PL-DINO: An Improved Transformer-Based Method for Plant Leaf Disease Detection
Abstract
1. Introduction
2. Materials and Methods
2.1. Datasets
2.2. DETR with Improved De-Noising Anchor Boxes
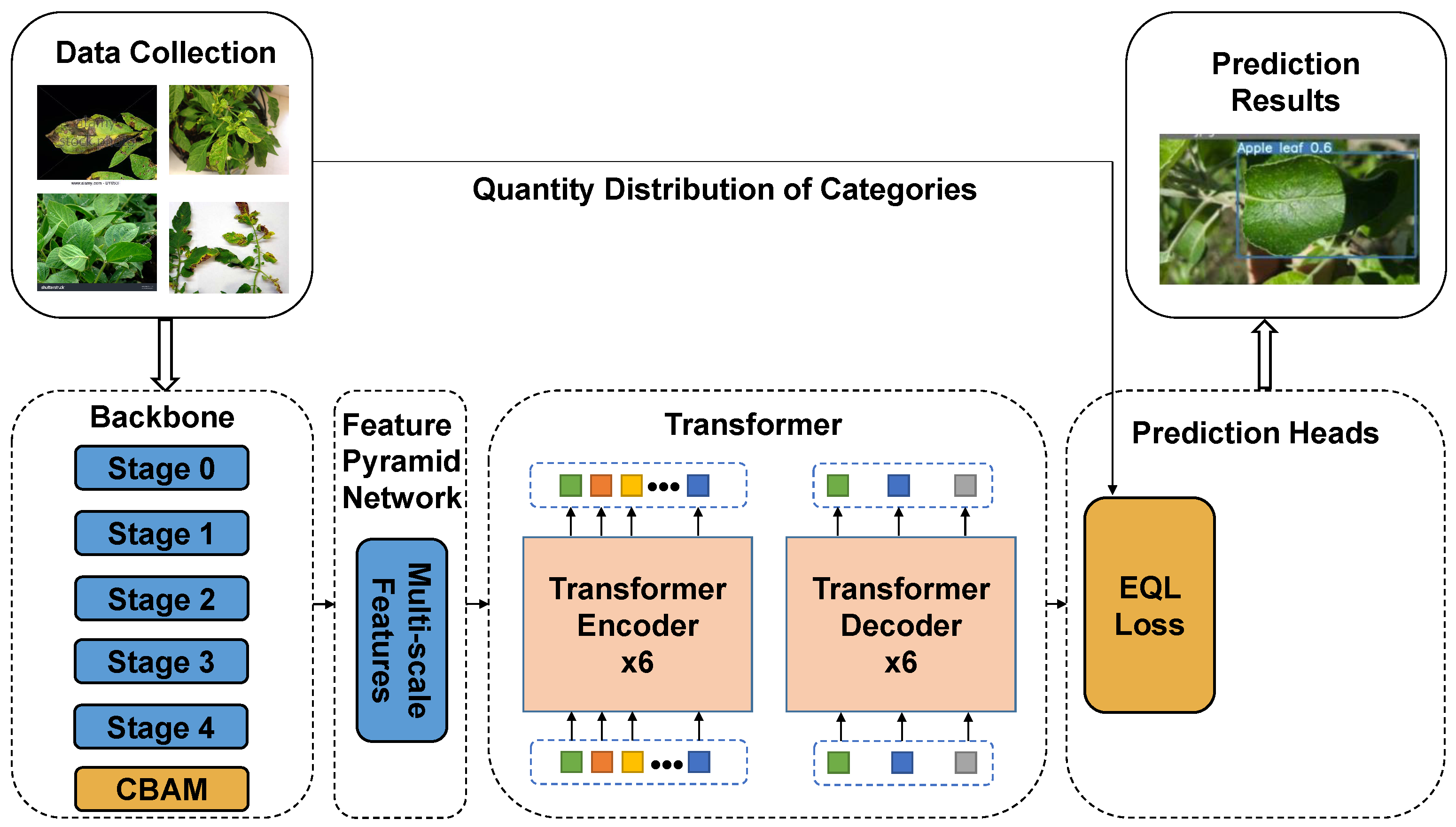
2.3. Framework of PL-DINO
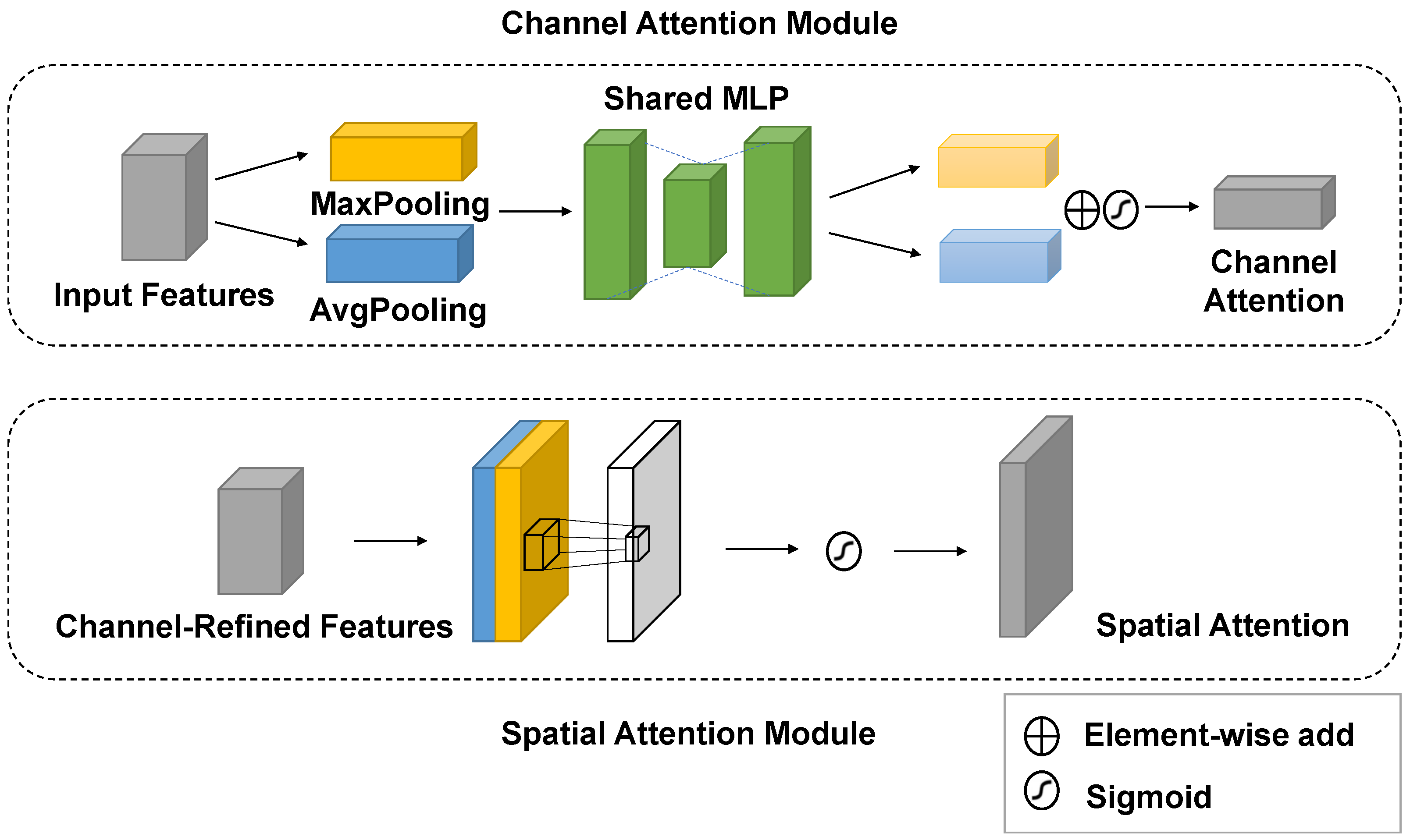
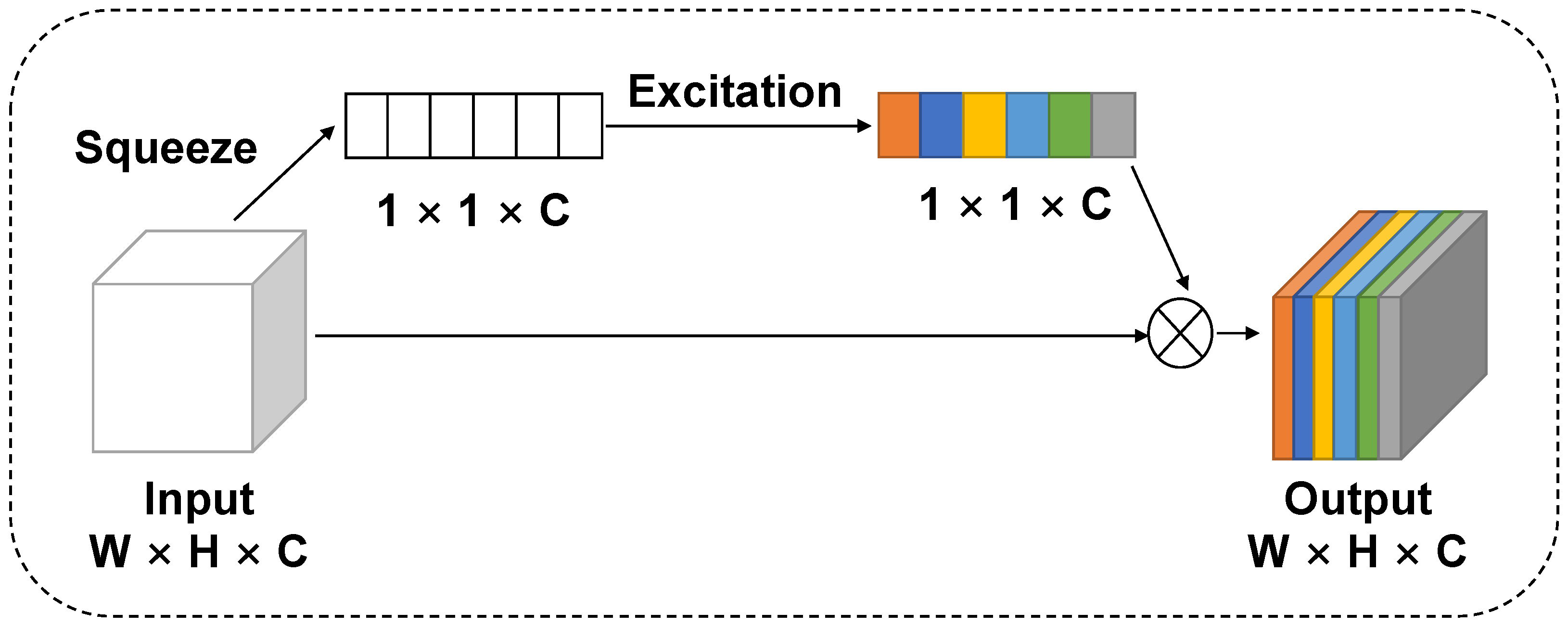
2.3.1. Convolutional Block Attention Module
2.3.2. Loss Function for Imbalanced Classification
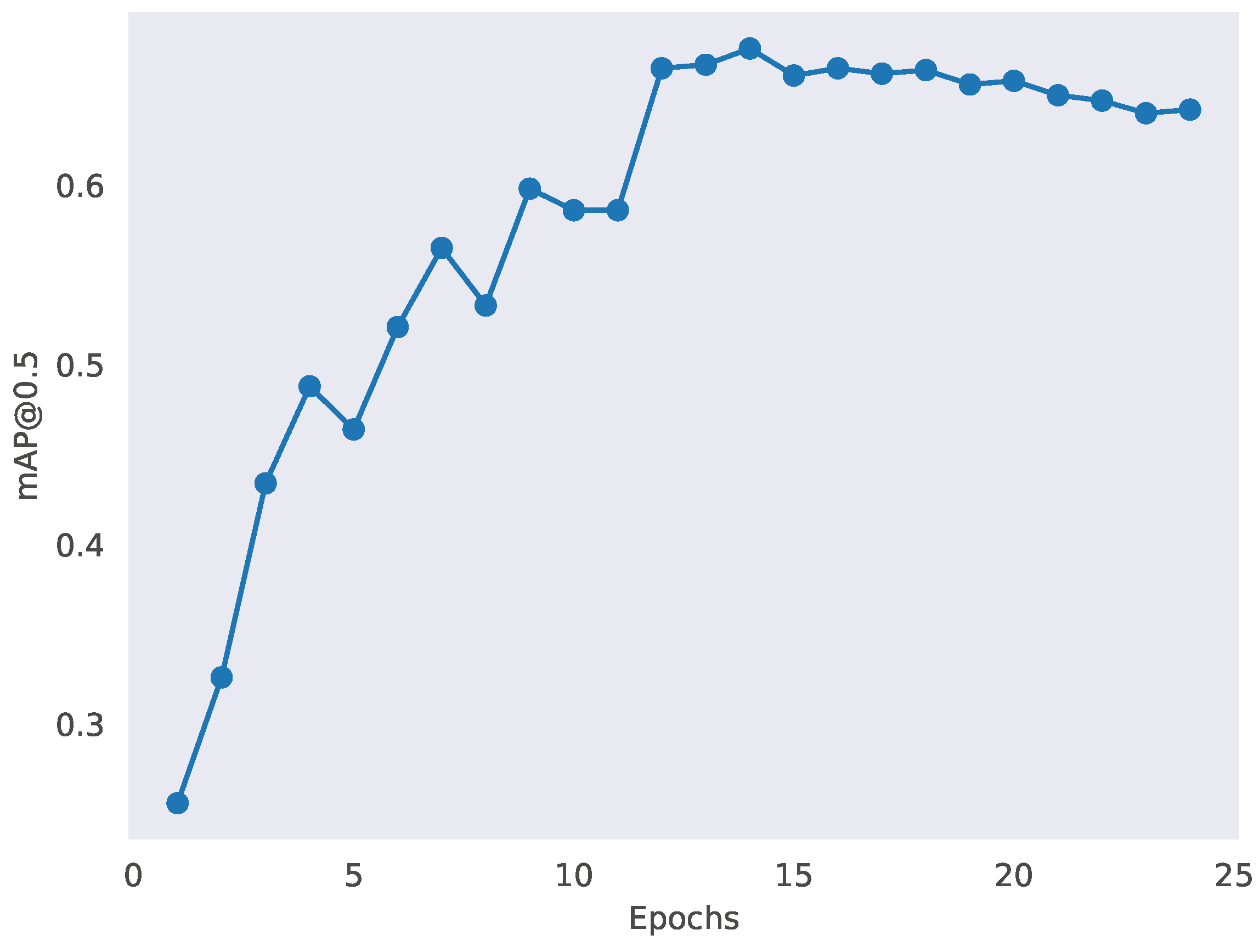
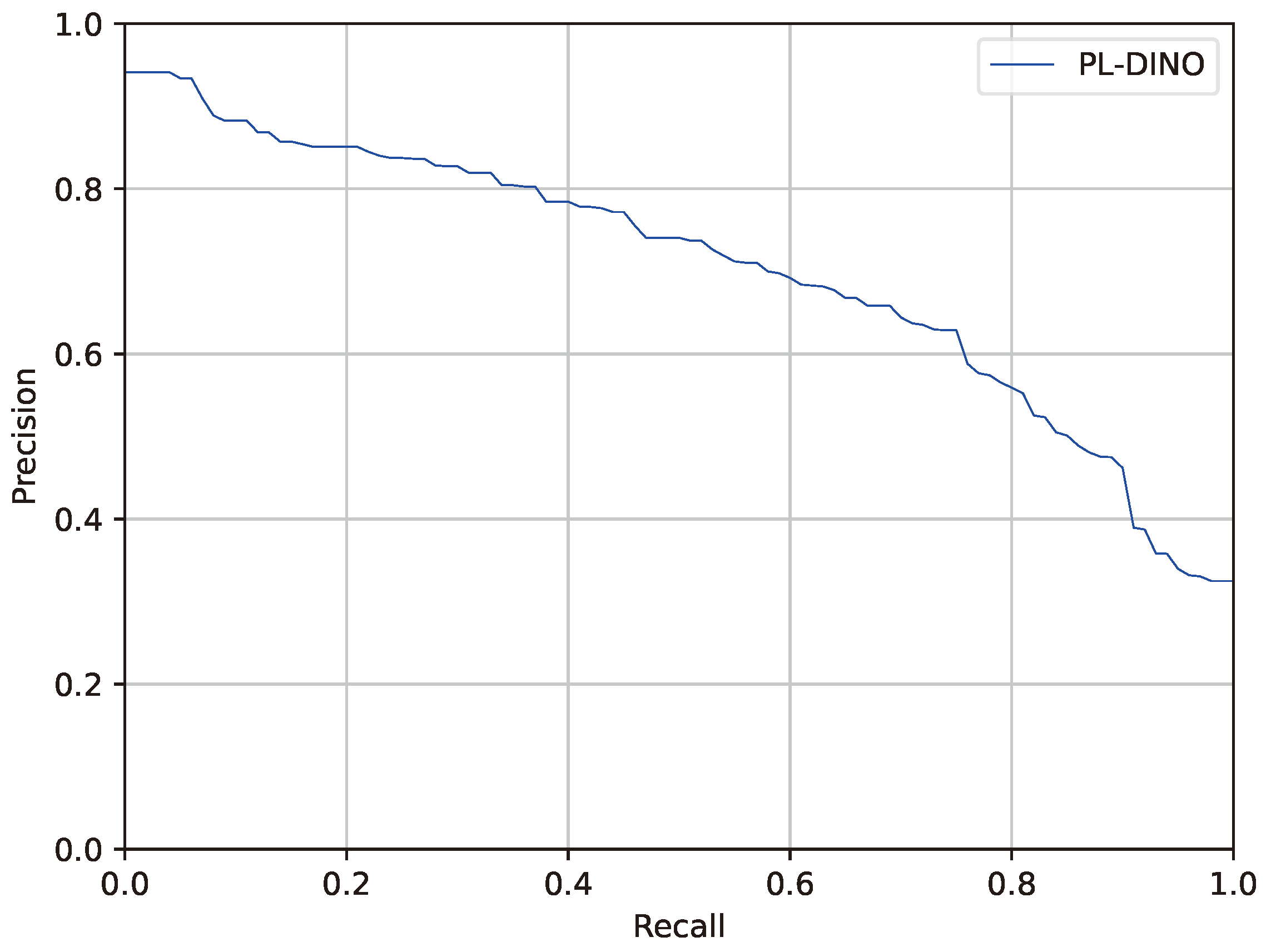
3. Results
3.1. Experimental Settings
3.2. Method Comparison
3.3. Model Ablation
3.4. Computational Expense
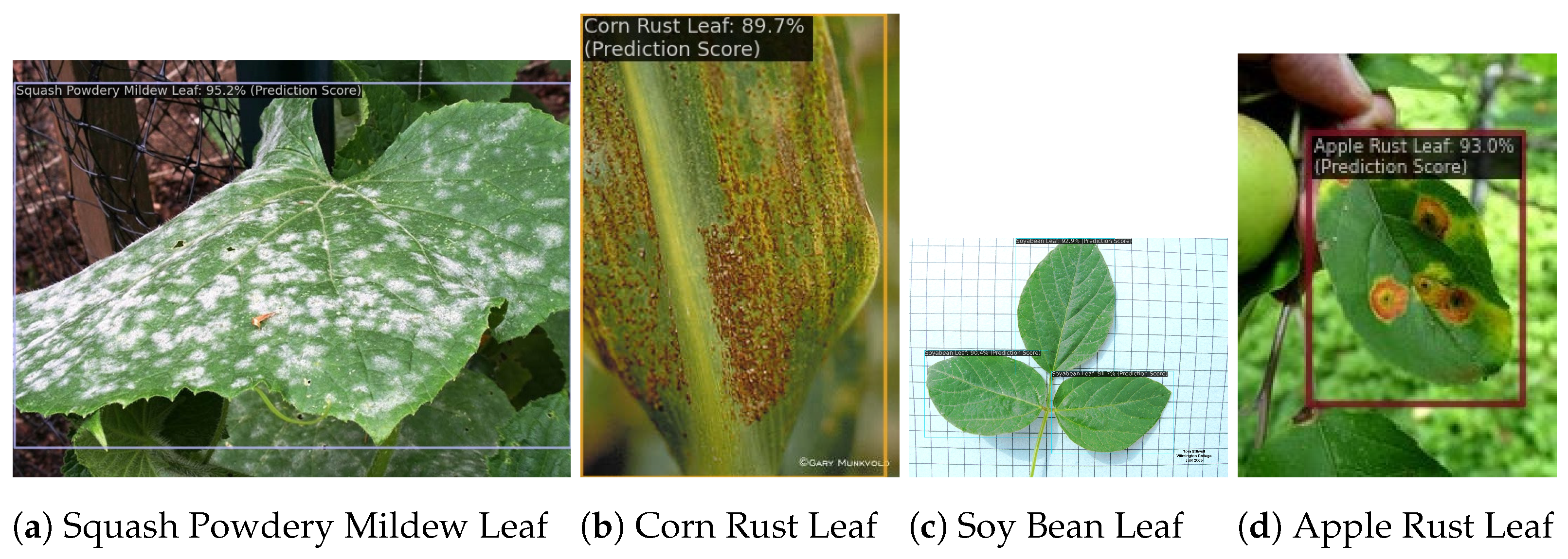
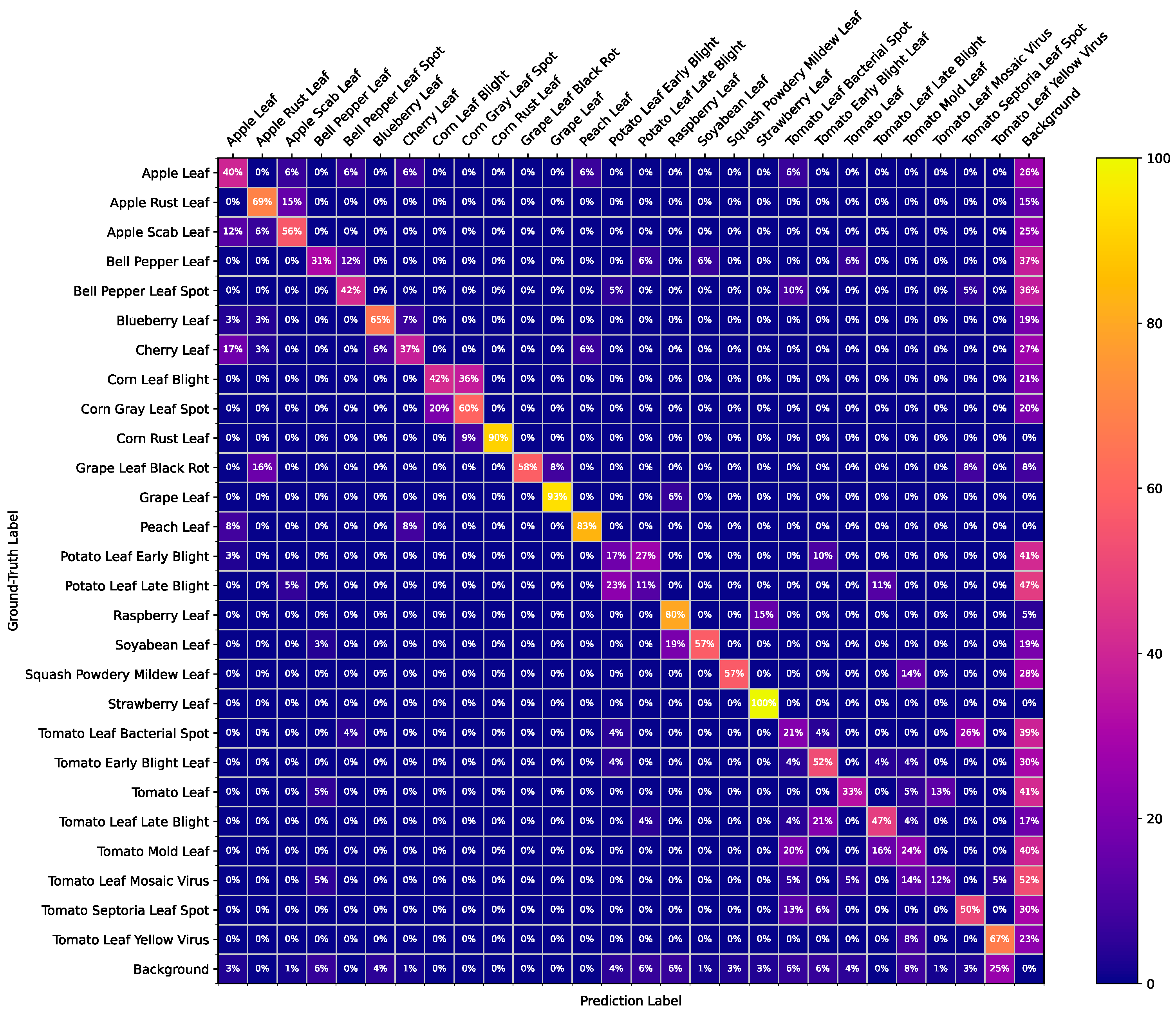
3.5. Result Visualization
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Xu, Y.; Li, J.; Wan, J. Agriculture and crop science in China: Innovation and sustainability. Crop J. 2017, 5, 95–99. [Google Scholar] [CrossRef]
- Shill, A.; Rahman, M.A. Plant disease detection based on YOLOv3 and YOLOv4. In Proceedings of the International Conference on Automation, Control and Mechatronics for Industry 4.0, IEEE, Rajshahi, Bangladesh, 8–9 July 2021; pp. 1–6. [Google Scholar]
- Atila, Ü.; Uçar, M.; Akyol, K.; Uçar, E. Plant leaf disease classification using EfficientNet deep learning model. Ecol. Inform. 2021, 61, 101182. [Google Scholar] [CrossRef]
- Bai, Y.; Hou, F.; Fan, X.; Lin, W.; Lu, J.; Zhou, J.; Fan, D.; Li, L. A lightweight pest detection model for drones based on transformer and super-resolution sampling techniques. Agriculture 2023, 13, 1812. [Google Scholar] [CrossRef]
