Novel Molecular Techniques for Identifying Agricultural Microorganisms
Abstract
1. Introduction
2. Nucleic Acids
2.1. Isothermal Amplification
2.2. High-Resolution Melting Analysis (HRMA)
2.3. Next-Generation Sequencing (NGS)
2.4. Fluorescence In Situ Hybridization (FISH)
3. Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS)
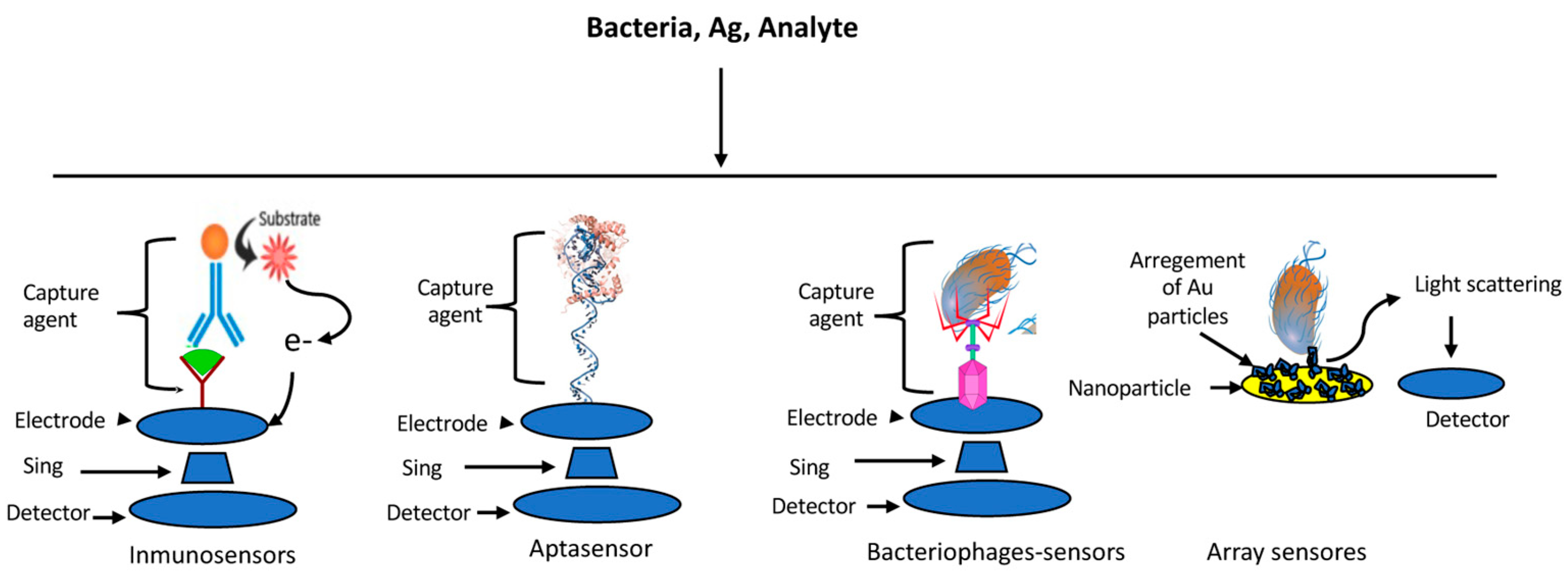
4. Sensors
4.1. Immune-Based Sensors
4.1.1. Electrochemical Immunosensors
4.1.2. Optical Immunosensors
4.1.3. Piezoelectric Immunosensors
4.2. Aptasensors
4.3. Bacteriophage-Based Sensors
4.4. Array-Based Sensors
4.5. Optoelectronic Nose
5. Optical Detection Methods
5.1. Vibrational Spectroscopy
5.1.1. Raman Spectrometry
5.1.2. SERS
5.2. Polarization
5.3. Laser Scattering
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ahmad, F.; Babalola, O.O.; Tak, H.I. Potential of MALDI-TOF mass spectrometry as a rapid detection technique in plant pathology: Identification of plant-associated microorganisms. Anal. Bioanal. Chem. 2012, 404, 1247–1255. [Google Scholar] [CrossRef]
- Larone, D.H. Medically Important Fungi: A Guide to Identification, 3rd ed.; ASM Press: Washington, DC, USA, 1995; pp. 190–192. [Google Scholar]
- Huang, W.E.; Ferguson, A.; Singer, A.C.; Lawson, K.; Thompson, I.P.; Kalin, R.M.; Larkin, M.J.; Bailey, M.J.; Whiteley, A.S. Resolving genetic functions within microbial populations: In situ analyses using rRNA and mRNA stable isotope probing coupled with single-cell Raman-fluorescence in situ hybridization. Appl. Environ. Microbiol. 2009, 75, 234–241. [Google Scholar] [CrossRef]
- Babalola, O.O.; Kirby, B.M.; Le Roes-Hill, M.; Cook, A.E.; Cary, S.C.; Burton, S.G.; Cowan, D.A. Phylogenetic analysis of actinobacterial populations associated with Antarctic dry valley mineral soils. Environ. Microbiol. 2009, 11, 566–576. [Google Scholar] [CrossRef]
- Babalola, O.O. Molecular techniques: An overview of methods for the detection of bacteria. Afr. J. Biotechnol. 2003, 2, 710–713. [Google Scholar] [CrossRef][Green Version]
- Huang, W.E.; Li, M.; Jarvis, R.M.; Goodacre, R.; Banwart, S.A. Shining light on the microbial world: The application of Raman microspectroscopy. Adv. Appl. Microbiol. 2010, 70, 153–186. [Google Scholar]
- Ali, M.M.; Li, F.; Zhiqing Zhang, F.L.; Zhang, K.; Kang, D.K.; Ankrum, J.A.; Le, X.C.; Zhao, W. Rolling circle amplification: A versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 2014, 43, 3324–3341. [Google Scholar] [CrossRef]
- Paez, J.G.; Lin, M.; Beroukhim, R.; Lee, J.C.; Zhao, X.; Richter, D.J.; Gabriel, S.; Herman, P.; Sasaki, H.; Altshuler, D.; et al. Genome coverage and sequence fidelity of phi29 polymerase-based multiple strand displacement whole genome amplification. Nucleic Acids Res. 2004, 32, e71. [Google Scholar] [CrossRef]
- Vincent, M.; Xu, Y.; Kong, H. Helicase-dependent isothermal DNA amplification. EMBO Rep. 2004, 5, 795–800. [Google Scholar] [CrossRef]
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. DNA detection using recombination proteins. PLoS Biol. 2006, 4, e204. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef]
- Misawa, Y.; Yoshida, A.; Saito, R.; Yoshida, H.; Okuzumi, K.; Ito, N.; Okada, M.; Moriya, K.; Koike, K. Application of loop-mediated isothermal amplification technique to rapid and direct detection of methicillin-resistant Staphylococcus aureus (MRSA) in blood cultures. J. Infect. Chemother. 2007, 13, 134–140. [Google Scholar] [CrossRef]
