [Google Scholar] [CrossRef]
- Touhara, K.; Vosshall, L. Sensing odorants and pheromones with chemosensory receptors. Annu. Rev. Physiol. 2009, 71, 307–332. [Google Scholar] [CrossRef]
- Wheelwright, M.; Whittle, C.R.; Riabinina, O. Olfactory systems across mosquito species. Cell Tissue Res. 2021, 383, 75–90. [Google Scholar] [CrossRef]
- Montagne, N.; Chertemps, T.; Brigaud, I.; Francois, A.; Francois, M.C.; de Fouchier, A.; Lucas, P.; Larsson, M.C.; Jacquin-Joly, E. Functional characterization of a sex pheromone receptor in the pest moth Spodoptera littoralis by heterologous expression in Drosophila. Eur. J. Neurosci. 2012, 36, 2588–2596. [Google Scholar] [CrossRef]
- de Fouchier, A.; Walker, W.B.; Montagne, N.; Steiner, C.; Binyameen, M.; Schlyter, F.; Chertemps, T.; Maria, A.; François, M.-C.; Monsempes, C.; et al. Functional evolution of Lepidoptera olfactory receptors revealed by deorphanization of a moth repertoire. Nat. Commun. 2017, 8, 15709. [Google Scholar] [CrossRef]
- Dobritsa, A.A.; van der Goes van Naters, W.; Warr, C.G.; Steinbrecht, R.A.; Carlson, J.R. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron 2003, 37, 827–841. [Google Scholar] [CrossRef] [PubMed]
- Hallem EAHo, M.G.; Carlson, J.R. The molecular basis of odor coding in the Drosophila antenna. Cell 2004, 117, 965–979. [Google Scholar] [CrossRef]
- Brand, A.H.; Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993, 118, 401–415. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, F.; Witzgall, P.; Walker, W.B. Protocol for heterologous expression of insect odourant receptors in Drosophila. Front. Ecol. Evol. 2016, 4, 24. [Google Scholar] [CrossRef]
- Walker, W.B., III; Gonzalez, F.; Garczynski, S.; Witzgall, P. The chemosensory receptors of codling moth Cydia pomonella–expression in larvae and adults. Sci. Rep. 2016, 6, 23518. [Google Scholar] [CrossRef]
- Bengtsson, J.M.; Gonzalez, F.; Cattaneo, A.M.; Montagné, N.; Walker, W.B.; Bengtsson, M.; Anfora, G.; Ignell, R.; Jacquin-Joly, E.; Witzgall, P. A predicted sex pheromone receptor of codling moth Cydia pomonella detects the plant volatile pear ester. Front. Ecol. Evol. 2014, 2, 33. [Google Scholar] [CrossRef]
- Cattaneo, A.M.; Kwadha, C.A.; Pullmann-Lindsley, H.; Erdei, A.L.; Pitts, R.J.; Walker, W.B., III. Functional Characterization of a Female-Biased Chemoreceptor of the Codling Moth (Cydia pomonella) Responding to Aldehydes and Other Volatile Compounds. J. Chem. Ecol. 2025, 51, 28. [Google Scholar] [CrossRef]
