1. Introduction
The cultivation of cotton is a vital component of China’s agricultural industry, and the presence of pests and diseases can significantly impact both the quantity and quality of cotton production. Therefore, it is imperative to effectively manage and control cotton pests and diseases [
1,
2]. There is a need for agricultural producers and operators to rapidly acquire accurate and specialized knowledge in pest and disease management. However, with the advancement of the Internet and the Internet of Things in agriculture, there is an ever-increasing accumulation of data in this field, including cotton pests and diseases. This has led to a progressively complex data structure, making manual processing of big data retrieval nearly impossible. Therefore, the adoption of Natural Language Processing (NLP) technology [
3] and deep learning technology becomes essential for extracting knowledge from agricultural big data and constructing a specialized knowledge base. However, when utilizing machine learning models for entity and relationship extraction (ERE), a substantial amount of annotated data is often required to train the models. Nevertheless, in the case of cotton pests and disease data, the abundance of data, high specialization requirements, and complex semantic structure leads to the high cost of manual annotation. Consequently, integrating small-sample learning with knowledge extraction models represents an appropriate approach to address this issue.
The primary characteristic of small-sample learning [
4] lies in its ability to distinguish between different entity and relationship models with only a limited number of labeled samples. This approach effectively reduces the burden of annotating large amounts of data and significantly accelerates the pace of knowledge mining [
5]. While small-sample learning has been extensively explored in computer vision and image classification, its progress in NLP has been relatively sluggish due to the complexity of textual data. However, with the continuous advancement of deep learning techniques, small-sample learning has gained traction in areas such as knowledge extraction [
6]. For instance, Han et al. [
7] used a small-sample learning model to evaluate the constructed relational extraction dataset FewRel with good results. Therefore, we investigate the use of fine-tuning models to achieve ERE on small-sample datasets.
ERE is one of the important tasks in knowledge extraction [
8,
9,
10]; the traditional ERE models rely heavily on the quality of manually annotated data, which in turn affects the effectiveness of the model’s ERE. These models improve the efficiency of the ERE task to a certain extent but require a large number of samples to pre-train the model. To improve the small-sample learning ability of the machine, the method for ERE is designed in this paper based on the UIE model. In particular, ERE prompt templates and schema in the field of cotton pests and diseases are designed to serve as inputs to the UIE model, such that only a small number of labeled samples are required to enable the UIE model to achieve significant results. Further, by fine-tuning the model, the ambiguity of words is effectively resolved, which reduces the errors in entity recognition and significantly improves the overall performance of the model.
2. Related Research
The existing methods for ERE [
11] mainly comprise rule-based approaches, traditional machine learning-based techniques, and deep learning-based methodologies. Rule-based methods for ERE primarily rely on experts to manually construct rule templates, which are less commonly used due to their reliance on specific languages, domains, and text styles. On the other hand, traditional machine learning-based approaches aim to learn statistical models from a large number of annotated corpora to extract entities and relationships. Such models include Hidden Markov Models, Conditional Random Fields (CRF), and maximum entropy models. For example, Kambhatla et al. [
12] used a maximum entropy model combined with feature vectors for relationship classification. Chunyu Wang et al. [
13] first applied CRF models to agricultural entity recognition in the field of agriculture.