- Zhao, X.; Li, Y.; Wang, L.; You, L.; Xu, Z.; Li, L.; He, X.; Liu, Y.; Wang, J.; Yang, L. Development and application of a loop-mediated isothermal amplification method on rapid detection Escherichia coli O157 strains from food samples. Mol. Biol. Rep. 2010, 37, 2183–2188. [Google Scholar] [CrossRef]
- Ho-Jong, J. Simple and rapid detection of potato leafroll virus by reverse transcription loop-mediated isothermal amplification. Plant Pathol. J. 2011, 27, 385–389. [Google Scholar] [CrossRef]
- Suzuki, R.; Fukuta, S.; Matsumoto, Y.; Hasegawa, T.; Kojima, H.; Hotta, M.; Miyake, N. Development of reverse transcription loop-mediated isothermal amplification assay as a simple detection method of Chrysanthemum stem necrosis virus in chrysanthemum and tomato. J. Virol. Methods 2016, 236, 29–34. [Google Scholar] [CrossRef]
- Selvaraj, V.; Maheshwari, Y.; Hajeri, S.; Yokomi, R. A rapid detection tool for VT isolates of Citrus tristeza virus by immunocapture-reverse transcriptase loop-mediated isothermal amplification assay. PLoS ONE 2019, 14, e0222170. [Google Scholar] [CrossRef]
- Montgomery, J.L.; Sanford, L.N.; Wittwer, C.T. High-resolution DNA melting analysis in clinical research and diagnostics. Expert. Rev. Mol. Diagn. 2010, 10, 219–240. [Google Scholar] [CrossRef]
- Yang, S.; Ramachandran, P.; Rothman, R.; Hsieh, Y.H.; Hardick, A.; Won, H.; Kecojevic, A.; Jackman, J.; Gaydos, C. Rapid identification of biothreat and other clinically relevant bacterial species by use of universal PCR coupled with high-resolution melting analysis. J. Clin. Microbiol. 2009, 47, 2252–2255. [Google Scholar] [CrossRef]
- Xanthopoulou, A.; Ganopoulos, I.; Tryfinopoulou, P.; Panagou, E.Z.; Osanthanunkul, M.; Madesis, P.; Kizis, D. Rapid and accurate identification of black aspergilli from grapes using high-resolution melting (HRM) analysis. J. Sci. Food Agric. 2019, 99, 309–314. [Google Scholar] [CrossRef]
- Cłapa, T.; Mikołajczak, K.; Błaszczyk, L.; Narożna, D. Development of high-resolution melting PCR (HRM-PCR) assay to identify native fungal species associated with the wheat endosphere. J. Appl. Genet. 2020, 61, 629–635. [Google Scholar] [CrossRef]
- van Dijk, E.L.; Jaszczyszyn, Y.; Naquin, D.; Thermes, C. The third revolution in sequencing technology. Trends Genet. 2018, 34, 666–681. [Google Scholar] [CrossRef]
- Thompson, J.F.; Steinmann, K.E. Single molecule sequencing with a HeliScope genetic analysis system. Curr. Protoc. Mol. Biol. 2010, 92, 7–10. [Google Scholar] [CrossRef]
- Eid, J.; Fehr, A.; Gray, J.; Luong, K.; Lyle, J.; Otto, G.; Peluso, P.; Rank, D.; Baybayan, P.; Bettman, B.; et al. Real-time DNA sequencing from single polymerase molecules. Science 2009, 323, 133–138. [Google Scholar] [CrossRef]
- Athanasopoulou, K.; Boti, M.A.; Adamopoulos, P.G.; Skourou, P.C.; Scorilas, A. Third-generation sequencing: The spearhead towards the radical transformation of modern genomics. Life 2021, 12, 30. [Google Scholar] [CrossRef]
- Ardui, S.; Ameur, A.; Vermeesch, J.R.; Hestand, M.S. Single molecule real-time (SMRT) sequencing comes of age: Applications and utilities for medical diagnostics. Nucleic Acids Res. 2018, 46, 2159–2168. [Google Scholar] [CrossRef]
- Ambardar, S.; Gupta, R.; Trakroo, D.; Lal, R.; Vakhlu, J. High Throughput sequencing: An overview of sequencing chemistry. Indian. J. Microbiol. 2016, 56, 394–404. [Google Scholar] [CrossRef]