- Yu, M.; Ma, X.; Guan, H. Recognition method of soybean leaf diseases using residual neural network based on transfer learning. Ecol. Inform. 2023, 76, 102096. [Google Scholar] [CrossRef]
- Cheng, S.; Cheng, H.; Yang, R.; Zhou, J.; Li, Z.; Shi, B.; Lee, M.; Ma, Q. A high performance wheat disease detection based on position information. Plants 2023, 12, 1191. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, J.; Cheng, W.; Chen, Z.; Zhou, J.; Cheng, H.; Lv, C. A high-precision plant disease detection method based on a dynamic pruning gate friendly to low-computing platforms. Plants 2023, 12, 2073. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wang, J.; Li, W.; Guan, P. T-CNN: Trilinear convolutional neural networks model for visual detection of plant diseases. Comput. Electron. Agric. 2021, 190, 106468. [Google Scholar] [CrossRef]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards real-time object detection with region proposal networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Vasconcelos, N. Cascade R-CNN: Delving into high quality object detection. In Proceedings of the Conference on Computer Vision and Pattern Recognition, IEEE, Salt Lake City, UT, USA, 18–23 June 2018; pp. 6154–6162. [Google Scholar]
- Zhu, X.; Lyu, S.; Wang, X.; Zhao, Q. TPH-YOLOv5: Improved YOLOv5 based on transformer prediction head for object detection on drone-captured scenarios. In Proceedings of the International Conference on Computer Vision Workshops, IEEE, Montreal, BC, Canada, 11–17 October 2021; pp. 2778–2788. [Google Scholar]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.Y.; Berg, A.C. SSD: Single shot multibox detector. In Proceedings of the European Conference on Computer Vision; Springer: Amsterdam, The Netherlands, 2016; pp. 21–37. [Google Scholar]