The effectiveness of traditional machine learning methods, however, is often hindered by intricate syntactic structures and lengthy texts. As a result, deep learning-based models for ERE have sprung up. For example, Guo et al. [
14] proposed the agricultural pest entity extraction model JMCA-ADP, which introduces a Convolutional Neural Network (CNN) on the basis of the Att-BiLSTM-CRF model and is able to extract the semantic feature information effectively. Song et al. [
15] introduced Word2Vec into the BiLSTM-CRF model and achieved good ERE results. Later, some researchers [
16,
17] demonstrated that a BiLSTM incorporating attention mechanism is able to extract complex semantic features of rice pests and weed texts. With the development of BERT pre-trained language models, Qiao et al. [
18] combined BERT with the joint extraction model LSTM-LSTM-Bias, which well extracted the entities and relationships from the agricultural dataset. However, all the above methods require the labeling of the experimental corpus. For this reason, remotely supervised learning applicable to large amounts of unlabeled data has attracted much attention. For example, Mintz et al. [
19] were the first to apply remote supervision to the task of relation extraction and extracted textual features with the help of a trained relational classifier. Lin et al. [
20] performed remote supervision based on a CNN with sentence-level attention to solve the problem of mislabeling arising from remote supervision. To effectively solve the data noise problem brought by remote supervision methods, Le et al. [
21] and Cui [
22] designed the segmented Convolutional Neural Network model MPCNN based on multiple heads of attention, which achieved certain results. Although remote supervision can perform ERE on unlabeled data, the method is susceptible to high mislabeling due to the interference of noisy data, and its accuracy is very unsatisfactory. To this end, this paper proposes a UIE-base model for ERE with a strong small-sample learning capability. By leveraging a limited number of labeled samples and employing fine-tuning techniques, the proposed model achieves satisfactory results on both dedicated and public datasets. The work in this paper has three main points of contribution:
(1) Since there are no publicly available datasets in the field of cotton pests and diseases, a corpus of cotton pest and disease data is first constructed in this paper. By analyzing and pre-processing the unstructured textual data within the corpus set, a professional-grade collection of 4475 instances pertaining to cotton pests and diseases is established;
(2) Based on the UIE model, the focus is on designing a specialized ERE prompt and schema on cotton pests and diseases, which are used as inputs to the UIE model to enhance the ability of ERNIE to extract specific features of cotton pests and diseases. The optimization and fine-tuning of the model by this method resulted in a great improvement in the small-sample learning capability of the UIE-base model, i.e., the model achieved an accuracy of 86.5% with only 40 training samples;
(3) The UIE model demonstrates strong information extraction capabilities in general, but its performance in specialized domains, especially in terms of few-shot learning capabilities, remains under-researched. In this paper, we address this gap by fine-tuning the UIE model using a limited amount of labeled data on cotton pests and diseases.
3. Materials and Methods
3.1. Dataset Construction
3.1.1. Data Acquisition and Pre-Processing
Due to the limited availability of publicly accessible corpus data on cotton pests and diseases, this study established a comprehensive knowledge corpus in the field by employing data acquisition, preprocessing, and data annotation techniques. The primary sources of data were diverse agricultural websites, including the National Agricultural Science Data Center, China Crop Germplasm Resource Information Network, China Agricultural Information Network, etc. The acquisition method primarily relied on utilizing the Python request library. After pre-processing the raw corpus using character format normalization and Python regular expressions, a normalized corpus of knowledge on cotton pests and diseases is formed, with a total of 4475 instances, which is used for subsequent experiments.