- Scarano, C.; Veneruso, I.; De Simone, R.R.; Di Bonito, G.; Secondino, A.; D’Argenio, V. The third-generation sequencing challenge: Novel insights for the omic sciences. Biomolecules 2024, 14, 568. [Google Scholar] [CrossRef]
- Lin, B.; Hui, J.; Mao, H. Nanopore technology and its applications in gene sequencing. Biosensors 2021, 11, 214. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, Y.; Ying, C.; Wang, D.; Du, C. Nanopore-based fourth-generation DNA sequencing technology. Genom. Proteom. Bioinform. 2015, 13, 4–16. [Google Scholar] [CrossRef]
- Handelsman, J.; Rondon, M.R.; Brady, S.F.; Clardy, J.; Goodman, R.M. Molecular biological access to the chemistry of unknown soil microbes: A new frontier for natural products. Chem. Biol. 1998, 5, 245–249. [Google Scholar] [CrossRef]
- Yang, B.; Wang, Y.; Qian, P.Y. Sensitivity and correlation of hypervariable regions in 16S rRNA genes in phylogenetic analysis. BMC Bioinform. 2016, 17, 135. [Google Scholar] [CrossRef]
- Winand, R.; Bogaerts, B.; Hoffman, S.; Lefevre, L.; Delvoye, M.; Braekel, J.V.; Fu, Q.; Roosens, N.H.; Keersmaecker, S.C.; Vanneste, K. Targeting the 16s rRNA gene for bacterial identification in complex mixed samples: Comparative evaluation of second (Illumina) and third (Oxford Nanopore Technologies) generation sequencing technologies. Int. J. Mol. Sci. 2019, 21, 298. [Google Scholar] [CrossRef]
- Kim, C.; Pongpanich, M.; Porntaveetus, T. Unraveling metagenomics through long-read sequencing: A comprehensive review. J. Transl. Med. 2024, 22, 111. [Google Scholar] [CrossRef]
- Hur, M.; Park, S.J. Identification of microbial profiles in heavy-metal-contaminated soil from full-length 16S rRNA reads sequenced by a PacBio System. Microorganisms 2019, 7, 357. [Google Scholar] [CrossRef]
- Edwards, A.; Debbonaire, A.R.; Sattler, B.; Mur, L.A.J.; Hodson, A.J. Extreme metagenomics using nanopore DNA sequencing: A field report from Svalbard, 78° N. BioRxiv 2016, 10, 073965. [Google Scholar] [CrossRef]
- Maguire, M.; Kase, J.A.; Roberson, D.; Muruvanda, T.; Brown, E.W.; Allard, M.; Musser, S.M.; Gonzalez-Escalona, N.G. Precision long-read metagenomics sequencing for food safety by detection and assembly of Shiga toxin-producing Escherichia coli in irrigation water. PLoS ONE 2021, 16, e0245172. [Google Scholar] [CrossRef]
- Jagadesh, M.; Dash, M.; Kumari, A.; Singh, S.K.; Verma, K.K.; Kumar, P.; Bhatt, R.; Sharma, S.K. Revealing the hidden world of soil microbes: Metagenomic insights into plant, bacteria, and fungi interactions for sustainable agriculture and ecosystem restoration. Microbiol. Res. 2024, 285, 127764. [Google Scholar] [CrossRef]
- Masenya, K.; Manganyi, M.C.; Dikobe, T.B. Exploring cereal metagenomics: Unravelling microbial communities for improved food security. Microorganisms 2024, 12, 510. [Google Scholar] [CrossRef] [PubMed]
- You, X.; Wang, S.; Du, L.; Chen, Y.; Wang, T.; Bo, X. Metagenomics reveals the variations in functional metabolism associated with greenhouse gas emissions during legume-vegetable rotation process. Ecotoxicol. Environ. Saf. 2024, 275, 116268. [Google Scholar] [CrossRef]
- Xiong, C.; Brajesh, K.; Singh, B.; Zhu, Y.G.; Hu, H.W.; Li, P.P.; Han, Y.L.; Han, L.L.; Zhang, Q.B.; Wang, J.T.; et al. Microbial species pool-mediated diazotrophic community assembly in crop microbiomes during plant development. mSystems 2024, 9, e0105523. [Google Scholar] [CrossRef]