- Light, D.M.; Knight, A.L.; Henrick, C.A.; Rajapaska, D.; Lingren, B.; Dickens, J.C.; Reynolds, K.M.; Buttery, R.G.; Merrill, G.; Roitman, J.; et al. A pear-derived kairomone with pheromonal potency that attracts male and female codling moth, Cydia pomonella (L.). Naturwissenschaften 2001, 88, 333–338. [Google Scholar] [CrossRef]
- Light, D.M.; Knight, A.L. Specificity of codling moth (Lepidoptera: Tortricidae) for the host plant kairomone, ethyl (2E,4Z)-2,4-decadienoate: Field bioassays with pome fruit volatiles, analogue, and isomeric compounds. J. Agric. Food Chem. 2005, 53, 4046–4053. [Google Scholar] [CrossRef]
- Tóth, M.; Jósvai, J.; Hári, K.; Pénzes, B.; Vuity Zs Holb, I.; Szarukán, I.; Kecskés Zs Dorgán-Zsuga, I.; Koczor, S.; Voigt, E. Pear ester based lures for the codling moth Cydia pomonella L.—A summary of research efforts in Hungary. Acta Phytopathol. Entomol. Hung. 2014, 49, 37–47. [Google Scholar] [CrossRef]
- Knight, A.L.; Preti, M.; Basoalto, E.; Mujica, V.; Favaro, R.; Angeli, S. Combining female removal with mating disruption for management of Cydia pomonella in apple. Entomol. Gen. 2022, 42, 309–321. [Google Scholar] [CrossRef]
- Tóth, M.; Landolt, P.; Szarukán, I.; Szólláth, I.; Vitányi, I.; Pénzes, B.; Hári, K.; Katalin Jósvai, J.; Koczor, S. Female-targeted attractant containing pear ester for Synanthedon myopaeformis. Entomol. Exp. Appl. 2012, 142, 27–35. [Google Scholar] [CrossRef]
- Erdei, A.L.; Sousa, M.; Gonzalez, F.; Bengtsson, M.; Witzgall, P. Host Plant Odour and Sex Pheromone are Integral to Mate Finding in Codling Moth. J. Chem. Ecol. 2025, 51, 13. [Google Scholar] [CrossRef] [PubMed]
- Cock, P.J.; Fields, C.J.; Goto, N.; Heuer, M.L.; Rice, P.M. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010, 38, 1767–1771. [Google Scholar] [CrossRef]
- Walker, W.B., III; Mori, B.A.; Cattaneo, A.M.; Gonzalez, F.; Witzgall, P.; Becher, P.G. Comparative transcriptomic assessment of the chemosensory receptor repertoire of Drosophila suzukii adult and larval olfactory organs. Comp. Biochem. Physiol. Part D Genom. Proteom. 2023, 45, 101049. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Seppey, M.; Manni, M.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness. In Gene Prediction: Methods and Protocols; Humana: New York, NY, USA, 2019; pp. 227–245. [Google Scholar] [CrossRef]
- Waterhouse, R.M.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. Using BUSCO to assess insect genomic resources. In Insect Genomics: Methods and Protocols; Humana Press: New York, NY, USA, 2019; pp. 59–74. [Google Scholar] [CrossRef]