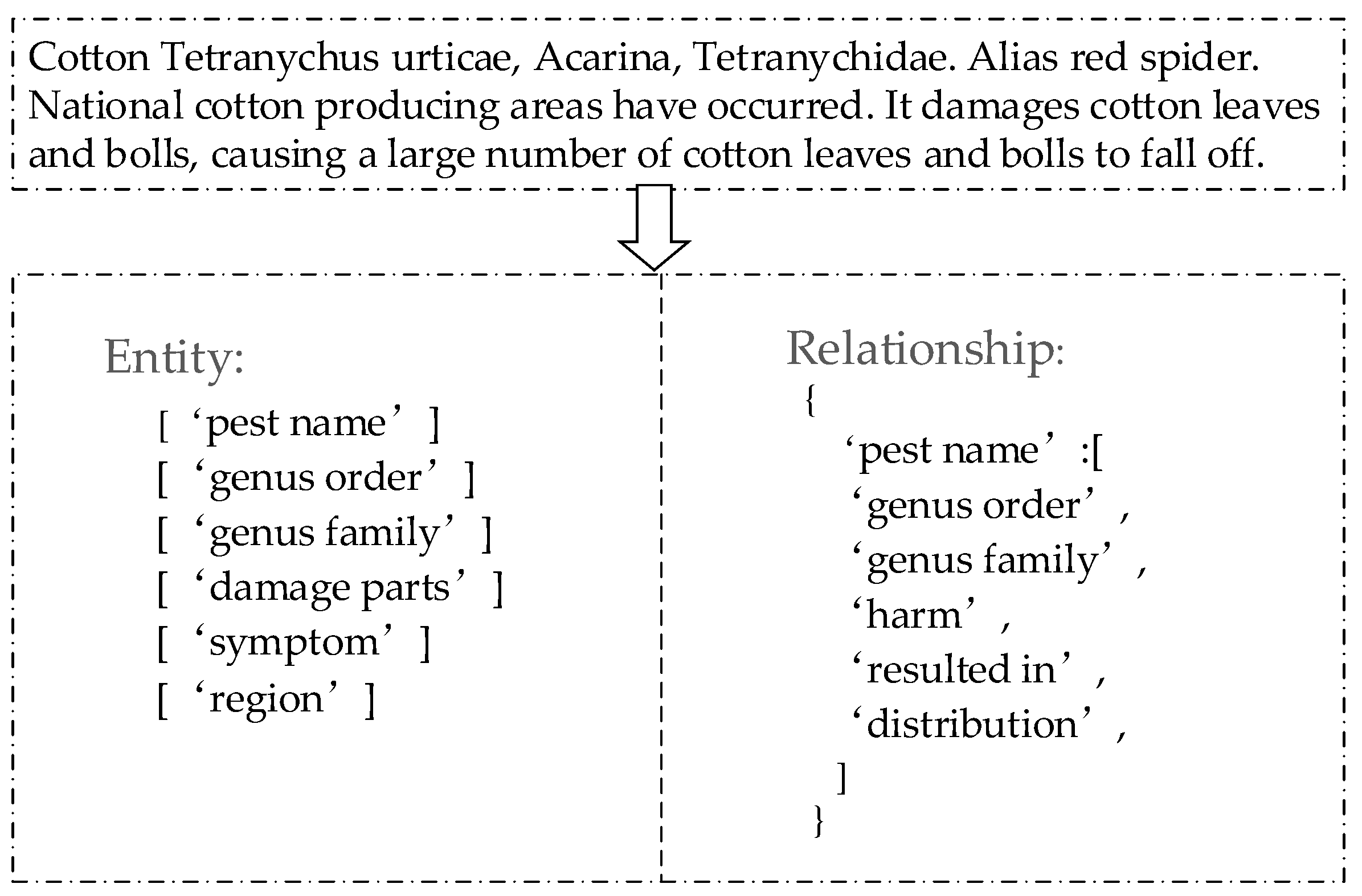
3.1.2. Classification of Entity and Relationship
Before using the constructed model to extract entities and relationships in the cotton pests and diseases corpus, consulting the professional books Chinese Agri-cultural Thesaurus and Special Classification for Agriculture, as well as consulting the plant protection experts, this paper first explores and defines the categories of entities and relationships related to pests and diseases [
23]. By conducting a comprehensive analysis of the corpus features, this study further subdivides the entity and relationship types within the cotton pests and diseases corpus, resulting in a total of 13 defined entity types and 12 relationship types. The specific categorization of entities and relationships is presented in
Table 1.
3.1.3. Data Annotation
In this paper, a small amount of the cotton pests and diseases corpus is manually annotated to fine-tune the training of the UIE model. In order to ensure the high quality of annotation data, this paper has formulated a comprehensive and explicit annotation policy based on the processed samples. Furthermore, expert consultation was sought for accurate data annotation. Finally, considering that the training data of this model is in JSON format, an existing advanced text annotation tool called Doccano was adopted in this study to achieve efficient data annotation. Random quality checks and audits were conducted on the annotated data to ensure adherence to the established guidelines. Specific examples of annotations are shown in
Figure 1.
The tool facilitates the direct conversion of annotated data into JSON format for fine-tuning the training of models. The resulting JSON data structure comprises two main components: entities and relationships. Entities encompass entity labels along with their corresponding start positions (start_offset and end_offset), while relationships consist of relationship types and their respective start positions (from_id and to_id). An illustrative example structure is presented in
Figure 2.
3.2. Entity and Relationship Extraction Model Construction
The overall structure of the UIE-base [
24] ERE model for cotton pests and diseases designed in this paper is shown in
Figure 3. The model mainly consists of three parts: the input layer, the coding layer, and the output layer. Among them, the input layer is mainly the input text content and the Structural Schema Instructor (SSI) mechanism. The coding layer mainly uses the ERNIE 3.0 [
25] knowledge-enhanced pre-training model to extract features from the input text. The output layer represents the extracted structures of different tasks in a unified encoding via Structured Extraction Language (SEL). The mechanics and relationships of the model modules are detailed below.
3.2.1. Input and Output Layers of the Model
The UIE model adaptively generates structures for different IE tasks mainly through the SSI, whose input layer relationships can be expressed as follows:
where
s denotes the defined structural extraction pattern,
x represents the input text sequence, and
y expresses the structured result generated. The parameters of Equation (1) can be specifically expanded:
where
s contains three types of tag segments: the name of the entity, the name of the relationship between entities, and special symbols ([
entity], [
association], and [
text]). These tokens collectively form the SSI, which is concatenated and placed ahead of the original text sequence to construct the final expression of the structural schema as represented by Equation (5):
For example, the SSI “[
entity] Cotton two-spotted leaf mite [
entity] Cotton red spider mite [
entity] All over the country [
association] Alias [
association] Distribution [
text]” represents a record of entity and relationship patterns extracted from the sentence “Cotton two-spotted leaf mite, alias cotton red spider mite, is distributed all over the country”. This record guides the UIE towards this particular task. It can be seen that the specialized design of schema in the SSI mechanism is critical. The schema construct design in this paper is shown in
Figure 4.