- Li, W.; Zhu, T.; Li, X.; Dong, J.; Liu, J. Recommending advanced deep learning models for efficient insect pest detection. Agriculture 2022, 12, 1065. [Google Scholar] [CrossRef]
- Jiang, P.; Chen, Y.; Liu, B.; He, D.; Liang, C. Real-time detection of apple leaf diseases using deep learning approach based on improved convolutional neural networks. IEEE Access 2019, 7, 59069–59080. [Google Scholar] [CrossRef]
- Liu, J.; Wang, X. Tomato diseases and pests detection based on improved YOlO V3 convolutional neural network. Front. Plant Sci. 2020, 11, 898. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Qiao, Y.; Liu, S.; Zhang, J.; Yang, Z.; Wang, M. An improved YOLOv5-based vegetable disease detection method. Comput. Electron. Agric. 2022, 202, 107345. [Google Scholar] [CrossRef]
- Qi, J.; Liu, X.; Liu, K.; Xu, F.; Guo, H.; Tian, X.; Li, M.; Bao, Z.; Li, Y. An improved YOLOv5 model based on visual attention mechanism: Application to recognition of tomato virus disease. Comput. Electron. Agric. 2022, 194, 106780. [Google Scholar] [CrossRef]
- Zhu, R.; Zou, H.; Li, Z.; Ni, R. Apple-Net: A model based on improved YOLOv5 to detect the apple leaf diseases. Plants 2023, 12, 169. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Wu, Q.; Chen, Y. Detecting soybean leaf disease from synthetic image using multi-feature fusion faster R-CNN. Comput. Electron. Agric. 2021, 183, 106064. [Google Scholar] [CrossRef]