- Nishimura, O.; Hara, Y.; Kuraku, S. gVolante for standardizing completeness assessment of genome and transcriptome assemblies. Bioinformatics 2017, 33, 3635–3637. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; De Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.O.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Briscoe, A.D.; Macias-Munoz, A.; Kozak, K.M.; Walters, J.R.; Yuan, F.R.; Jamie, G.A.; Martin, S.H.; Dasmahapatra, K.K.; Ferguson, L.C.; Mallet, J.; et al. Female Behaviour Drives Expression and Evolution of Gustatory Receptors in Butterflies. PLoS Genet. 2013, 9, e1003620. [Google Scholar] [CrossRef]
- Wanner, K.W.; Anderson, A.R.; Trowell, S.C.; Theilmann, D.A.; Robertson, H.M.; Newcomb, R.D. Female-biased expression of odourant receptor genes in the adult antennae of the silkworm, Bombyx mori. Insect Mol. Biol. 2007, 16, 107–119. [Google Scholar] [CrossRef]
- Walker, W.B., III; Cattaneo, A.M.; Stout, J.L.; Evans, M.L.; Garczynski, S.F. Chemosensory Receptor Expression in the Abdomen Tip of the Female Codling Moth, Cydia pomonella L. (Lepidoptera: Tortricidae). Insects 2023, 14, 948. [Google Scholar] [CrossRef]
- Walker, W.B., III; Roy, A.; Anderson, P.; Schlyter, F.; Hansson, B.S.; Larsson, M.C. Transcriptome Analysis of Gene Families Involved in Chemosensory Function in Spodoptera littoralis (Lepidoptera: Noctuidae). BMC Genom. 2019, 20, 428. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Longueville, J.E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef] [PubMed]
- Anisimova, M.; Gascuel, O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 2006, 55, 539–552. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Peterson, D.; Tamura, K. MEGA-CC: Computing core of molecular evolutionary genetics analysis program for automated and iterative data analysis. Bioinformatics 2012, 28, 2685–2686. [Google Scholar] [CrossRef]
- Bengtsson, M.; Bäckman, A.-C.; Liblikas, I.; Ramirez, M.I.; Borg-Karlson, A.-K.; Ansebo, L.; Anderson, P.; Löfqvist, J.; Witzgall, P. Plant odor analysis of apple: Antennal response of codling moth females to apple volatiles during phenological development. J. Agric. Food Chem. 2001, 49, 3736–3741. [Google Scholar] [CrossRef]
- Cattaneo, A.M.; Gonzalez, F.; Bengtsson, J.M.; Corey, E.A.; Jacquin-Joly, E.; Montagné, N.; Salvagnin, U.; Walker, W.B.; Witzgall, P.; Anfora, G.; et al. Candidate pheromone receptors of codling moth Cydia pomonella respond to pheromones and kairomones. Sci. Rep. 2017, 7, 41105. [Google Scholar] [CrossRef]
- Gardette, M.; Alexakis, A.; Normant, J.F. Synthesis of (Z,Z)-3, 13-octadecadien-1-yl acetate component of the sex pheromone of Synanthedon tenuis. J. Chem. Ecol. 1983, 9, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Münch, D.; Galizia, C.G. DoOR: The Database of Odorant Responses. ChemoSense 2011, 13, 1–6. [Google Scholar]
