Characterizing Bacterial Communities in Agroecosystems of the UNESCO Global Geopark Mixteca Alta, Oaxaca
Abstract
1. Introduction
2. Materials and Methods
2.1. Soil Sampling
2.2. DNA Extraction and Sequencing
2.3. Data Analysis
3. Results
3.1. Diversity and Composition of Bacterial Communities of Agroecosystems of GMA
3.2. Diversity Estimates
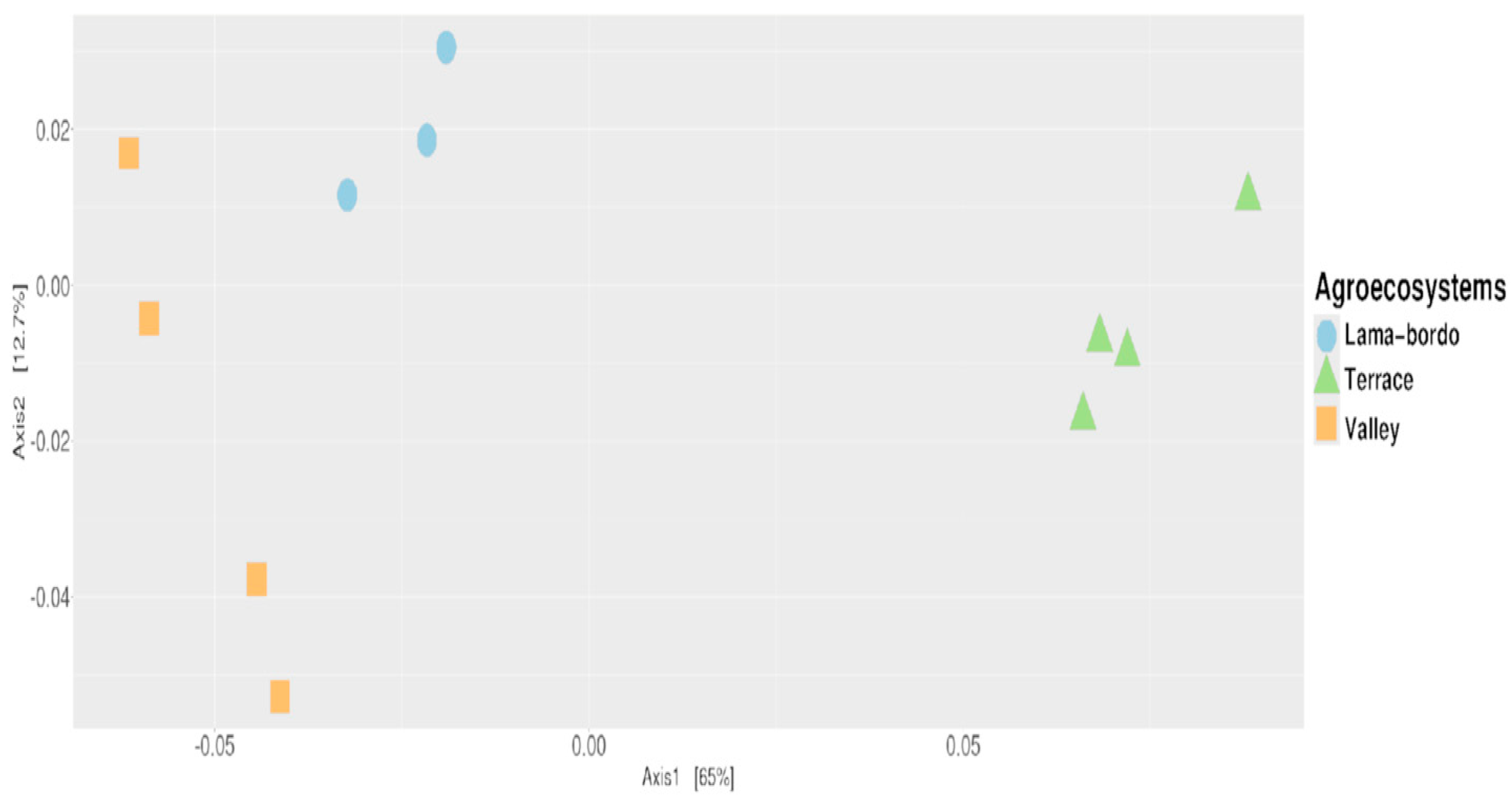
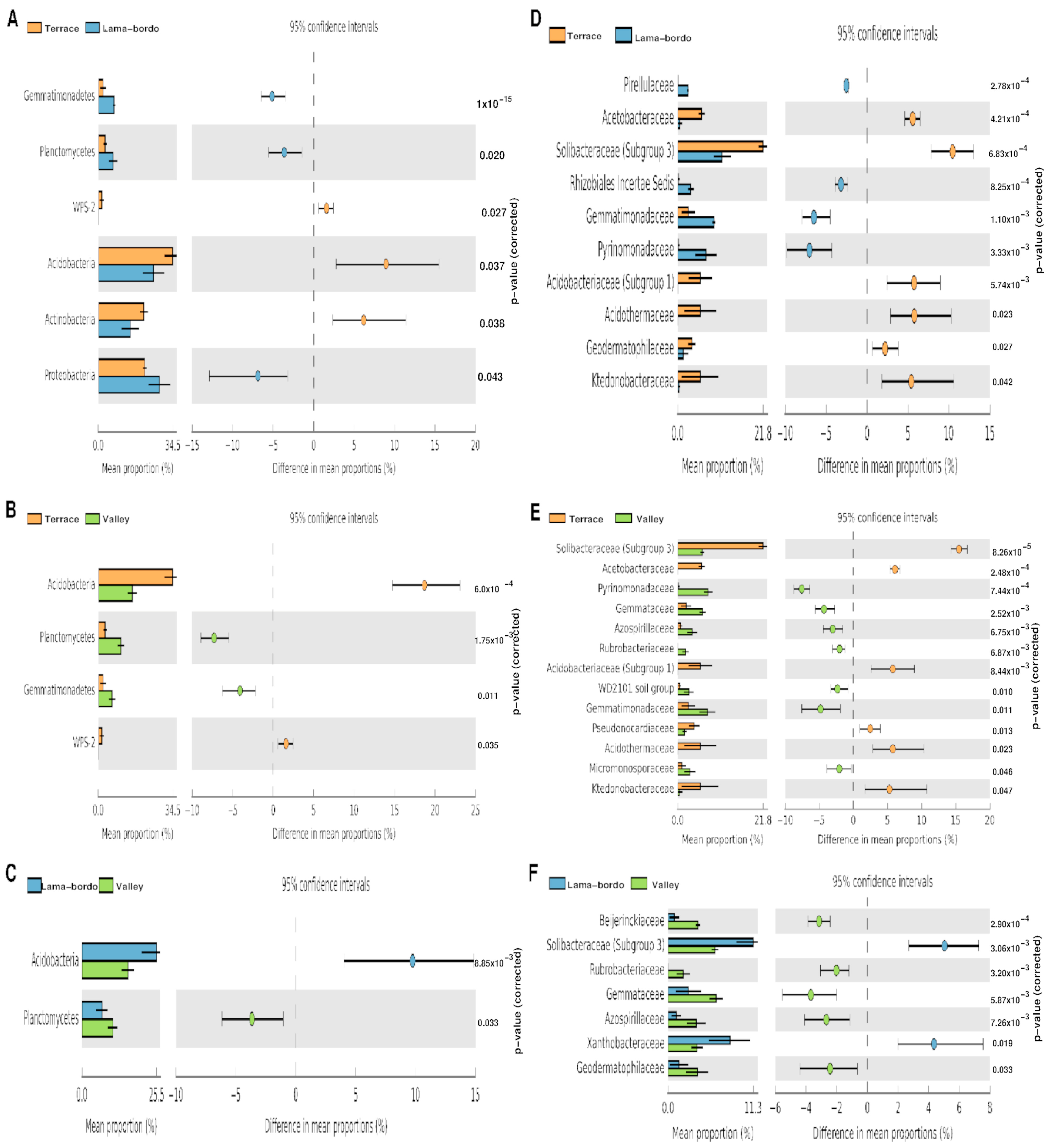
3.3. Agroecosystem–Taxonomy Association
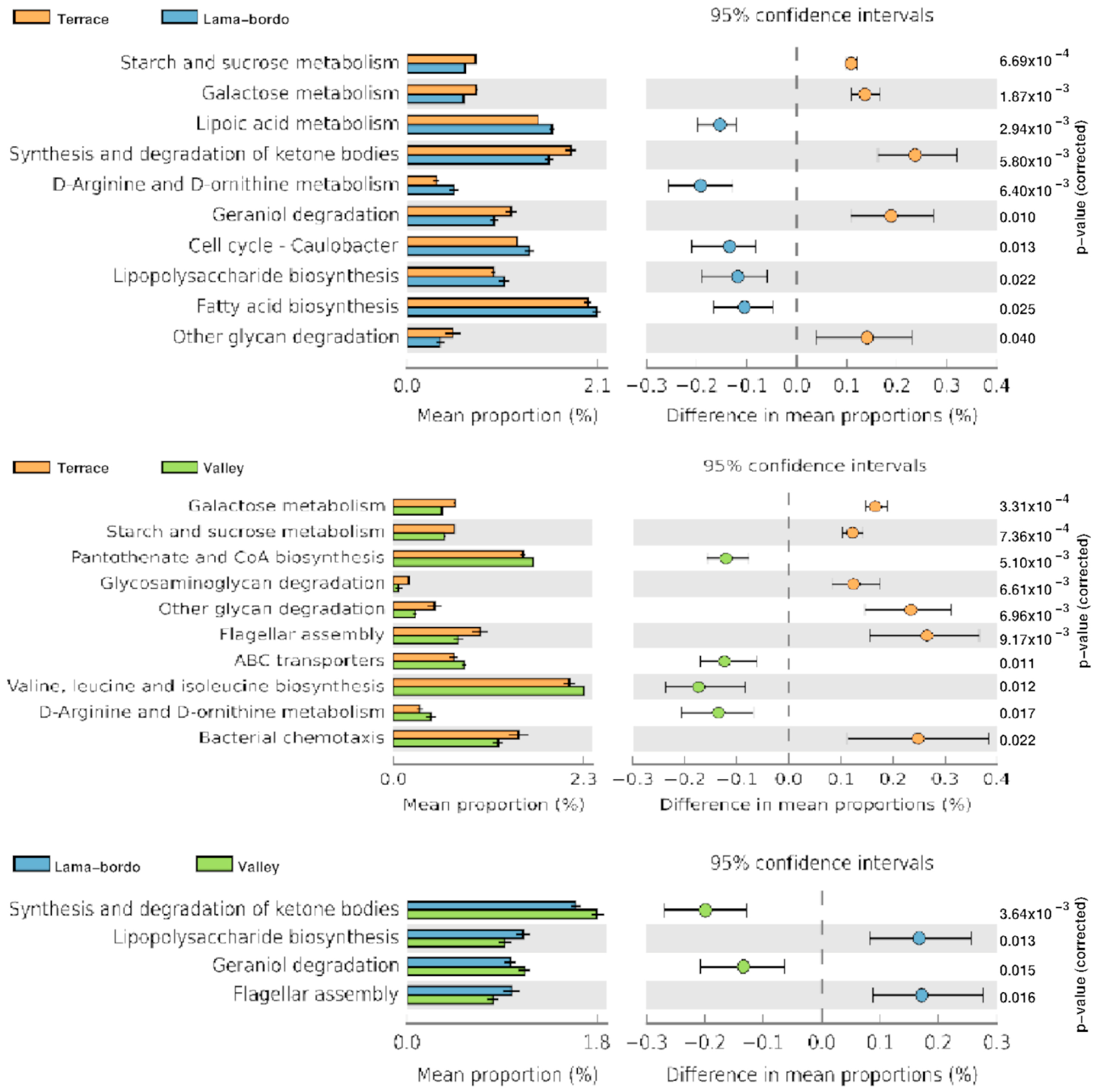
3.4. Functional Profile Prediction of Agroecosystems
4. Discussion
4.1. Microbiomes of Ancient Agroecosystems of Mixteca Alta Geopark
4.2. Ecological Adaptations in Agroecosystems of Mixteca Alta Geopark
4.3. Exploring Major Metabolic Pathways of Prokaryotic Communities in MAG Agroecosystems
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mueller, R.G.; Joyce, A.A.; Borejsza, A. Alluvial archives of the Nochixtlan valley, Oaxaca, Mexico: Age and significance for reconstructions of environmental change. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2012, 321–322, 121–136. [Google Scholar] [CrossRef]
- Vazquez-Castro, G.; Solís-Castillo, B. Late Pleistocene-Holocene paleoclimatic implications in the Mixteca Alta, Oaxaca, Mexico, by using rock magnetism and micromorphological techniques. J. S. Am. Earth Sci. 2021, 108, 103186. [Google Scholar] [CrossRef]
- Orozco-Ramírez, Q.; Lorenzen, M.; Fernández de Castro Martínez, G.; Ramírez, C. Social and Biophysical Factors of the Forest Transition in the UNESCO Mixteca Alta Global Geopark. Investig. Geográficas 2022, 108, e60465. [Google Scholar]
- Palacio-Prieto, J.L.; Rosado-González, E.; Ramírez-Miguel, X.; Oropeza-Orozco, O.; Cram-Heydrich, S.; Ortiz-Pérez, M.A.; Figueroa-Mah-Eng, J.M.; Fernández de Castro-Martínez, G. Erosion, Culture and Geoheritage; the Case of Santo Domingo Yanhuitlán, Oaxaca, México. Geoheritage 2016, 8, 359–369. [Google Scholar] [CrossRef]
- Leigh, D.S.; Kowalewski, S.A.; Holdridge, G. 3400 years of agricultural engineering in Mesoamerica: Lama-bordos of the Mixteca Alta, Oaxaca, Mexico. J. Archaeol. Sci. 2013, 40, 4107–4111. [Google Scholar] [CrossRef]
- Spores, R. Settlement, farming technology, and environment in the Nochixtlan Valley. Science 1969, 166, 557–569. [Google Scholar] [CrossRef]
- Solis Castillo, B.; Bocco, G. Terraces and landscape in Mixteca Alta, Oaxaca, Mexico: Micromorphological indicators. Span. J. Soil Sci. 2018, 8, 194–213. [Google Scholar] [CrossRef]
- Orozco-Ramírez, Q.; Bocco, G.; Solís-Castillo, B. Cajete maize in the Mixteca Alta region of Oaxaca, Mexico: Adaptation, transformation, and permanence. Agroecol. Sustain. Food Syst. 2020, 44, 1162–1184. [Google Scholar] [CrossRef]