- Gonin, M.; Salas-González, I.; Gopaulchan, D.; Frene, J.P.; Roden, S.; Van de Poel, B.; Salt, D.E.; Castrillo, G. Plant microbiota controls an alternative root branching regulatory mechanism in plants. Proc. Nat. Acad. Sci. USA 2023, 120, e2301054120. [Google Scholar] [CrossRef]
- Zheng, S.; Qi, J.; Fu, T.; Chen, Y.; Qiu, X. Novel mechanisms of cadmium tolerance and Cd-induced fungal stress in wheat: Transcriptomic and metagenomic insights. Ecotoxicol. Environ. Saf. 2023, 256, 114842. [Google Scholar] [CrossRef]
- Moter, A.; Göbel, U.B. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J. Microbiol. Methods 2000, 41, 85–112. [Google Scholar] [CrossRef] [PubMed]
- Moussata, D.; Goetz, M.; Gloeckner, A.; Kerner, M.; Campbell, B.; Hoffman, A.; Biesterfeld, S.; Flourie, B.; Saurin, J.C.; Galle, P.R. Confocal laser endomicroscopy is a new imaging modality for recognition of intramucosal bacteria in inflammatory bowel disease in vivo. Gut 2011, 60, 26–33. [Google Scholar] [CrossRef]
- Harris, E.H. Chlamydomonas as a model organism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2021, 52, 363–406. [Google Scholar] [CrossRef]
- Uniacke, J.; Colón-Ramos, D.; Zerges, W. FISH and immunofluorescence staining in Chlamydomonas. Methods Mol. Biol. 2011, 714, 15–29. [Google Scholar] [CrossRef] [PubMed]
- Selosse, M.A.; Setaro, S.; Glatard, F.; Richard, F.; Urcelay, C.; Weiß, M. Sebacinales are common mycorrhizal associates of Ericaceae. New Phytol. 2007, 174, 864–878. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, S.; Hückelhoven, R.; Schäfer, P.; Imani, J.; Sharma, M.; Weiss, M.; Waller, F.; Kogel, K.H. The root endophytic fungus Piriformospora indica requires host cell death for proliferation during mutualistic symbiosis with barley. Proc. Natl. Acad. Sci. USA 2006, 103, 18450–18457. [Google Scholar] [CrossRef]
- Sharma, M.; Schmid, M.; Rothballer, M.; Hause, G.; Zuccaro, A.; Imani, J.; Kämpfer, P.; Domann, E.; Schäfer, P.; Hartmann, A.; et al. Detection and identification of bacteria intimately associated with fungi of the order Sebacinales. Cell Microbiol. 2008, 10, 2235–2246. [Google Scholar] [CrossRef]
- Scheid, D.; Stubner, S.; Conrad, R. Identification of rice root associated nitrate, sulfate and ferric iron reducing bacteria during root decomposition. FEMS Microbiol. Ecol. 2004, 50, 101–110. [Google Scholar] [CrossRef][Green Version]
- Roy, R.; Conrad, R. Effect of methanogenic precursors (acetate, hydrogen, propionate) on the suppression of methane production by nitrate in anoxic rice field soil. FEMS Microbiol. Ecol. 1999, 28, 49–61. [Google Scholar] [CrossRef]
- Schwab, S.; Hirata, E.S.; Amaral, J.C.A.; da Silva, C.G.N.; Ferreira, J.P.; da Silva, L.V.; Rouws, J.R.C.; Rouws, L.F.M.; Baldani, J.I.; Reis, V.M. Quantifying and visualizing Nitrospirillum amazonense strain CBAmC in sugarcane after using different inoculation methods. Plant Soil 2023, 48, 197–216. [Google Scholar] [CrossRef]
- Giebel, R.; Worden, C.; Rust, S.M.; Kleinheinz, G.T.; Robbins, M.; Sandrin, T.R. Microbial fingerprinting using matrixassisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) applications and challenges. Adv. Appl. Microbiol. 2010, 71, 149–184. [Google Scholar] [PubMed]
- Ryzhov, V.; Fenselau, C. Characterization of the protein subset desorbed by MALDI from whole bacterial cells. Anal. Chem. 2001, 73, 746–750. [Google Scholar] [CrossRef] [PubMed]