- Wang, M.; Fu, B.; Fan, J.; Wang, Y.; Zhang, L.; Xia, C. Sweet potato leaf detection in a natural scene based on faster R-CNN with a visual attention mechanism and DIoU-NMS. Ecol. Inform. 2023, 73, 101931. [Google Scholar] [CrossRef]
- Zhou, G.; Zhang, W.; Chen, A.; He, M.; Ma, X. Rapid detection of rice disease based on FCM-KM and faster R-CNN fusion. IEEE Access 2019, 7, 143190–143206. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiao, D.; Liu, Y.; Wu, H. An algorithm for automatic identification of multiple developmental stages of rice spikes based on improved Faster R-CNN. Crop J. 2022, 10, 1323–1333. [Google Scholar] [CrossRef]
- Pan, J.; Xia, L.; Wu, Q.; Guo, Y.; Chen, Y.; Tian, X. Automatic strawberry leaf scorch severity estimation via faster R-CNN and few-shot learning. Ecol. Inform. 2022, 70, 101706. [Google Scholar] [CrossRef]
- Zhang, X.; Dong, H.; Gong, L.; Cheng, X.; Ge, Z.; Guo, L. Multiple paddy disease recognition methods based on deformable transformer attention mechanism in complex scenarios. Int. J. Comput. Appl. 2023, 45, 660–672. [Google Scholar] [CrossRef]
- Dananjayan, S.; Tang, Y.; Zhuang, J.; Hou, C.; Luo, S. Assessment of state-of-the-art deep learning based citrus disease detection techniques using annotated optical leaf images. Comput. Electron. Agric. 2022, 193, 106658. [Google Scholar] [CrossRef]
- Zhang, Y.; Kang, B.; Hooi, B.; Yan, S.; Feng, J. Deep long-tailed learning: A survey. IEEE Trans. Pattern Anal. Mach. Intell. 2023, 45, 10795–10816. [Google Scholar] [CrossRef]