- Orozco-Ramírez, Q.; Bocco, G. Agricultural landscapes diversity in the Global Geopark UNESCO Mixteca Alta, Oaxaca, Mexico. Rev. Geogr. Agrícola 2021, 66, 29–51. [Google Scholar]
- Orozco-Ramírez, Q.; Velasco Santiago, A.; Ramos Ortiz, J. La agrobiodiversidad de la Mixteca Alta, patrimonio biocultural no reconocido: El caso del Geoparque Mixteca Alta. In Hallazgos Del Patrimonio Natural, Cultural y Derechos Humanos en México. Un enfoque Participativo y Multidisciplinario; Sedas Larios, E., Martinez, M.V., Eds.; Congreso de la Unión, Universidad Anáhuac: Naucalpan, México, 2020; pp. 107–125. [Google Scholar]
- Pérez Rodríguez, V. Terrace Agriculture in the Mixteca Alta Region, Oaxaca, Mexico: Ethnographic and Archeological Insights on Terrace Construction and Labor Organization. Cult. Agric. Food Environ. 2016, 38, 18–27. [Google Scholar] [CrossRef]
- Pomboza Tamaquiza, P.P.; Navarro Garza, H.; Pérez Olvera, M.A.; Flores Sanchez, D. Prácticas organizativas mixtecas asociadas con la seguridad alimentaria y su patrimonio. Revista Mexicana de Ciencias Agrícolas 2017, 8, 3697–3710. [Google Scholar] [CrossRef][Green Version]
- Marcos-Rivera, U.; Ugalde-Lezama, S.; Valdés-Velarde, E.; Tarango-Arámbula, L.A.; Buendía-Espinoza, J.C. Multifunctionality in maize production systems in the Mixteca Alta region of Oaxaca. Agro Product. 2023, 16, 83–96. [Google Scholar]
- Liu, X.; Floate, K.D.; Gorzelak, M.A.; Holman, D.B.; Hrycauk, S.; Kubota, H.; Lupwayi, N.; Neilson, J.A.D.; Ortega Polo, R.; Petri, R.M.; et al. Prairie Agroecosystems: Interconnected Microbiomes of Livestock, Soil and Insects. Agriculture 2023, 13, 326. [Google Scholar] [CrossRef]
- Lehmann, A.; Zheng, W.; Rillig, M.C. Soil biota contributions to soil aggregation. Nat. Ecol. Evol. 2017, 1, 1828–1835. [Google Scholar] [CrossRef]
- Trognitz, F.; Hackl, E.; Widhalm, S.; Sessitsch, A. The role of plant-microbiome interactions in weed establishment and control. FEMS Microbiol. Ecol. 2016, 92, 1–15. [Google Scholar] [CrossRef]
- Zheng, W.; Zeng, S.; Bais, H.; Lamanna, J.M. Plant growth-promoting rhizobacteria (PGPR) reduce evaporation and increase soil water retention. Water Resour. Res. 2018, 54, 3673–3687. [Google Scholar] [CrossRef]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant–microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef]
- Fitzpatrick, C.R.; Copeland, J.; Wang, P.W.; Guttman, D.S.; Kotanen, P.M.; Johnson, M.T.J. Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl. Acad. Sci. USA 2018, 115, E1157. [Google Scholar] [CrossRef]
- Matthews, A.; Pierce, S.; Hipperson, H.; Raymond, B. Rhizobacterial Community Assembly Patterns Vary Between Crop Species. Front. Microbiol. 2019, 10, 581. [Google Scholar] [CrossRef]