- Galizia, C.G.; Münch, D.; Strauch, M.; Nissler, A.; Ma, S. Integrating Heterogeneous Odor Response Data into a Common Response Model: A DoOR to the Complete Olfactome. Chem. Senses 2010, 35, 551–563. [Google Scholar] [CrossRef]
- Bengtsson, M.; Liljefors, T.; Hansson, B.S.; Löfstedt, C.; Copaja, S.V. Structure-activity relationships for chain-shortened analogs of (Z)-5-decenyl acetate, a pheromone component of the turnip moth, Agrotis segetum. J. Chem. Ecol. 1990, 16, 667–684. [Google Scholar] [CrossRef]
- Cattaneo, A.M.; Witzgall, P.; Kwadha, C.A.; Becher, P.G.; Walker, W.B., III. Heterologous expression and functional characterization of Drosophila suzukii OR69a transcript variants unveiled response to kairomones and to a candidate pheromone. J. Pest Sci. 2022, 96, 1149–1171. [Google Scholar] [CrossRef]
- Pettersson, J.H.; Cattaneo, A.M. Heterologous investigation of metabotropic and ionotropic odorant receptors in ab3A neurons of Drosophila melanogaster. Front. Mol. Biosci. Sec. Protein Biochem. Basic Appl. Sci. 2024, 10, 1275901. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef]
- Garczynski, S.F.; Martin, J.A.; Griset, M.; Willett, L.S.; Cooper, W.R.; Swisher, K.D.; Unruh, T.R. CRISPR/Cas9 Editing of the Codling Moth (Lepidoptera: Tortricidae) CpomOR1 Gene Affects Egg Production and Viability. J. Econ. Entomol. 2017, 110, 1847–1855. [Google Scholar] [CrossRef]
- Tsirigos, K.D.; Peters, C.; Shu, N.; Kall, L.; Elofsson, A. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 2015, 43, W401–W407. [Google Scholar] [CrossRef] [PubMed]
- Omasits, U.; Ahrens, C.H.; Muller, S.; Wollscheid, B. Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 2014, 30, 884–886. [Google Scholar] [CrossRef]
- Bobkov, Y.V.; Walker, W.B.; Cattaneo, A.M. Altered functional properties of the codling moth Orco mutagenized in the intracellular loop-3. Sci. Rep. 2021, 11, 3893. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.; Tu, Z. Odorant receptor c-terminal motifs in divergent insect species. J. Insect Sci 2008, 8, 53. [Google Scholar] [CrossRef]
- Montagné, N.; Wanner, K.; Jacquin-Joly, E. Olfactory genomics within the Lepidoptera. In Insect Pheromone Biochemistry and Molecular Biology; Academic Press: New York, NY, USA, 2021; pp. 469–505. [Google Scholar] [CrossRef]
- Bengtsson, J.M.; Trona, F.; Montagné, N.; Anfora, G.; Ignell, R.; Witzgall, P.; Jacquin-Joly, E. Putative chemosensory receptors of the codling moth, Cydia pomonella, identified by antennal transcriptome analysis. PLoS ONE 2012, 7, e31620. [Google Scholar] [CrossRef]