- Kang, B.; Xie, S.; Rohrbach, M.; Yan, Z.; Gordo, A.; Feng, J.; Kalantidis, Y. Decoupling representation and classifier for long-tailed recognition. In Proceedings of the International Conference on Learning Representations, ICLR, Addis Ababa, Ethiopia, 26–30 April 2020; pp. 1–16. [Google Scholar]
- Zhou, B.; Cui, Q.; Wei, X.S.; Chen, Z.M. BBN: Bilateral-branch network with cumulative learning for long-tailed visual recognition. In Proceedings of the Conference on Computer Vision and Pattern Recognition, IEEE, Seattle, WA, USA, 13–19 June 2020; pp. 9719–9728. [Google Scholar]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal loss for dense object detection. In Proceedings of the International Conference on Computer Vision, IEEE, Venice, Italy, 22–29 October 2017; pp. 2980–2988. [Google Scholar]
- Li, B.; Yao, Y.; Tan, J.; Zhang, G.; Yu, F.; Lu, J.; Luo, Y. Equalized focal loss for dense long-tailed object detection. In Proceedings of the Conference on Computer Vision and Pattern Recognition, IEEE, New Orleans, LA, USA, 18–24 June 2022; pp. 6990–6999. [Google Scholar]
- Cui, Y.; Jia, M.; Lin, T.Y.; Song, Y.; Belongie, S. Class-balanced loss based on effective number of samples. In Proceedings of the Conference on Computer Vision and Pattern Recognition, IEEE, Long Beach, CA, USA, 15–20 June 2019; pp. 9268–9277. [Google Scholar]
- Cao, K.; Wei, C.; Gaidon, A.; Arechiga, N.; Ma, T. Learning imbalanced datasets with label-distribution-aware margin loss. In Proceedings of the Advances in Neural Information Processing Systems, NeurIPS, Vancouver, BC, Canada, 8–14 December 2019; pp. 1567–1578. [Google Scholar]