- Schlemper, T.R.; van Veen, J.A.; Kuramae, E.E. Co-Variation of Bacterial and Fungal Communities in Different Sorghum Cultivars and Growth Stages is Soil Dependent. Microb. Ecol. 2018, 76, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Toju, H.; Peay, K.G.; Yamamichi, M.; Narisawa, K.; Hiruma, K.; Naito, K.; Fukuda, S.; Ushio, M.; Nakaoka, S.; Onoda, Y.; et al. Core microbiomes for sustainable agroecosystems. Nat. Plants 2018, 4, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Xiong, C.; Lu, Y. Microbiomes in agroecosystem: Diversity, function and assembly mechanisms. Environ. Microbiol. 2022, 14, 833–849. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, M.; Six, J. Soil structure and microbiome functions in agroecosystems. Nat. Rev. Earth Environ. 2023, 4, 4–18. [Google Scholar] [CrossRef]
- Hariharan, J.; Sengupta, A.; Grewal, P.; Dick, W.A. Functional Predictions of Microbial Communities in Soil as Affected by Long-term Tillage Practices. Agric. Environ. Lett. 2017, 2, 170031. [Google Scholar] [CrossRef]
- Navarrete-Euan, H.; Rodríguez-Escamilla, Z.; Pérez-Rueda, E.; Escalante-Herrera, K.; Martínez-Núñez, M.A. Comparing Sediment Microbiomes in Contaminated and Pristine Wetlands along the Coast of Yucatan. Microorganisms 2021, 9, 877. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Pavoine, S.; Dufour, A.B.; Chessel, D. From dissimilarities among species to dissimilarities among communities: A double principal coordinate analysis. J. Theor. Biol. 2004, 228, 523–537. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef]
- Jacoby, R.; Peukert, M.; Succurro, A.; Koprivova, A.; Kopriva, S. The Role of Soil Microorganisms in Plant Mineral Nutrition-Current Knowledge and Future Directions. Front. Plant Sci. 2017, 19, 1617. [Google Scholar] [CrossRef]
- Jansson, J.K.; Hofmockel, K.S. Soil microbiomes and climate change. Nat. Rev. Microbiol. 2020, 18, 35–46. [Google Scholar] [CrossRef]
- Trivedi, P.; Mattupalli, C.; Eversole, K.; Leach, J.E. Enabling sustainable agriculture through understanding and enhancement of microbiomes. New Phytol. 2021, 230, 2129–2147. [Google Scholar] [CrossRef]
- Hermans, S.M.; Lear, G.; Case, B.S.; Buckley, H.L. The soil microbiome: An essential, but neglected, component of regenerative agroecosystems. iScience 2023, 26, 106028. [Google Scholar] [CrossRef]
- Górska, E.B.; Stępień, W.; Hewelke, E.; Lata, J.-C.; Gworek, B.; Gozdowski, D.; Sas-Paszt, L.; Bazot, S.; Lisek, A.; Gradowski, M.; et al. Response of soil microbiota to various soil management practices in 100-year-old agriculture field and identification of potential bacterial ecological indicator. Ecol. Indic. 2024, 158, 111545. [Google Scholar] [CrossRef]