SEL expressions at the model output layer encompass specific semantic units tailored to a particular task. For instance, in the ERE task, the node name represents a distinct node type within the text, i.e., an entity category, whereas the relationship name signifies a specific information unit present in the corpus, i.e., an associative link between entities. Specific examples are shown in
Figure 5. In
Figure 5, the entities (spots) are highlighted in blue, while the relationship names are marked in red.
3.2.2. Prompt Template
The notable advantage of UIE lies in its capability to swiftly adapt to diverse IE tasks through fine-tuning. In order to enhance the model’s ability to capture entity and relationship features within the cotton pests and diseases corpus, this study proposes specific prompt templates as inputs to the model, as illustrated in Equations (6) and (7):
In the equation above, t represents the type of task to be extracted, such as ERE tasks. The
consists of two parts, language explanation (explain) and syntax (grammar), which indicate the entity and relationship type of the extraction definition, such as “pest name” for entity type and “Alias” for relationship type. The prompt template can be customized to extract various types of entities and relationships. In other words, a corpus can be divided into multiple prompts, each containing different entity and relationship types.
Figure 6 illustrates an example prompt format.
3.2.3. ERNIE Module
The ERNIE Module serves as the central component of the UIE framework [
26], responsible for encoding and decoding tasks. Its network architecture consists of two types of modules: a generic representation module and a task-specific representation module, as illustrated in
Figure 3. The generic representation module employs a multilayer Transformer-XL [
27] structure as its backbone network. Transformer-XL incorporates an auxiliary recursive memory module to effectively model longer text sequences, enabling enhanced learning of associative relations between pre- and post-textual content within such sequences. Moreover, it leverages diverse pre-training tasks from different paradigms to better capture generic lexical and syntactic−semantic information present in the training data. On the other hand, the task-specific representation module also adopts a Transformer-XL but focuses on learning high-level semantic features specific to particular task types. Further, we fine-tuned the model using the cross-entropy loss function so that it accurately identifies the entities and relationships of cotton pests and diseases. The calculation process of fine-tuning is shown in Formula (8):
where
represents the loss in the given task dataset
, and
denotes the input
x of sample
s and the corresponding entity and relationship label
y.
indicates the predicted probability of the model under parameters
,
for entity and relationship label
y given input
x and the condition
s logarithmically. By minimizing this loss function, it enables the model to enhance its ability to predict entity and relationship labels.
The decoding layer of the model is implemented using double pointers (
). The implementation principle involves utilizing the full link layer to calculate scores for each entity relation’s start pointer
and end pointer
, converting these scores into probabilities using SoftMax, and then predicting start and end positions of the entity and relationship based on these probabilities. The specific calculation is shown in Formulas (9)–(11):
where the weight pointer and bias vector of the start pointer are represented as
and
, while the weight pointer and bias vector of the end pointer are denoted as
and
.
represents the probability of the
ith element,
i denotes the index of the category,
n represents the dimension of the input vector, i.e., the number of entity and relationship categories,
refers to the
ith element of the input vector S, and
refers to the
jth element in the input vector. The prediction of the entity and relationship in the input sentence can be achieved by utilizing probabilities calculated through this process.
4. Experimental Results and Discussion
4.1. Experimental Environment and Experimental Parameters
The specific configuration of the experimental environment in this paper is shown in
Table 2. The training parameters of the model are set as shown in
Table 3.
4.2. Indicators for Model Evaluation
To evaluate the ERE effectiveness of the model, three evaluation criteria are used in this paper: precision, recall, and
F1, which are calculated as shown in Formulas (12)–(14).
where
represents the count of correctly identified positive samples in the model’s output sequence,
signifies the count of incorrectly predicted positive samples in the output sequence,
denotes the count of accurately predicted negative samples in the model’s output sequence,
P indicates the ratio of correctly predicted and positive samples to all predicted positives, and
R represents the proportion of truly positive samples that are correctly predicted by the model, i.e., it is a measure of correct predictions among all true positive. The
F1 score combines accuracy and recall evaluations, which is particularly useful for imbalanced categories, where higher values indicate better overall performance of the model.