- Singh, D.; Jain, N.; Jain, P.; Kayal, P.; Kumawat, S.; Batra, N. PlantDoc: A dataset for visual plant disease detection. In Proceedings of the ACM India Joint International Conference on Data Science and Management of Data, Hyderabad, India, 5–7 January 2020; ACM: New York, NY, USA, 2020; pp. 249–253. [Google Scholar]
- Lin, T.Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft COCO: Common objects in context. In Proceedings of the European Conference on Computer Vision, Zurich, Switzerland, 6–12 September 2014; Springer: Berlin/Heidelberg, Germany, 2014; pp. 740–755. [Google Scholar]
- Carion, N.; Massa, F.; Synnaeve, G.; Usunier, N.; Kirillov, A.; Zagoruyko, S. End-to-end object detection with transformers. In Proceedings of the European Conference on Computer Vision, Glasgow, UK, 23–28 August 2020; pp. 213–229. [Google Scholar]
- Zhu, X.; Su, W.; Lu, L.; Li, B.; Wang, X.; Dai, J. Deformable DETR: Deformable transformers for end-to-end object detection. In Proceedings of the International Conference on Learning Representations, ICLR, Vienna, Austria, 3–7 May 2021; pp. 1–16. [Google Scholar]
- Zhang, H.; Li, F.; Liu, S.; Zhang, L.; Su, H.; Zhu, J.; Ni, L.M.; Shum, H.Y. DINO: DETR with improved denoising anchor boxes for end-to-end object detection. arXiv 2022, arXiv:2203.03605. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. CBAM: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision, Munich, Germany, 8–14 September 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 3–19. [Google Scholar]