- Bonnelly, R.; Calderon, V.V.; Ortiz, I.; Ovando, A.; Pinales, C.; Lara, W.; Mateo-Perez, S.E.; Cardenas-Alegría, O.; Ramos, R.T.; Rodriguez-Rodriguez, Y.; et al. Comparison of two bacterial characterization techniques for the genomic analysis of river microbiomes. Appl. Microbiol. 2023, 3, 1037–1045. [Google Scholar] [CrossRef]
- Sura-de Jong, M.; Reynolds, R.J.B.; Richterova, K.; Musilova, L.; Staicu, L.C.; Chocholata, I.; Cappa, J.J.; Taghavi, S.; van der Lelie, D.; Frantik, T.; et al. Selenium hyperaccumulators harbor a diverse endophytic bacterial community characterized by high selenium resistance and plant growth promoting properties. Front. Plant Sci. 2015, 6, 113. [Google Scholar] [CrossRef] [PubMed]
- Wensing, A.; Zimmermann, S.; Geider, K. Identification of the corn pathogen Pantoea stewartii by mass spectrometry of whole-cell extracts and its detection with novel PCR primers. Appl. Environ. Microbiol. 2010, 76, 6248–6256. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, Q.; Li, B.; Liu, B.; Wu, G.; Ibrahim, M.; Xie, G.; Li, H.; Sun, G. Differentiation in MALDI-TOF MS and FTIR spectra between two closely related species Acidovorax oryzae and Acidovorax citrulli. BMC Microbiol. 2012, 12, 182. [Google Scholar] [CrossRef] [PubMed]
- Šalplachta, J.; Kubesová, A.; Horký, J.; Matoušková, H.; Tesařová, M.; Horká, M. Characterization of Dickeya and Pectobacterium species by capillary electrophoretic techniques and MALDI-TOF MS. Anal. Bioanal. Chem. 2015, 407, 7625–7635. [Google Scholar] [CrossRef]
- Inglis, P.W.; Mello, S.C.M.; Martins, I.; Silva, J.B.T.; Macêdo, K.; Sifuentes, D.N.; Valadares-Inglis, M.C. Trichoderma from brazilian garlic and onion crop soils and description of two new species: Trichoderma azevedoi and Trichoderma peberdyi. PLoS ONE 2020, 15, e0228485. [Google Scholar] [CrossRef]
- Martínez-Hidalgo, P.; Flores-Félix, J.D.; Sánchez-Juanes, F.; Rivas, R.; Mateos, P.F.; Regina, I.S.; Martínez-Molina, E.; Igual, J.M.; Velázquez, E. Identification of canola roots endophytic bacteria and analysis of their potential as biofertilizers for canola crops with special emphasis on sporulating bacteria. Agronomy 2021, 11, 1796. [Google Scholar] [CrossRef]
- Nunes, A.R.; Sánchez-Juanes, F.; Gonçalves, A.C.; Alves, G.; Silva, L.R.; Flores-Félix, J.D. Evaluation of raw cheese as a novel source of biofertilizer with a high level of biosecurity for blueberry. Agronomy 2022, 12, 1150. [Google Scholar] [CrossRef]
- Singh, R.; Mukherjee, M.D.; Sumana, G.; Gupta, R.K.; Sood, S.; Malhotra, B.D. Biosensors for pathogen detection: A smart approach towards clinical diagnosis. Sens. Actuators B Chem. 2014, 197, 385–404. [Google Scholar] [CrossRef]
- Chen, Y.; Qian, C.; Liu, C.; Shen, H.; Wang, Z.; Ping, J.; Wu, J.; Chen, H. Nucleic acid amplification free biosensors for pathogen detection. Biosens. Bioelectron. 2020, 153, 112049. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.D. (Bio)sensors for measurement of analytes implicated in food safety: A review. Trac-Trends Anal. Chem. 2002, 21, 96–115. [Google Scholar] [CrossRef]
- Mello, L.D.; Kubota, L.T. Review of the use of biosensors as analytical tools in the food and drink industries. Food Chem. 2002, 77, 237–256. [Google Scholar] [CrossRef]