4.3. Experiments and Analysis of Results
In this paper, six sets of experiments are designed to verify the effect of the model. The first experiment is the hyperparameter setting of the model. Secondly, the performance of the UIE multi-model was verified using a self-constructed dataset and a public dataset, respectively. Then, the better performance was identified as the UIE-base model. To further validate the generalization ability of the UIE-base model, experiments were designed to compare the performance of the UIE-base model with other classical models, such as the BERT-BiLSTM-CRF on the self-constructed dataset and the public dataset. Further, the small-sample learning ability of the UIE-base model was verified by refining the number of samples, and the most suitable small sample size for model learning was identified.
4.3.1. Experiments on Hyperparameter Settings of the Model
Hyperparameter settings play a crucial role in the model, and their appropriateness directly impacts the convergence speed and generalization ability of the model. The UIE model encompasses several hyperparameters, with learning rate, batch size, and number of training rounds (epochs) being the key ones. Among these, the learning rate primarily controls the step size during parameter updates; a larger learning rate may hinder convergence, while a smaller one can result in slower training progress. Additionally, batch size determines both training speed and stability of the model. To determine optimal hyperparameters for the UIE model, this study conducts multiple experiments on our dataset using different values for learning rate and batch size when 50 training samples are used over 30 epochs. The experimental results are presented in
Table 4. In the end, five sets of learning rates and batch sizes were selected, respectively, based on fine-tuning experience and the results of multiple rounds of testing (in
Table 4). From
Table 4, the best overall performance of the UIE model was achieved when a learning rate of 5 × 10
−5 and a batch size of 10 were chosen.
4.3.2. UIE Multi-Model Experiments
Due to the intricate and diverse nature of information extraction tasks, as well as the varying requirements for different models in processing these tasks, UIE systems typically incorporate multiple models. To select a suitable UIE model for ERE tasks, this study conducts experimental tests on various UIE models using both a self-constructed dataset and a publicly available industrial dataset. The experimental results are presented in
Table 5 and
Table 6. From these tables, it can be observed that the UIE-base model achieves the highest
F1 value in the multi-model test across both datasets, indicating superior overall performance compared to other UIE models. From the comparison of
Table 5 and
Table 6, it can be seen that the UIE model is overall better tested on the self-constructed dataset than on the public dataset (industry). The reason may be related to the quality and quantity of the dataset. Compared with public datasets, self-built datasets exhibit superior label consistency. Inconsistent labeling can perplex the model and result in performance degradation. Secondly, the samples within the self-built dataset are more relevant to the specific task that the model aims to perform, with a lower noise level compared to public datasets, enabling the better capture of data characteristics by the model. Furthermore, while large public datasets provide more information, they also introduce additional noise and extraneous details easily. Conversely, self-built datasets are relatively smaller yet meticulously organized and marked with high-quality labels, offering clearer guiding principles for improved model performance.
Finally, to accurately reflect the performance of UIE multiple models on different datasets, the
F1 values are represented using
Figure 7. As can be seen in
Figure 7, the
F1 values of the UIE-base model on the two datasets reach 79.17% and 62.26%, respectively, which is the best among all models. Therefore, the subsequent experiments in this paper adopt the UIE-base model as the entity–relationship extraction model.
4.3.3. Comparative Experiments of Different Models
To further verify the effectiveness of the UIE-base model in the ERE task, in this subsection, classical models such as the BERT-CRF are chosen to conduct experiments on the self-constructed dataset and the Chinese medical public dataset, respectively, which can also verify the generalization ability of the model in this paper. The experimental results are shown in
Table 7. It should be noted that the data format used by the UIE model and other models, such as the BERT-CRF, is different; the UIE model usually adopts the JSON data format, while other models usually adopt the BIO data format. Therefore, it is necessary to convert the format of JSON and BIO when comparing the experiments. As can be seen in
Table 7, the UIE-base model has the best ERE for both the self-built and public datasets. On the self-constructed dataset, the F1 value of the UIE model is far higher than that of most of the models and 1.4% higher than the next best model, BERT-BiLSTM-CRF. Since the cotton dataset has fewer samples than the public dataset as a whole, and the BERT-BiLSTM-CRF is more suitable for large sample sets, this also leads to their performance on the cotton dataset being inferior to that of the UIE-base model. This suggests that the UIE-base model is better suited to light sample size sets and initially sets the tone for the UIE-base in subsequent small-sample learning tests. Finally, to show the results more intuitively, the comparison results are visualized, as shown in
Figure 8a,b. It can be seen in
Figure 8 that the comprehensive performance of the UIE-base model is significantly better than the others.