- Tan, J.; Wang, C.; Li, B.; Li, Q.; Ouyang, W.; Yin, C.; Yan, J. Equalization loss for long-tailed object recognition. In Proceedings of the Conference on Computer Vision and Pattern Recognition, IEEE, Seattle, WA, USA, 13–19 June 2020; pp. 11662–11671. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-Excitation networks. In Proceedings of the Conference on Computer Vision and Pattern Recognition, IEEE, Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar]
- Wang, C.Y.; Bochkovskiy, A.; Liao, H.Y.M. YOLOv7: Trainable bag-of-freebies sets new state-of-the-art for real-time object detectors. In Proceedings of the Conference on Computer Vision and Pattern Recognition, IEEE, Vancouver, BC, Canada, 17–24 June 2023; pp. 7464–7475. [Google Scholar]
- Wang, Q.; Wu, B.; Zhu, P.; Li, P.; Zuo, W.; Hu, Q. ECA-Net: Efficient channel attention for deep convolutional neural networks. In Proceedings of the Conference on Computer Vision and Pattern Recognition, IEEE, Seattle, WA, USA, 13–19 June 2020; pp. 11531–11539. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the Conference on Computer Vision and Pattern Recognition, IEEE, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Zhang, H.; Wu, C.; Zhang, Z.; Zhu, Y.; Lin, H.; Zhang, Z.; Sun, Y.; He, T.; Mueller, J.; Manmatha, R.; et al. ResNeSt: Split-attention networks. In Proceedings of the Conference on Computer Vision and Pattern Recognition, IEEE, New Orleans, LA, USA, 18–24 June 2022; pp. 2736–2746. [Google Scholar]

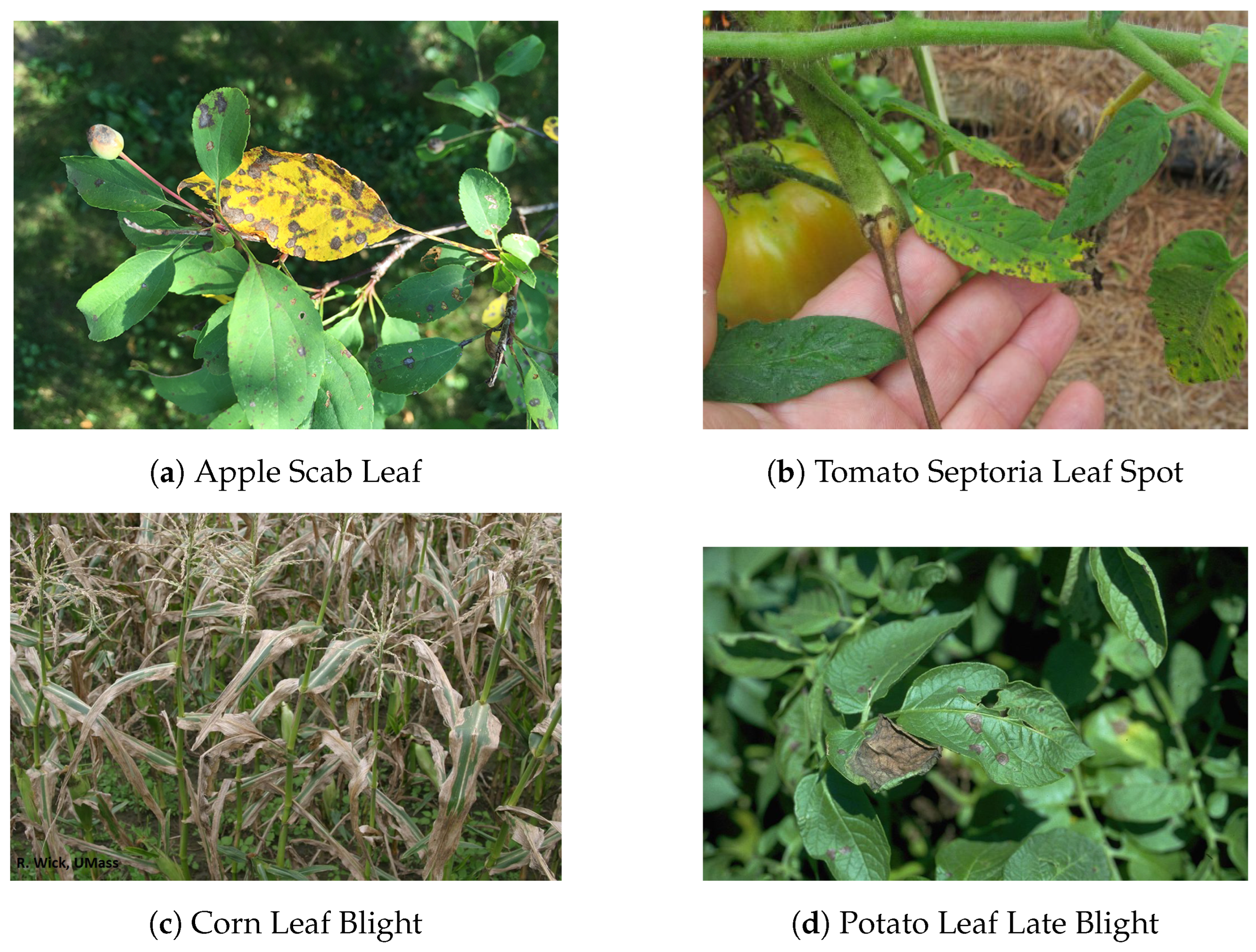


| Methods | Precision (%) | Recall (%) | F1-Score (%) | mAP@0.5 (%) |
|---|---|---|---|---|
| Faster R-CNN | 38.0 | 61.4 | 42.3 | 45.6 |
| YOLOv5s | 50.2 | 57.6 | 53.5 | 52.6 |
| YOLOv5m | 56.6 | 60.8 | 56.1 | 55.4 |
| YOLOv7 | 57.9 | 69.7 | 62.0 | 67.0 |
| DETR | 40.7 | 67.1 | 46.3 | 48.9 |
| PL-DINO | 62.9 | 75.0 | 63.2 | 70.3 |