- Qaisrani, M.M.; Zaheer, A.; Mirza, M.S.; Naqqash, T.; Qaisrani, T.B.; Hanif, M.K.; Rasool, G.; Malik, K.A.; Ullah, S.; Jamal, M.S.; et al. A comparative study of bacterial diversity based on culturable and culture-independent techniques in the rhizosphere of maize (Zea mays L.). Saudi J. Biol. Sci. 2019, 26, 1344–1351. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.; Singh, B.; Patra, A.; Tripathi, A.; Easwaran, M.; Choudhary, J.R.; Choudhary, M.; Aggarwal, S.K. Maize microbiome: Current insights for the sustainable agriculture. In Microbiomes and Plant Health: Panoply and Their Applications; Solanki, M.K., Kashyap, P.L., Ansari, R.A., Kumari, B., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 267–297. [Google Scholar]
- Palaniyandi, S.A.; Yang, S.H.; Zhang, L.; Suh, J.W. Effects of actinobacteria on plant disease suppression and growth promotion. Appl. Microbiol. Biotechnol. 2013, 97, 9621–9636. [Google Scholar] [CrossRef] [PubMed]
- Bruto, M.; Prigent-Combaret, C.; Muller, D.; Moënne-Loccoz, Y. Analysis of genes contributing to plant-beneficial functions in Plant Growth-Promoting Rhizobacteria and related Proteobacteria. Sci. Rep. 2014, 4, 6261. [Google Scholar] [CrossRef] [PubMed]
- Lanzavecchia, G.; Frascarelli, G.; Rocchetti, L.; Bellucci, E.; Bitocchi, E.; Di Vittori, V.; Sillo, F.; Ferraris, I.; Carta, G.; Delledonne, M.; et al. Genotype Combinations Drive Variability in the Microbiome Configuration of the Rhizosphere of Maize/Bean Intercropping System. Int. J. Mol. Sci. 2024, 25, 1288. [Google Scholar] [CrossRef]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Meier-Kolthoff, J.P.; Klenk, H.P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2015, 80, 1–43. [Google Scholar] [CrossRef]
- Górska, E.B.; Steien, W.; Cunha, A.; Garcia, N.A.S.; Szyszkowska, K.; Gozdowski, D.; Gworek, B.; Sas-Paszt, L.; Lisek, A.; Hewelke, E.; et al. Microbial diversity as an indicator of a diversified cropping system for luvisoils in a moderate climate case study-long term experiments from poland. Ecol. Indic. 2022, 141, 109133. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Garrido-Oter, R.; Münch, P.C.; Weiman, A.; Dröge, J.; Pan, Y.; McHardy, A.C.; Schulze-Lefert, P. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 2015, 17, 392–403. [Google Scholar] [CrossRef] [PubMed]
- Xiong, C.; Zhu, Y.G.; Wang, J.T.; Singh, B.; Han, L.L.; Shen, J.P.; Li, P.P.; Wang, G.B.; Wu, C.F.; Ge, A.H.; et al. Host selection shapes crop microbiome assembly and network complexity. New Phytol. 2021, 229, 1091–1104. [Google Scholar] [CrossRef]
- Zhang, H.; Priemé, A.; Faucherre, S.; Elberling, B.; Jia, Z. Soil microbiomes modulate distinct patterns of soil respiration and methane oxidation in arctic active layer and permafrost. Acta Microbiol. Sin. 2017, 57, 839–855. [Google Scholar]