- Ferreira, L.S.; De Souza, M.B.; Trierweiler, J.O.; Broxtermann, O.; Folly, R.O.M.; Hitzmann, B. Aspects concerning the use of biosensors for process control: Experimental and simulation investigations. Comput. Chem. Eng. 2003, 27, 1165–1173. [Google Scholar] [CrossRef]
- Wilson, R. The use of gold nanoparticles in diagnostics and detection. Chem. Soc. Rev. 2008, 37, 2028–2045. [Google Scholar] [CrossRef]
- You, Y.; Lim, S.; Gunasekaran, S. Streptavidin-coated Au nanoparticles coupled with biotinylated antibody-based bifunctional linkers as plasmon-enhanced immunobiosensors. ACS Appl. Nano Mater. 2020, 3, 1900–1909. [Google Scholar] [CrossRef]
- Felix, F.S.; Angnes, L. Electrochemical immunosensors-a powerful tool for analytical applications. Biosens. Bioelectron. 2018, 102, 470–478. [Google Scholar] [CrossRef]
- Cho, I.H.; Lee, J.; Kim, J.; Kang, M.S.; Paik, J.K.; Ku, S.; Cho, H.M.; Irudayaraj, J.; Kim, D.H. Current technologies of electrochemical immunosensors: Perspective on signal amplification. Sensors 2018, 18, 207. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Wen, L.; Tian, J.; Wu, Y.; Liu, F.; Lin, Y.; Hua, W.; Wu, G.A. Portable electrochemical immunosensor for highly sensitive point-of-care testing of genetically modified crops. Biosens. Bioelectron. 2019, 142, 111504. [Google Scholar] [CrossRef] [PubMed]
- Arévalo, F.J.; Granero, A.M.; Fernández, H.; Raba, J.; Zón, M.A. Citrinin (CIT) determination in rice samples using a micro fluidic electrochemical immunosensor. Talanta 2011, 83, 966–973. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Baldo, M.A.; Bertolino, F.A.; Messina, G.A.; Sanz, M.I.; Raba, J. Modified magnetic nanoparticles in an electrochemical method for the ochratoxin A determination in Vitis vinifera red grapes tissues. Talanta 2010, 83, 651–657. [Google Scholar] [CrossRef] [PubMed]
- Ogert, R.A.; Brown, J.E.; Singh, B.R.; Shriver-Lake, L.C.; Ligler, F.S. Detection of Clostridium botulinum toxin a using a fiber optic-based biosensor. Anal. Biochem. 1992, 205, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Oztuna, A.; Nazir, H.; Baysallar, M. Simultaneous Bacillus anthracis spores detection via aminated-poly(vinyl chloride) coated piezoelectric crystal immunosensor. J. Coat. 2014, 2014, 256168. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef]
- Prieto-Simón, B.; Samitier, J. Signal off aptasensor based on enzyme inhibition induced by conformational switch. Anal. Chem. 2014, 86, 1437–1444. [Google Scholar] [CrossRef] [PubMed]
- Alhamoud, Y.; Li, Y.; Zhou, H.; Al-Wazer, R.; Gong, Y.; Zhi, S.; Yang, D. Label-free and highly-sensitive detection of ochratoxin A using one-pot synthesized reduced graphene oxide/gold nanoparticles-based impedimetric aptasensor. Biosensors 2021, 11, 87. [Google Scholar] [CrossRef]
- Lu, X.; Wang, L.; He, B.; Zhao, R.; Bai, C.; Zhang, Y.; Ren, W.; Jiang, L.; Suo, Z.; Xu, Y. AgPdNFs and AuNOs@GO nanocomposites for T-2 toxin detection by catalytic hairpinassembly. Mikrochim. Acta 2023, 190, 120. [Google Scholar] [CrossRef]
- Peng, H.; Chen, I.A. Rapid colorimetric detection of bacterial species through the capture of gold nanoparticles by chimeric phages. ACS Nano 2018, 13, 1244–1252. [Google Scholar] [CrossRef]
- Abdelhamied, N.; Abdelrahman, F.; El-Shibiny, A.; Hassan, R.Y.A. Bacteriophage-based nano-biosensors for the fast impedimetric determination of pathogens in food samples. Sci. Rep. 2023, 13, 3498. [Google Scholar] [CrossRef]