- Chahda, J.S.; Soni, N.; Sun, J.S.; Ebrahim, S.A.M.; Weiss, B.L.; Carlson, J.R. The molecular and cellular basis of olfactory response to tsetse fly attractants. Chem. Senses 2019, 44, E67–E68. [Google Scholar] [CrossRef]
- Jennings, W.G.; Sevenants, M.R. Volatile Esters of Bartlett Pear III. J. Food Sci. 1964, 29, 123–240. [Google Scholar] [CrossRef]
- Prinz, H. Hill coefficients, dose–response curves and allosteric mechanisms. J. Chem. Biol. 2010, 3, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Preti, M.; Favaro, R.; Knight, A.L.; Angeli, S. Remote monitoring of Cydia pomonella adults among an assemblage of nontargets in sex pheromone-kairomone-baited smart traps. Pest Manag. Sci. 2021, 77, 4084–4090. [Google Scholar] [CrossRef] [PubMed]
- De Cristofaro, A.; Ioriatti, C.; Pasqualini, E.; Anfora, G.; Germinara, G.S.; Villa, M.; Rotundo, G. Electrophysiological responses of Cydia pomonella to codlemone and pear ester ethyl (E,Z)-2,4-decadienoate: Peripheral interactions in their perception and evidences for cells responding to both compounds. Bull. Insectology 2004, 57, 137–144. [Google Scholar]
- Matich, A.J.; Rowan, D.D.; Banks, N.H. Solid phase microextraction for quantitative headspace sampling of apple volatiles. Anal. Chem. 1996, 68, 4114–4118. [Google Scholar] [CrossRef]
- Vinczer, P.; Baán, G.; Novák, L.; Szántay, C. A novel stereocontrolled synthesis of (z,z)-3,13-octadecadien-1-yl acetate, the sex pheromone of Synanthedon species. Tetrahedron Lett. 1984, 25, 2701–2704. [Google Scholar] [CrossRef]
- Roelofs, W.L.; Comeau, A.; Hill, A.; Milicevic, G. Sex attractant of the codling moth: Characterization with electroantennogram technique. Science 1971, 174, 297–299. [Google Scholar] [CrossRef]
- Beroza, M.; Bierl, B.A.; Moffitt, H.R. Sex pheromones: (E8,E10)-Dodecadien-1-ol in the codling moth. Science 1974, 183, 89–90. [Google Scholar] [CrossRef]
- McDonough, L.M.; Moffitt, H.R. Sex pheromone of the codling moth. Science 1974, 183, 978. [Google Scholar] [CrossRef][Green Version]
- Frérot, B.; Priesner, E.; Gallois, M. A sex attractant for the green budworm moth, Hedya nubiferana. Z. Naturforsch. 1979, 34c, 1248–1252. [Google Scholar] [CrossRef]
- Roelofs, W.L.; Brown, R.L. Pheromones and evolutionary relationships of Tortricidae. Ann. Rev. Ecol. Syst. 1982, 13, 395–422. [Google Scholar] [CrossRef]