- Barnard, R.L.; Osborne, C.A.; Firestone, M.K. Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J. 2013, 7, 2229–2241. [Google Scholar] [CrossRef]
- Glaeser, S.P.; Kämpfer, P. The Family Sphingomonadaceae. In The Prokaryotes; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 641–707. [Google Scholar]
- Company, J.; Valiente, N.; Fortesa, J.; García-Comendador, J.; Lucas-Borja, M.E.; Ortega, R.; Miralles, I.; Estrany, J. Secondary succession and parent material drive soil bacterial community composition in terraced abandoned olive groves from a Mediterranean hyper-humid mountainous area. Agric. Ecosyst. Environ. 2022, 332, 107932. [Google Scholar] [CrossRef]
- Chen, Y.; Neilson, J.W.; Kushwaha, P.; Maier, R.M.; Barberán, A. Life-history strategies of soil microbial communities in an arid ecosystem. ISME J. 2021, 15, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Stone, B.W.G.; Dijkstra, P.; Finley, B.K.; Fitzpatrick, R.; Foley, M.M.; Hayer, M.; Hofmockel, K.S.; Koch, B.J.; Li, J.; Liu, X.J.A.; et al. Life history strategies among soil bacteria-dichotomy for few, continuum for many. ISME J. 2023, 17, 611–619. [Google Scholar] [CrossRef] [PubMed]
- López Castañeda, N. Lamabordos en la Mixteca Alta: Características de Suelos, Abandono y Dinámica Erosiva. Master Dissertation/Thesis, Universidad Nacional Autónoma de Mexico, CDMX, Mexico, 2019. [Google Scholar]
- Santiago-Mejía, B.E.; Martínez-Menez, M.R.; Rubio-Granados, E.; Vaquera-Huerta, H.; Sánchez-Escudero, J. Variabilidad espacial de propiedades físicas y químicas del suelo en un sistema lama-bordo en la Mixteca Alta de Oaxaca, México. Agric. Soc. Desarro. 2018, 15, 275–288. [Google Scholar] [CrossRef]
- Moubareck, C.A.; Alawlaqi, B.; Alhajeri, S. Characterization of physicochemical parameters and bacterial diversity of composted organic food wastes in Dubai. Heliyon 2023, 9, e16426. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Wang, C.; Liu, R.; Cai, R.; Sun, C. Physiological and Metabolic Insights into the First Cultured Anaerobic Representative of Deep-Sea Planctomycetes Bacteria. eLife 2024, 12, RP89874. [Google Scholar] [CrossRef]
- Johnston, J.; Vilardi, K.; Cotto, I.; Sudarshan, A.; Bian, K.; Klaus, S.; Bachmann, M.; Parsons, M.; Wilson, C.; Bott, C.; et al. Metatranscriptomic Analysis Reveals Synergistic Activities of Comammox and Anammox Bacteria in Full-Scale Attached Growth Nitrogen Removal System. Environ. Sci. Technol. 2024, 58, 13023–13034. [Google Scholar] [CrossRef]