4.3.4. Few-Shot Learning Test Experiments for Models
To verify the small-sample learning ability of the UIE model, this paper designs two sets of experiments based on the cotton pests and diseases dataset. One is to select 5, 15, and 30 samples in the UIE multi-model to verify its performance at several small-sample numbers in order to find the optimal model when samples are small, with the experimental results shown in
Table 8. Secondly, the number of training samples most suitable for the UIE-base model is further determined through testing, and the results are shown in
Table 9. Analyzing
Table 8, overall, the comprehensive performance of UIE models is improved when the number of samples increased by 10 or 15 entries; thus, it can be initially proved that the UIE model has some small-sample learning ability. In addition, with 5 samples, the UIE-base has the worst performance, while the UIE-m-base has the best overall performance. However, continuing to increase the number of samples, the performance of the UIE-base model improves significantly, while the performance of the other UIE models has some ups and downs. For example, the F1 value of the UIE-m-base improves, but its accuracy decreases significantly. This suggests that the UIE-base model is more stable in small-sample learning than the other UIE models. Two points can be summarised from
Table 9: (1) In general, increasing the number of training samples usually improves the training effect of the model. (2) The model works best with 40 training samples, indicating that choosing the right number of training samples is important.
Meanwhile, to better evaluate the comprehensive performance of the model, this paper visualizes the change in the loss rate of the UIE-base model at different numbers of samples, and the results are shown in
Figure 9. As can be seen from
Figure 9, the loss rate of the model is the lowest at 40 samples, which indicates that the model training effect is the best at this number of samples.
4.4. Discussion
Based on the above experiments and analyses, this paper demonstrates a method to implement small-sample learning based on the UIE model and applies it well to the ERE task of cotton pests and diseases, which effectively solves the problem that the existing model is difficult to capture semantic information under limited labeled data. However, the development of small-sample learning in this field still has some limitations.
Firstly, small-sample learning suffers from the risk of overfitting due to the insufficient amount of data, which, in turn, limits the model’s performance in learning and extracting feature representations. The currently existing solutions to address the risk of overfitting are as follows:
(1) Data enhancement: increasing data diversity through artificial methods such as rotation and flipping to help models learn more generalized features;
(2) Regularization technique: using Dropout to enable the model to randomly discard a portion of neurons during the training process, reducing the model’s dependence on specific training samples and enhancing the model’s generalization ability;
(3) Model selection and simplification: to address the risk of overfitting in small-sample learning, selecting or designing simple model structures will be more effective than complex models;
(4) Theoretical research and experimental validation: we need to deeply understand the causes of overfitting and then conduct multiple rounds of experiments on different small-sample learning tasks to verify the effectiveness of different strategies.
In addition, there are other limitations in the small-sample learning approach proposed in this paper. These include constraints on the model’s generalization ability due to the limited number of samples, challenges related to domain adaptability, difficulties in model selection and tuning, computational resource requirements, and the impact of labeled data quality on the feature learning of the model. All these limitations are the future challenges of small-sample learning development. Therefore, future work will mainly be carried out in terms of training strategies and optimization techniques for models and how to obtain high-quality labeled data.
5. Conclusions
In order to effectively alleviate the data scarcity problem caused by the need for a large amount of labeled data for NLP tasks, a small-sample learning method based on the UIE model is proposed to achieve the entity and relationship extraction of cotton pests and diseases. It aims to solve the problem of existing models in knowledge extraction (requiring a large amount of manually labeled training data), and the main work is summarized as follows:
(1) Dataset construction: Crawler technology is used to obtain unstructured data related to cotton pests and diseases to constitute the corpus in this paper, and after pre-processing the data in this corpus, such as removing residuals, the entity and relationship types of cotton pests and diseases are defined through an in-depth analysis of the characteristics of the data and combining it with the guidance of agricultural experts. Subsequently, a limited amount of data annotation is conducted on the corpus using the Doccano annotation tool. Ultimately, a dataset comprising 13 entity types and 12 relationship types is formed for fine-tuning learning purposes in the UIE model;
(2) A method for extracting entities and relationships from cotton pests and diseases, based on the UIE, is proposed. Considering the characteristics of the cotton pests and diseases corpus, specialized prompt templates and schema constructs are designed for fine-tuning the mode, which greatly improves the ability to learn from small samples, resulting in the UIE-base model reaching an accuracy of 86.5% when there are only 40 training samples;
(3) The paper presents six sets of training strategies to experimentally evaluate the model. The UIE multi-model performance test demonstrates that the UIE-base model better satisfies the accuracy and speed requirements for ERE tasks. By comparing the performance of the UIE-base model with models like BERT-CRF on our constructed dataset, we observe a 1.4% improvement in the F1 value over the BERT-BiLSTM-CRF model. This confirms that the UIE-base model exhibits superior overall performance with lightweight samples. Furthermore, small-sample learning experiments reveal that increasing the number of training samples generally enhances model training effectiveness; however, selecting an appropriate number of training samples leads to optimal learning outcomes for the UIE-base model. Specifically, when trained with 40 samples, this model efficiently captures semantic features and effectively identifies entity categories and their associative relationships within a corpus.