- Davis, H.G.; McDonough, L.M.; Burditt, A.K.; Bieri-Leonhardt, B.A. Filbertworm sex pheromone. Identification and field tests of (E,E)- and (E,Z)-8,10 dodecadien-1-ol acetates. J. Chem. Ecol. 1984, 10, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Witzgall, P.; Chambon, J.P.; Bengtsson, M.; Unelius, C.R.; Appelgren, M.; Makranczy, G.; Muraleedharan, N.; Reed, D.W.; Hellrigl, K.; Buser, H.-R.; et al. Sex pheromones and attractants in the Eucosmini and Grapholitini (Lepidoptera, Tortricidae). Chemoecology 1996, 7, 13–23. [Google Scholar] [CrossRef]
- Chambers, U.; Walton, V.M.; Mehlenbacher, S.A. Susceptibility of hazelnut cultivars to filbertworm, Cydia latiferreana. Hortic. Sci. 2011, 46, 1377–1380. [Google Scholar] [CrossRef]
- Lebreton, S.; Borrero-Echeverry, F.; Gonzalez, F.; Solum, M.; Wallin, E.A.; Hedenström, E.; Hansson, B.S.; Gustavsson, A.L.; Bengtsson, M.; Birgersson, G.; et al. A Drosophila female pheromone elicits species-specific long-range attraction via an olfactory channel with dual specificity for sex and food. BMC Biol. 2017, 15, 88. [Google Scholar] [CrossRef]
- Jumean, Z.; Gries, R.; Unruh, T.; Rowland, E.; Gries, G. Identification of the larval aggregation pheromone of Codling moth, Cydia pomonella. J. Chem. Ecol. 2005, 31, 911–924. [Google Scholar] [CrossRef] [PubMed]
- Parthasarathy, V.; Chempakam, B.; Zachariah, T.J. Chemistry of Spices; CAB International: Oxfordshire, UK, 2008; ISBN 978-1-84593-405-7. [Google Scholar]
- Rajeshwari, U.; Andallu, B. Medicinal benefits of coriander (Coriandrum sativum L.). Spatula DD 2011, 1, 51–58. [Google Scholar] [CrossRef]
- Bobkov, Y.V.; Ache, B.W. Block by amiloride derivatives of odor-evoked discharge in lobster olfactory receptor neurons through action on a presumptive TRP channel. Chem. Senses 2007, 32, 149–159. [Google Scholar] [CrossRef]
- Pask, G.M.; Bobkov, Y.V.; Corey, E.A.; Ache, B.W.; Zwiebel, L.J. Blockade of insect odorant receptor currents by amiloride derivatives. Chem. Senses 2013, 38, 221–229. [Google Scholar] [CrossRef]
- Röllecke KWerner, M.; Ziemba, P.M.; Neuhaus, E.M.; Hatt, H.; Gisselmann, G. Amiloride derivatives are effective blockers of insect odorant receptors. Chem. Senses 2013, 38, 231–236. [Google Scholar] [CrossRef]
- Bobkov, Y.V.; Corey, E.A.; Ache, B.W. An inhibitor of Na+/Ca2+ exchange blocks activation of insect olfactory receptors. Biochem. Biophys. Res. Commun. 2014, 450, 1104–1109. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, T.D. Pheromones and Animal Behavior: Chemical Signals and Signatures; Cambridge University Press: New York, NY, USA, 2014. [Google Scholar] [CrossRef]