| Models | mAP@0.5 (%) | Precision (%) | Recall (%) |
|---|---|---|---|
| Baseline | 67.2 | 63.4 | 64.0 |
| Baseline + CBAM | 68.2 | 63.1 | 69.0 |
| Baseline + EQL | 68.6 | 65.6 | 64.0 |
| Baseline + CBAM + EQL | 70.3 | 62.9 | 75.0 |
| Backbone Network | Attention Module | mAP@0.5 (%) | Precision (%) | Recall (%) |
|---|---|---|---|---|
| ResNet50 | - | 67.2 | 63.4 | 64.0 |
| ResNet50 | SE | 66.7 | 62.4 | 63.0 |
| ResNet50 | ECA | 66.1 | 59.1 | 69.0 |
| ResNet50 | CBAM | 68.2 | 63.1 | 69.0 |
| ResNeSt50 | - | 64.3 | 60.3 | 66.0 |
| ResNeSt50 | SE | 65.1 | 64.2 | 63.0 |
| ResNeSt50 | ECA | 63.8 | 61.5 | 62.0 |
| ResNeSt50 | CBAM | 64.9 | 59.1 | 68.0 |
| Models | Time (h:min:s) |
|---|---|
| Faster R-CNN | 4:40:14 |
| YOLOv5 | 9:03:25 |
| YOLOv7 | 8:46:33 |
| DETR | 12:10:47 |
| PL-DINO | 5:43:05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Zhu, L.; Liu, J. PL-DINO: An Improved Transformer-Based Method for Plant Leaf Disease Detection. Agriculture 2024, 14, 691. https://doi.org/10.3390/agriculture14050691
Li W, Zhu L, Liu J. PL-DINO: An Improved Transformer-Based Method for Plant Leaf Disease Detection. Agriculture. 2024; 14(5):691. https://doi.org/10.3390/agriculture14050691
Chicago/Turabian StyleLi, Wei, Lizhou Zhu, and Jun Liu. 2024. "PL-DINO: An Improved Transformer-Based Method for Plant Leaf Disease Detection" Agriculture 14, no. 5: 691. https://doi.org/10.3390/agriculture14050691
APA StyleLi, W., Zhu, L., & Liu, J. (2024). PL-DINO: An Improved Transformer-Based Method for Plant Leaf Disease Detection. Agriculture, 14(5), 691. https://doi.org/10.3390/agriculture14050691