In summary, the method proposed in this paper can serve as a valuable point of reference for ERE in diverse domains.
Author Contributions
Conceptualization, W.Y. (Weiwei Yuan); methodology, W.Y. (Weiwei Yuan); software, W.Y. (Wanxia Yang) and L.H.; validation, W.Y. (Wanxia Yang), L.H. and T.Z.; formal analysis W.Y. (Weiwei Yuan); investigation, Y.H., J.L. and W.Y. (Wenbo Yan); resources, W.Y. (Weiwei Yuan); data curation, T.Z., Y.H. and J.L.; writing—original draft preparation, W.Y. (Weiwei Yuan); writing—review and editing, W.Y. (Wanxia Yang) and L.H.; visualization, W.Y. (Weiwei Yuan); funding acquisition, W.Y. (Wanxia Yang) and L.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key R&D Program of China (grant number: 2022ZD0115801), funding from the Data Acquisition and Processing and Testing and Analyzing the Knowledge Graph of Smart Farm Brain (GSAU-JSFW-2023-97).
Data Availability Statement
Data presented in this study are available on request from the author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Song, R.; He, Q.; Liu, X. Occurrence Characteristics and Control Technology of Main Pests and Diseases in Cotton Cultivation in Xinjiang. Agric. Eng. Technol. 2020, 40, 44–45. [Google Scholar] [CrossRef]
- Bai, J. High yield cultivation and pest control technology of cotton in Xinjiang. Seed Sci. 2022, 40, 22–24+36. [Google Scholar] [CrossRef]
- Zhao, T.; Xu, M.; Chen, A. Survey of Natural Language Processing. J. Xinjiang Norm. Univ. Philos. Soc. Sci. 2024, 2, 1–23. [Google Scholar] [CrossRef]
- Ge, Y.; Guo, Y.; Das, S.; Al-Garadi, M.A.; Sarker, A. Few-shot learning for medical text: A review of advances, trends, and opportunities. J. Biomed. Inform. 2023, 144, 104458. [Google Scholar] [CrossRef]
- Ji, Y.; Zhang, W.; Liu, Q.; Liu, S.; Hong, C.; Qiu, C.; Zhu, J.; Hui, Y.; Xiao, W. Based on few-shot learning of relation extraction method for Chinese Text. J. Nanjing Univ. Posts Telecommun. (Nat. Sci. Ed.) 2023, 43, 64–71. [Google Scholar] [CrossRef]
- Hou, J.; Li, X.; Yao, H.; Sun, H.; Mai, T.; Zhu, R. BERT-Based Chinese Relation Extraction for Public Security. IEEE Access 2020, 8, 132367–132375. [Google Scholar] [CrossRef]
- Han, X.; Zhu, H.; Yu, P.; Wang, Z.; Yao, Y.; Liu, Z.; Sun, M. Fewrel: A large-scale supervised few-shot relation classification dataset with state-of-the-art evaluation. arXiv 2018, arXiv:1810.10147. [Google Scholar]
- Bao, T. Research on Entity Recognition and Relation Extraction in The Field of Tomato Pests and Diseases. Master’s Thesis, Jiangsu University, Zhenjiang, China, 2022. [Google Scholar] [CrossRef]
- Miao, Z. Research on Knowledge Map Construction and Knowledge Extraction Method of Agricultural Diseases and Pests; Jilin Agricultural University: Changchun, China, 2023. [Google Scholar]
- Zhuang, H. Research on Knowledge Extraction Technology Based on Deep Learning. Master’s Thesis, Guilin University of Electronic Technology, Guilin, China, 2021. [Google Scholar] [CrossRef]
- Hu, X.Y.; Zhang, W.; Xiao, S.; Cheng, R.; Hu, Y.; Zhou, Y.; Niu, P. Survey of Entity Relationship Extraction Based on Deep Learning. J. Softw. 2019, 30, 1793–1818. [Google Scholar] [CrossRef]
- Kambhatla, N. Combining lexical, syntactic, and semantic features with maximum entropy models for information extraction. In Proceedings of the ACL Interactive Poster and Demonstration Sessions, Barcelona, Spain, 21–27 July 2004; pp. 178–181. [Google Scholar]