- Andersson, M.N.; Löfstedt, C.; Newcomb, R.D. Insect olfaction and the evolution of receptor tuning. Front. Ecol. Evol.—Chem. Ecol. 2015, 3, 53. [Google Scholar] [CrossRef]
- Robertsson, H.M.; Warr, C.G.; Carlson, J.R. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2003, 100 (Suppl. S2), 14537–14542. [Google Scholar] [CrossRef]
- Couto, A.; Alenius, M.; Dickson, B.J. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr. Biol. 2005, 15, 1535–1547. [Google Scholar] [CrossRef]
- Martin, F.; Boto, T.; Gomez-Diaz, C.; Alcorta, E. Elements of olfactory reception in adult Drosophila melanogaster. Anat. Rec. 2013, 296, 1477–1488. [Google Scholar] [CrossRef]
- Münch, D.; Galizia, C.G. DoOR 2.0-Comprehensive mapping of Drosophila melanogaster odorant responses. Sci. Rep. 2016, 6, 21841. [Google Scholar] [CrossRef] [PubMed]
- Task, D.; Lin, C.-C.; Vulpe, A.; Afify, A.; Ballou, S.; Brbic, M.; Schlegel, P.; Raji, J.; Jefferis, G.S.X.E.; Li, H.; et al. Chemoreceptor co-expression in Drosophila melanogaster olfactory neurons. eLife 2022, 11, e72599. [Google Scholar] [CrossRef]
- Vulpe, A.; Menuz, K. Ir76b is a Co-receptor for Amine Responses in Drosophila Olfactory Neurons. Front. Cell. Neurosci. 2021, 15, 759238. [Google Scholar] [CrossRef]
- Butterwick, J.A.; Del Mármol, J.; Kim, K.H.; Kahlson, M.A.; Rogow, J.A.; Walz, T.; Ruta, V. Cryo-EM structure of the insect olfactory receptor Orco. Nature 2018, 560, 447–452. [Google Scholar] [CrossRef]
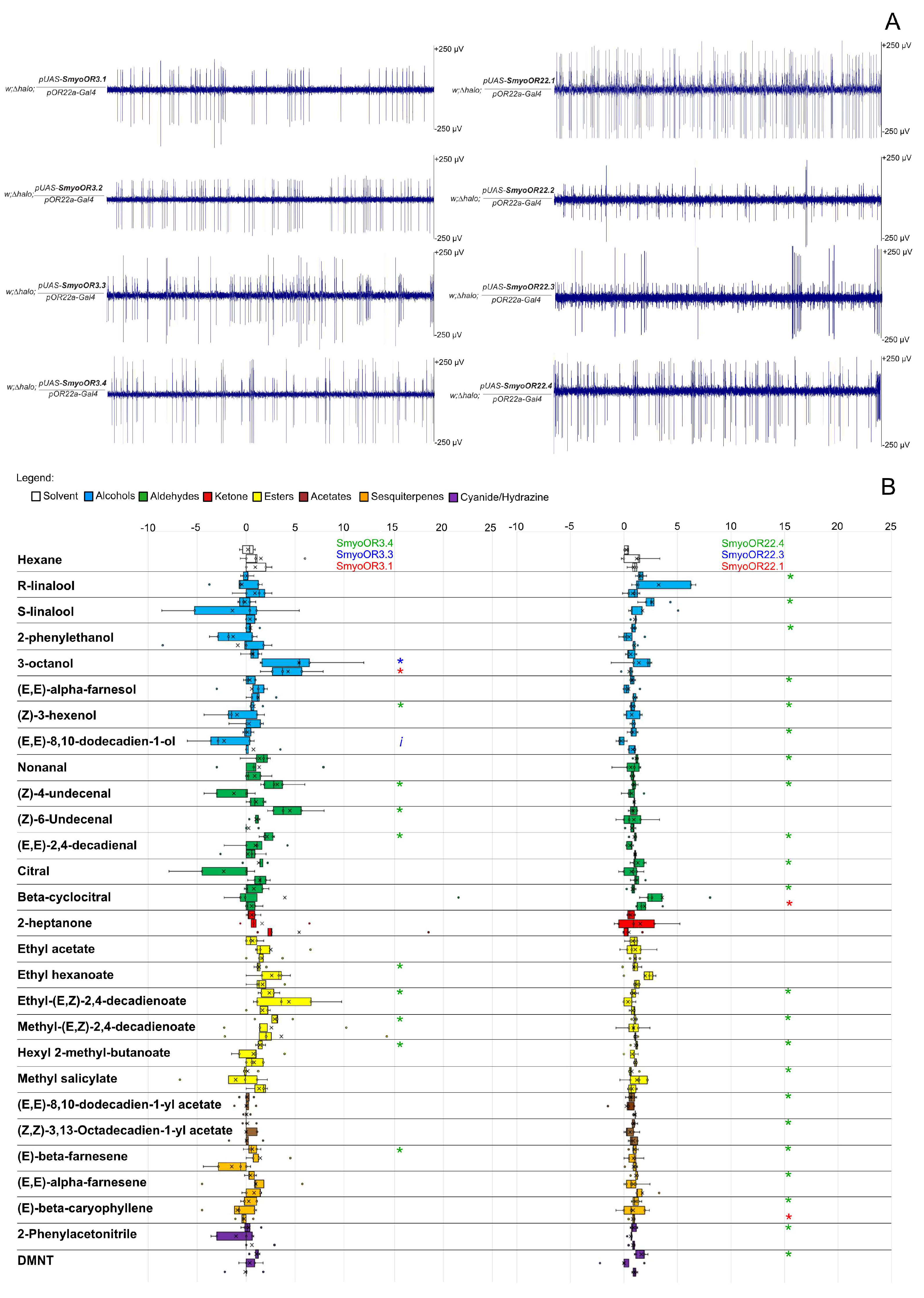
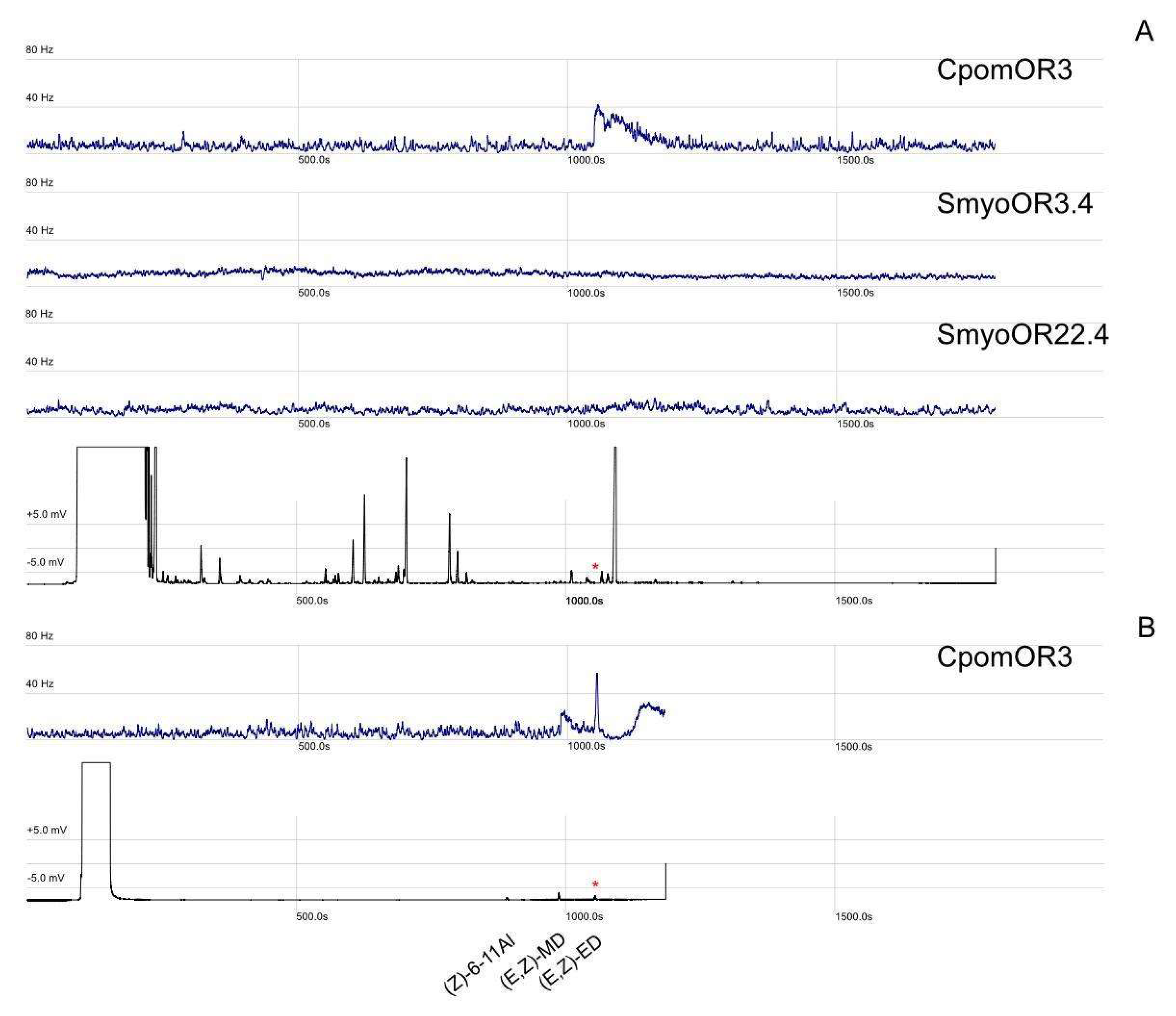
| Class | Compound Name | CAS | MW (g/mol) | Vp (mmHg @25 °C) | OR3.4 | OR3.3 | OR3.1 | OR22.4 | OR22.3 | OR22.1 |
|---|---|---|---|---|---|---|---|---|---|---|
| Alkane | Hexane | 110-54-3 | 86.17758 | 151.000000 | * | |||||
| Terpene alcohol | R-linalool | 126-90-9 | 154.25266 | 0.091000 | * | |||||
| Terpene alcohol | S-linalool | 126-91-0 | 154.25266 | 0.091000 | * | |||||
| Aromatic alcohol | 2-phenylethanol | 60-12-8 | 122.16690 | 0.086800 | * | |||||
| Aliphatic alcohol | 3-octanol | 589-98-0 | 130.23066 | 0.512000 | * | * | ||||
| Polyinsaturated alcohol | (E,E)-α-farnesol | 4602-84-0 | 222.37142 | 0.000370 | * | |||||
| Monoinsaturated alcohol | (Z)-3 hexen-1-ol | 928-96-1 | 100.16084 | 1.039000 | * | * | ||||
| Polyinsaturated alcohol | (E,E)-8,10-dodecadien-1-ol | 33956-49-9 | 182.30654 | 0.001000 | i | * | ||||
| Unsaturated aldehyde | nonanal | 124-19-6 | 142.24166 | 0.532000 | * | |||||
| Monoinsaturated aldehyde | (Z)-4-undecenal | 68820-32-6 | 168.27960 | 0.045000 | * | * | ||||
| Monoinsaturated aldehyde | (Z)-6-undecenal | - | 168.27960 | 0.045400 | * | |||||