- Cao, Q.; Liu, B.; Wu, J.; Zhang, X.; Ma, W.; Cui, D. Soil organic carbon fraction accumulation and bacterial characteristics in curtilage soil: Effects of land conversion and land use. PLoS ONE 2023, 18, e0283802. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Dong, Q.; Zhang, K.; Sha, D.; Jiang, C.; Yang, X.; Liu, X.; Zhang, H.; Wang, X.; Guo, F.; et al. Maize-peanut rotational strip intercropping improves peanut growth and soil properties by optimizing microbial community diversity. PeerJ 2022, 10, e13777. [Google Scholar] [CrossRef]
- Chee-Sanford, J.; Tian, D.; Sanford, R. Consumption of N2O and other N-cycle intermediates by Gemmatimonas aurantiaca strain T-27. Microbiol. SGM 2019, 165, 1345–1354. [Google Scholar] [CrossRef]
- Mujakić, I.; Piwosz, K.; Koblížek, M. Phylum Gemmatimonadota and Its Role in the Environment. Microorganisms 2022, 10, 151. [Google Scholar] [CrossRef]
- Spain, A.; Krumholz, L.; Elshahed, M. Abundance, composition, diversity and novelty of soil Proteobacteria. ISME J. 2009, 3, 992–1000. [Google Scholar] [CrossRef]
- Fu, X.; Huang, Y.; Fu, Q.; Qiu, Y.; Zhao, J.; Li, J.; Wu, X.; Yang, Y.; Liu, H.; Yang, X.; et al. Critical transition of soil microbial diversity and composition triggered by plant rhizosphere effects. Front. Plant Sci. 2023, 14, 1252821. [Google Scholar] [CrossRef] [PubMed]
- Yao, F.; Yang, S.; Wang, Z.; Wang, X.; Ye, J.; Wang, X.; DeBruyn, J.M.; Feng, X.; Jiang, Y.; Li, H. Microbial Taxa Distribution Is Associated with Ecological Trophic Cascades along an Elevation Gradient. Front. Microbiol. 2017, 8, 2071. [Google Scholar] [CrossRef] [PubMed]
- De Castro, V.H.; Schroeder, L.F.; Quirino, B.F.; Kruger, R.H.; Barreto, C.C. Acidobacteria from oligotrophic soil from the Cerrado can grow in a wide range of carbon source concentrations. Can. J. Microbiol. 2013, 59, 746–753. [Google Scholar] [CrossRef] [PubMed]
- Kielak, A.M.; Barreto, C.C.; Kowalchuk, G.A.; van Veen, J.A.; Kuramae, E.E. The Ecology of Acidobacteria: Moving beyond Genes and Genomes. Front. Microbiol. 2016, 7, 744. [Google Scholar] [CrossRef]
- Larsbrink, J.; McKee, L.S. Bacteroidetes bacteria in the soil: Glycan acquisition, enzyme secretion, and gliding motility. Adv. Appl. Microbiol. 2020, 110, 63–98. [Google Scholar]
- Lapébie, P.; Lombard, V.; Drula, E.; Terrapon, N.; Henrissat, B. Bacteroidetes use thousands of enzyme combinations to break down glycans. Nat. Commun. 2019, 10, 2043. [Google Scholar] [CrossRef]
- Lidbury, I.D.E.A.; Borsetto, C.; Murphy, A.R.J.; Bottrill, A.; Jones, A.M.E.; Bending, G.D.; Hammond, J.P.; Chen, Y.; Wellington, E.M.H.; Scanlan, D.J. Niche-adaptation in plant-associated Bacteroidetes favours specialisation in organic phosphorus mineralisation. ISME J. 2021, 15, 1040–1055. [Google Scholar] [CrossRef]
- Takahashi, Y.; Fujitáni, H.; Hirono, Y.; Tago, K.; Wang, Y.; Hayatsu, M.; Tsuneda, S. Enrichment of Comammox and Nitrite-Oxidizing Nitrospira from Acidic Soils. Front. Microbiol. 2020, 11, 1737. [Google Scholar] [CrossRef]