- Wang, C.; Wang, F. Study on recognition of Chinese agricultural named entity with conditional random fields. J. Agric. Univ. Hebei 2014, 37, 132–135. [Google Scholar] [CrossRef]
- Guo, X.; Hao, X.; Tang, Z.; Diao, L.; Bai, Z.; Lu, S.; Li, L. ACE-ADP: Adversarial Contextual Embeddings Based Named Entity Recognition for Agricultural Diseases and Pests. Agriculture 2021, 11, 912. [Google Scholar] [CrossRef]
- Song, L.; Liu, S.; Wang, C. Text entity extraction of agricultural technology demand based on word vector + BiLSTM + CRF. Jiangsu Agric. Sci. 2021, 49, 186–193. [Google Scholar] [CrossRef]
- Wu, Y.; Ding, G.; Hu, B. Research on agricultural financial text relation extraction based on attention mechanism. Data Anal. Knowl. Discov. 2019, 3, 86–92. [Google Scholar]
- Shen, L. Research on Entity Relationship Extraction ang Knowledge Graph Construction Method for Rice Cultivation Program. Master’s Thesis, Nanjing Agriculural University, Nanjing, China, 2019. [Google Scholar] [CrossRef]
- Qiao, B.; Zou, Z.; Huang, Y.; Fang, K.; Zhu, X.; Chen, Y. A joint model for entity and relation extraction based on BERT. Neural Comput. Appl. 2022, 34, 3471–3481. [Google Scholar] [CrossRef]
- Mintz, M.; Bills, S.; Snow, R.; Jurafsky, D. Distant supervision for relation extraction without labeled data. In Proceedings of the Joint Conference of the 47th Annual Meeting of the ACL and the 4th International Joint Conference on Natural Language Processing of the AFNLP, Singapore, 1 August 2009; pp. 1003–1011. [Google Scholar]
- Lin, Y.; Shen, S.; Liu, Z.; Luan, H.; Sun, M. Neural relation extraction with selective attention over instances. In Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Berlin, Germany, 16 August 2016; pp. 2124–2133. [Google Scholar]
- Le, Y.; Wang, w.; Zhang, K.; Liang, Z.; Liu, F.; Chen, Y.; Wu, Y.; Zhang, Y. Agricultural pest and disease relation extraction based on multi-attention mechanism and distant supervision. J. Anhui Agric. Univ. 2020, 47, 682–686. [Google Scholar] [CrossRef]
- Cui, Z. Research and Implementation of Plant Relationship Extraction in Tibetan Plateau Based on Distant Supervision; Xizang Minzu University: Xianyang, China, 2022. [Google Scholar] [CrossRef]
- Jiang, J.; Guan, C.; Liu, J.; Guan, Y.; Ke, S. Annotation Scheme and Corpus Construction for Agricultural Knowledge Based on Active Learning and Crowdsourcing. J. Chin. Inf. Process. 2023, 37, 33–45. [Google Scholar]
- Lu, Y.; Liu, Q.; Dai, D.; Xiao, X.; Lin, H.; Han, X.; Sun, L.; Wu, H. Unified structure generation for universal information extraction. arXiv 2022, arXiv:2203.12277. [Google Scholar]
- Sun, Y.; Wang, S.; Feng, S.; Ding, S.; Pang, C.; Shang, J.; Wang, H. Ernie 3.0: Large-scale knowledge enhanced pre-training for language understanding and generation. arXiv 2021, arXiv:2107.02137. [Google Scholar]
- Ping, Z.; Qi, S.; Sun, W.; Lou, X.; Wo, J.; Zhang, Y.; Jiang, T.; Shan, L. An Entity and Event Recognition Method for Power Grid Fault Handling Plan Based on UIE Framework. Electr. Power 2023, 56, 138–146. [Google Scholar]
- Dai, Z.; Yang, Z.; Yang, Y.; Carbonell, J.; Le, Q.V.; Salakhutdinov, R. Transformer-xl: Attentive language models beyond a fixed-length context. arXiv 2019, arXiv:1901.02860. [Google Scholar]
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).