| Polyinsaturated aldehyde | (E,E)-2,4-decadienal | 25152-84-5 | 152.23672 | 0.030000 | * | * | ||||
| Monoterpenoid aldehyde | Citral | 5392-40-5 | 152.23672 | 0.200000 | * | |||||
| Monoterpenoid aldehyde | β-cyclocitral | 432-25-7 | 152.23672 | 0.176000 | * | * | ||||
| Aliphatic ketone | 2-heptanone | 110-43-0 | 114.18778 | 4.732000 | ||||||
| Fatty acid ester | Ethyl acetate | 141-78-6 | 88.10616 | 111.716003 | ||||||
| Fatty acid ester | Ethyl hexanoate | 123-66-0 | 144.21392 | 1.665000 | * | |||||
| Aliphatic ester | Ethyl-(E,Z)-2,4-decadienoate | 3025-30-7 | 196.28980 | 0.010000 | * | * | ||||
| Aliphatic ester | Methyl-(E,Z)-2,4-decadienoate | 4493-42-9 | 182.26286 | 0.028000 | * | * | ||||
| Aliphatic ester | hexyl 2-methyl-butanoate | 10032-15-2 | 186.29474 | 0.158000 | * | * | ||||
| Aromatic ester | Methyl salicylate | 119-36-8 | 152.14936 | 0.034300 | * | |||||
| Polyinsaturated aliphatic acetate | (E,E)-8,10-dodecadien-1-yl acetate | 53880-51-6 | 224.34368 | 0.001000 | * | |||||
| Polyinsaturated aliphatic acetate | (Z,Z)-3,13-octadecadien-1-yl acetate | 53120-27-7 | 308.50532 | - | * | |||||
| Sesquiterpene | (E)-β-farnesene | 18794-84-8 | 204.35628 | 0.010000 | * | * | ||||
| Sesquiterpene | (E,E)-α-farnesene | 502-61-4 | 204.35628 | 0.010000 | * | |||||
| Sesquiterpene | (E)-β-caryophyllene | 87-44-5 | 204.35628 | 0.013000 | * | * | ||||
| Aromatic nitrile | Phenylacetonitrile | 140-29-4 | 117.15079 | 0.056000 | * | |||||
| Alkatriene | 4,8-dimethyl-1,3(E),7-nonatriene (DMNT) | 19945-61-0 | 150.26446 | - | * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cattaneo, A.M.; Walker, W.B., III. Candidate Pheromone Receptors of the Red-Belted Clearwing Moth Synanthedon myopaeformis Bind Pear Ester and Other Semiochemicals. Agriculture 2025, 15, 1112. https://doi.org/10.3390/agriculture15101112
Cattaneo AM, Walker WB III. Candidate Pheromone Receptors of the Red-Belted Clearwing Moth Synanthedon myopaeformis Bind Pear Ester and Other Semiochemicals. Agriculture. 2025; 15(10):1112. https://doi.org/10.3390/agriculture15101112
Chicago/Turabian StyleCattaneo, Alberto Maria, and William B. Walker, III. 2025. "Candidate Pheromone Receptors of the Red-Belted Clearwing Moth Synanthedon myopaeformis Bind Pear Ester and Other Semiochemicals" Agriculture 15, no. 10: 1112. https://doi.org/10.3390/agriculture15101112
APA StyleCattaneo, A. M., & Walker, W. B., III. (2025). Candidate Pheromone Receptors of the Red-Belted Clearwing Moth Synanthedon myopaeformis Bind Pear Ester and Other Semiochemicals. Agriculture, 15(10), 1112. https://doi.org/10.3390/agriculture15101112