- Chisholm, C.; Di, H.J.; Cameron, K.; Podolyan, A.; Shah, A.; Hsu, L.; Shen, J. Soil moisture is a primary driver of comammox Nitrospira abundance in New Zealand soils. Sci. Total Environ. 2023, 858, 159961. [Google Scholar] [CrossRef] [PubMed]
- Sheremet, A.; Jones, G.M.; Jarett, J.; Bowers, R.M.; Bedard, I.; Culham, C.; Eloe-Fadrosh, E.A.; Ivanova, N.; Malmstrom, R.R.; Grasby, S.E.; et al. Ecological and genomic analyses of candidate phylum WPS-2 bacteria in an unvegetated soil. Environ. Microbiol. 2020, 22, 3143–3157. [Google Scholar] [CrossRef] [PubMed]
- Ji, M.; Williams, T.J.; Montgomery, K.; Wong, H.L.; Zaugg, J.; Berengut, J.F.; Bissett, A.; Chuvochina, M.; Hugenholtz, P.; Ferrari, B.C. Candidatus Eremiobacterota, a metabolically and phylogenetically diverse terrestrial phylum with acid-tolerant adaptations. ISME J. 2021, 15, 2692–2707. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Zhang, S.; Li, H.; Teng, K.; Wu, S.; Liu, Y.; Yu, F.; He, Z.; Li, L.; Li, L.; et al. Metagenomic insights into the response of soil microbial communities to pathogenic Ralstonia solanacearum. Front. Plant Sci. 2024, 16, 1325141. [Google Scholar] [CrossRef]
- Nazari, M.T.; Machado, B.S.; Marchezi, G.; Crestani, L.; Ferrari, V.; Colla, L.M.; Piccin, J.S. Use of soil actinomycetes for pharmaceutical, food, agricultural, and environmental purposes. 3 Biotech. 2022, 12, 232. [Google Scholar] [CrossRef]
- Ward, N.L.; Challacombe, J.F.; Janssen, P.H.; Henrissat, B.; Coutinho, P.M.; Wu, M.; Xie, G.; Haft, D.H.; Sait, M.; Badger, J.; et al. Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microbiol. 2009, 75, 2046–2056. [Google Scholar] [CrossRef]
- Moe, L.A. Amino acids in the rhizosphere: From plants to microbes. Am. J. Bot. 2013, 100, 1692–1705. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wilson, A.J.; Han, J.; Hui, A.; O’Sullivan, L.; Huan, T.; Haney, C.H. Amino Acid Availability Determines Plant Immune Homeostasis in the Rhizosphere Microbiome. MBio 2023, 14, e0342422. [Google Scholar] [CrossRef]
- González-López, J.; Rodelas, B.; Pozo, C.; Salmerón-López, V.; Martínez-Toledo, M.V.; Salmerón, V. Liberation of amino acids by heterotrophic nitrogen fixing bacteria. Amino Acids 2005, 28, 363–367. [Google Scholar] [CrossRef]
- López-Sámano, M.; Beltrán, L.F.L.; Sánchez-Thomas, R.; Dávalos, A.; Villaseñor, T.; García-García, J.D.; García-de Los Santos, A. A novel way to synthesize pantothenate in bacteria involves β-alanine synthase present in uracil degradation pathway. Microbiologyopen 2020, 9, e1006. [Google Scholar] [CrossRef]