- Li, B.; Li, X.; Dong, Y.; Wang, B.; Li, D.; Shi, Y.; Wu, Y. Colorimetric sensor array based on gold nanoparticles with diverse surface charges for microorganisms identification. Anal. Chem. 2017, 9, 10639–10643. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Yang, T.; Wang, X.Y.; Wang, Y.T.; Liu, M.X.; Chen, M.L.; Yu, Y.L.; Wang, J.H. A novel three-dimensional nanosensing array for the discrimination of sulfur-containing species and sulfur bacteria. Anal. Chem. 2019, 91, 6012–6018. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhang, J.; Hu, X.; Huang, X.; Zhang, X.; Zou, X.; Shi, J. A visible colorimetric sensor array based on chemo-responsive dyes and chemometric algorithms for real-time potato quality monitoring systems. Food Chem. 2023, 405, 134717. [Google Scholar] [CrossRef] [PubMed]
- Arslan, M.; Zareef, M.; Tahir, H.E.; Guo, Z.; Rakha, A.; Xuetao, H.; Shi, J.; Zhihua, L.; Xiaobo, Z.; Khan, M.R. Discrimination of rice varieties using smartphone-based colorimetric sensor arrays and gas chromatography techniques. Food Chem. 2022, 368, 130783. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Askim, J.R.; Suslick, K.S. The optoelectronic nose: Colorimetric and fluorometric sensor arrays. Chem. Rev. 2019, 119, 231–292. [Google Scholar] [CrossRef] [PubMed]
- Carey, J.R.; Suslick, K.S.; Hulkower, K.I.; Imlay, J.A.; Imlay, K.R.; Ingison, C.K.; Ponder, J.B.; Sen, A.; Wittrig, A.E. Rapid identification of bacteria with a disposable colorimetric sensing array. J. Am. Chem. Soc. 2011, 133, 7571–7576. [Google Scholar] [CrossRef]
- Maquelin, K.; Choo-Smith, L.P.; Kirschner, C.; Ngo-Thi, N.; Naumann, D.; Puppels, G. Vibrational spectroscopic studies of microorganisms. In Handbook of Vibrational Spectroscopy; Chalmers, J.M., Griffit, P.R., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2006. [Google Scholar] [CrossRef]
- Maquelin, K.; Kirschner, C.; Choo-Smith, L.P.; Ngo-Thi, N.; Van Vreeswijk, T.; Stämmler, M.; Endtz, H.; Bruining, H.; Naumann, D.; Puppels, G. Prospective study of the performance of vibrational spectroscopies for rapid identification of bacterial and fungal pathogens recovered from blood cultures. J. Clin. Microbiol. 2003, 41, 324–329. [Google Scholar] [CrossRef]
- Harz, M.; Rösch, P.; Popp, J. Vibrational spectroscopy-a powerful tool for the rapid identification of microbial cells at the single-cell level. Cytometry A 2009, 75, 104–113. [Google Scholar] [CrossRef]
- Vallejo-Pérez, M.R.; Navarro-Contreras, H.R.; Sosa-Herrera, J.A.; Lara-Ávila, J.P.; Ramírez-Tobías, H.M.; Díaz-Barriga Martínez, F.D.; Flores-Ramírez, R.; Rodríguez-Vázquez, Á.G. Detection of Clavibacter michiganensis subsp. michiganensis assisted by micro-Raman spectroscopy under laboratory conditions. Plant Pathol. J. 2018, 34, 381–392. [Google Scholar] [CrossRef]
- Vallejo-Pérez, M.R.; Sosa-Herrera, J.A.; Navarro-Contreras, H.R.; Álvarez-Preciado, L.G.; Rodríguez-Vázquez, Á.G.; Lara-Ávila, J.P. Raman spectroscopy and machine-learning for early detection of bacterial cancer of tomato: The asympthomatic disease condition. Plants 2021, 10, 1542. [Google Scholar] [CrossRef]
- Vo-Dinh, T.; Liu, Y.; Fales, A.M.; Ngo, H.; Wang, H.N.; Register, J.K.; Yuan, H.; Norton, S.J.; Griffin, G.D. SERS nanosensors and nanoreporters: Golden opportunities in biomedical applications. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2015, 7, 17–33. [Google Scholar] [CrossRef]
- Premasiri, W.R.; Moir, D.T.; Klempner, M.S.; Krieger, N.; Jones, G.; Ziegler, L.D. Characterization of the surface enhanced Raman scattering (SERS) of bacteria. J. Phys. Chem. B 2005, 109, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, M.; Zhao, H.; Ren, X.; Lin, T.; Zhang, P.; Zheng, D. Rapid detection and identification of fungi in grain crops using colloidal Au nanoparticles based on surface-enhanced Raman scattering and multivariate statistical analysis. World J. Microbiol. Biotechnol. 2022, 39, 26. [Google Scholar] [CrossRef]
- Zhao, H.; Cui, X.; Zhang, P.; Zhou, M.; Liu, C.; Shi, X.; Ma, J. Surface-enhanced Raman spectroscopy detection for fenthion pesticides based on gold molecularly imprinted polymer solid-state substrates. Appl. Spectrosc. 2024, 37028241253860. [Google Scholar] [CrossRef]
- Lv, M.; Pu, H.; Sun, D.W. A tailored dual core-shell magnetic SERS substrate with precise shell-thickness control for trace organophosphorus pesticides residues detection. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2024, 316, 124336. [Google Scholar] [CrossRef]
- Xu, S.; Guo, Y.; Liang, X.; Lu, H. Intelligent rapid detection techniques for low-content components in fruits and vegetables: A comprehensive review. Foods 2024, 13, 1116. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, J.A. The significances of bacterial colony patterns. Bioessays 1995, 17, 597–607. [Google Scholar] [CrossRef]
- Badieyan, S.; Dilmaghani-Marand, A.; Hajipour, M.J.; Ameri, A.; Razzaghi, M.R.; Rafii-Tabar, H.; Mahmoudi, M.; Sasanpour, P. Detection and discriminate on of bacterial colonies with Mueller matrix imaging. Sci. Rep. 2018, 8, 10815. [Google Scholar] [CrossRef]
- Chen, Y.; Chu, J.; Xin, B.; Qi, J. Mechanical stability of polarization signatures in biological tissue characterization. Biomed. Opt. Express 2024, 15, 2652–2665. [Google Scholar] [CrossRef]
- Krafft, D.; Scarboro, C.G.; Hsieh, W.; Doherty, C.; Balint-Kurti, P.; Kudenov, M. Mitigating illumination-, leaf-, and view-angle dependencies in hyperspectral imaging using polarimetry. Plant Phenomics 2024, 6, 0157. [Google Scholar] [CrossRef]
- Kim, K.P.; Singh, A.K.; Bai, X.; Leprun, L.; Bhunia, A.K. Novel PCR assays complement laser biosensor-based method and facilitate listeria species detection from food. Sensors 2015, 15, 22672–22691. [Google Scholar] [CrossRef]
- Singh, A.K.; Sun, X.; Bai, X.; Kim, H.; Abdalhaseib, M.U.; Bae, E.; Bhunia, A.K. Label-free, non-invasive light scattering sensor for rapid screening of Bacillus colonies. J. Microbiol. Methods 2015, 109, 56–66. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jan-Roblero, J.; Cruz-Maya, J.A.; Cancino-Diaz, J.C. Novel Molecular Techniques for Identifying Agricultural Microorganisms. Agriculture 2024, 14, 987. https://doi.org/10.3390/agriculture14070987
Jan-Roblero J, Cruz-Maya JA, Cancino-Diaz JC. Novel Molecular Techniques for Identifying Agricultural Microorganisms. Agriculture. 2024; 14(7):987. https://doi.org/10.3390/agriculture14070987
Chicago/Turabian StyleJan-Roblero, Janet, Juan A. Cruz-Maya, and Juan C. Cancino-Diaz. 2024. "Novel Molecular Techniques for Identifying Agricultural Microorganisms" Agriculture 14, no. 7: 987. https://doi.org/10.3390/agriculture14070987
APA StyleJan-Roblero, J., Cruz-Maya, J. A., & Cancino-Diaz, J. C. (2024). Novel Molecular Techniques for Identifying Agricultural Microorganisms. Agriculture, 14(7), 987. https://doi.org/10.3390/agriculture14070987