- Zheng, Q.; Hu, Y.; Kosina, S.M.; Van Goethem, M.W.; Tringe, S.G.; Bowen, B.P.; Northen, T.R. Conservation of beneficial microbes between the rhizosphere and the cyanosphere. New Phytol. 2023, 240, 1246–1258. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Wu, Y.; Hong, M.; Yun, Z.; Li, T.; Jiang, Y. α-Lipoic acid treatment alleviates postharvest pericarp browning of litchi fruit by regulating antioxidant ability and energy metabolism. Postharvest Biol. Technol. 2021, 180, 111629. [Google Scholar] [CrossRef]
- Sezgin, A.; Altuntaş, C.; Demiralay, M.; Cinemre, S.; Terzi, R. Exogenous alpha lipoic acid can stimulate photosystem II activity and the gene expressions of carbon fixation and chlorophyll metabolism enzymes in maize seedlings under drought. J. Plant Physiol. 2019, 232, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Vuko, M.; Cania, B.; Vogel, C.; Kublik, S.; Schloter, M.; Schulz, S. Shifts in reclamation management strategies shape the role of exopolysaccharide and lipopolysaccharide-producing bacteria during soil formation. Microb. Biotechnol. 2020, 13, 584–598. [Google Scholar] [CrossRef] [PubMed]
- Hewavitharana, S.S.; Klarer, E.; Reed, A.J.; Leisso, R.; Poirier, B.; Honaas, L.; Rudell, D.R.; Mazzola, M. Temporal Dynamics of the Soil Metabolome and Microbiome During Simulated Anaerobic Soil Disinfestation. Front. Microbiol. 2019, 10, 2365. [Google Scholar] [CrossRef]
- Veum, K.S.; Lorenz, T.; Kremer, R.J. Phospholipid Fatty Acid Profiles of Soils under Variable Handling and Storage Conditions. Agron. J. 2019, 111, 1090–1096. [Google Scholar] [CrossRef]
- Gunina, A.; Kuzyakov, Y. Sugars in soil and sweets for microorganisms: Review of origin, content, composition and fate. Soil Biol. Biochem. 2015, 90, 87–100. [Google Scholar] [CrossRef]
- Keegstra, J.M.; Carrara, F.; Stocker, R. The ecological roles of bacterial chemotaxis. Nat. Rev. Microbiol. 2022, 20, 491–504. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Núñez, M.A.; Orozco-Ramírez, Q. Characterizing Bacterial Communities in Agroecosystems of the UNESCO Global Geopark Mixteca Alta, Oaxaca. Agriculture 2024, 14, 2180. https://doi.org/10.3390/agriculture14122180
Martínez-Núñez MA, Orozco-Ramírez Q. Characterizing Bacterial Communities in Agroecosystems of the UNESCO Global Geopark Mixteca Alta, Oaxaca. Agriculture. 2024; 14(12):2180. https://doi.org/10.3390/agriculture14122180
Chicago/Turabian StyleMartínez-Núñez, Mario Alberto, and Quetzalcoátl Orozco-Ramírez. 2024. "Characterizing Bacterial Communities in Agroecosystems of the UNESCO Global Geopark Mixteca Alta, Oaxaca" Agriculture 14, no. 12: 2180. https://doi.org/10.3390/agriculture14122180
APA StyleMartínez-Núñez, M. A., & Orozco-Ramírez, Q. (2024). Characterizing Bacterial Communities in Agroecosystems of the UNESCO Global Geopark Mixteca Alta, Oaxaca. Agriculture, 14(12), 2180. https://doi.org/10.3390/agriculture14122180





