1. Introduction
Rice (
Oryza sativa), which provides people with the necessary energy, nutrients, and trace elements, is one of the staple foods of the world‘s population [
1]. However, the quality and nutrient content of rice changes with storage time. Rice that is stored for an extended period of time loses some of its edible quality and could pose a great threat to food safety and human health [
2]. Aged rice has a lower commodity price; some unscrupulous elements use it to sell it as fresh rice, seriously disrupting the normal trading of the rice market. One year of storage produced a minor decline in quality and less of a change than fresh rice. However, it is not simple to detect because there is no significant change in surface color [
3]. Storage for about three years leads to noticeable changes in surface properties and effects on the edible flavor, with the rice appearing slightly darker in color than fresh rice [
4]. Five years will cause the rice to taste harsh. Long-term storage also raises the risk of microbial contamination, which can result in the production of poisonous compounds (like
Aflatoxins) [
5]. Currently, the two primary methods used to assess rice quality are chemical analysis and manual tasting. Because manual tasting is subjective, the findings can differ based on the taster’s age, gender, and location [
6]. Chemical procedures evaluate the rice’s pH, fatty acid content, and other characteristics to determine the optimal storage duration. These techniques are dependable, but meeting the testing requirements of a large number of samples is challenging, and the analytical and testing phases take a lot of time. Thus, in order to ensure food safety and control the market for staple grains, a quick, precise, and practical way to identify the rice storage year is required.
In an effort to address this issue, a growing body of research was published on the quantitative determination of several target rice components using [
7,
8] spectroscopic techniques. Near-infrared (NIRS) spectroscopy methods are utilized for the inspection of agricultural product quality. Near-infrared spectroscopy technology is used in the quality inspection of agricultural products by collecting and analyzing the near-infrared spectra of agricultural products; the internal composition and structure information of agricultural products can be obtained, and their quality can be evaluated. Huang, Fuping, and others [
9] coupled near-infrared spectroscopy (NIRS) with three machine learning techniques to rapidly distinguish between rice stored for one year and two years. This resulted in 90% test accuracy and a 5.0% error between model training and testing. When paired with machine learning techniques, NIRS can serve as a valuable chemometric tool. Raman spectroscopy has also been used in the identification of agricultural products for qualitative or quantitative analysis, as well as for the identification of varieties, origins, and vintages of agricultural products [
10,
11]. Min Sha et al. [
12] looked at 72 rice samples (28 indica rice, 25 japonica rice, and19 sticky rice) from the main producing areas using principal component analysis (PCA), window analysis, Hierarchical cluster analysis (HCA), and support vector machine. They cut the time in half and achieved 91.71% prediction accuracy. This approach shows promise as a successful feature extraction technique to enhance the accuracy of rice variety identification. Jian Yang [
13] et al. used three different excitation wavelengths of lasers at 355, 460, and 556 nm to excite leaf fluorescence and measured the fluorescence spectra using the built-in LIF LiDAR system in the laboratory, after which the fluorescence spectra were analyzed in conjunction with principal component analysis (PCA) and support vector machines (SVM), and the overall recognition rate of the six plant species under the three excitation wavelengths of the light source was 80%, 83.3% and 90%, respectively. The final results showed that the 556 nm excitation light source was better than the 355 and 460 nm excitation light sources for the classification of the same plant. In terms of species differentiation, Long Wanjun et al. [
14] successfully differentiated 45 geographical origins, achieving a 100% classification effect. They used excitation-emission matrix (EEM) fluorescence to obtain fluorescence spectra of geographical origin from different parts of the world, along with three chemometrics methods to create a model that could tell the difference.
Brown rice refers to rice that is peeled and has no other grinding treatment. Compared with polished rice, it has higher nutrient and trace element content [
15]. Therefore, this study mainly aims to obtain information on the structure and properties of the main nutrients of brown rice by detecting the fluorescence spectra of brown rice in different years. This is due to the fact that the year differences are largely the result of specific components in the brown rice samples gradually changing, which, in turn, causes the spectra to differ. Therefore, the original fluorescence spectra of brown rice and main nutrients analyzed purely from different years were spectroscopically analyzed, and the identification of rice storage time, as well as the rapid differentiation of the storage year and quality of brown rice, were realized by comparing the classification accuracy between the original fluorescence spectral data, the preprocessed fluorescence spectral data, and the fused fluorescence spectral data classified by combining with the classification modeling method. The application of this technique will significantly impact agricultural output and food quality monitoring, promote rational application and waste reduction, and aid producers, operators, and consumers in accurately assessing the quality of agricultural products.
2. Materials and Methods
2.1. Experimental Materials
The samples were selected from mature japonica rice varieties provided by the Rice Research Institute of the Jilin Academy of Agricultural Sciences (JASA), and the variety was “Dongdao 12” [
16]. The origin of the rice was the research land of Gongzhuling Rice Research Institute (43°30′16.85″ N, 124°49′22.08″ E), Jilin Province, and the harvest years of the samples were 2018, 2019, 2020, 2021, 2022, and 2023, and the rice samples of each year were dried and preserved at low temperatures.
Jin Song brand hulling machine is used to hull japonica rice paddy, model JLGJ-45, motor voltage 220 V, power 120 W. The hulling rate is more than 99%, which meets the requirements of the newly promulgated national standards GB 1350-1999 [
17] and GB/T 17891-1999 [
18]. Japonica rice paddy is brown rice after hulling. Select the brown rice with complete and full grains without damage, and put the selected brown rice into a breathable mesh bag to be stored under low temperature for subsequent experiments. The experimental samples are shown in
Figure 1.
The analytical pure samples of nutrient elements in brown rice discussed in this paper are gluten from wheat, CAS No. 8002-80-0, amylopectin from maize, CAS No. 9037-22-3, Riboflavine, CAS No. 83-88-5, and Lignin, CAS No. 8068-05-11. All materials are from Shanghai, China, Aladdin Biochemical Technology Co.
2.2. Experimental Instruments
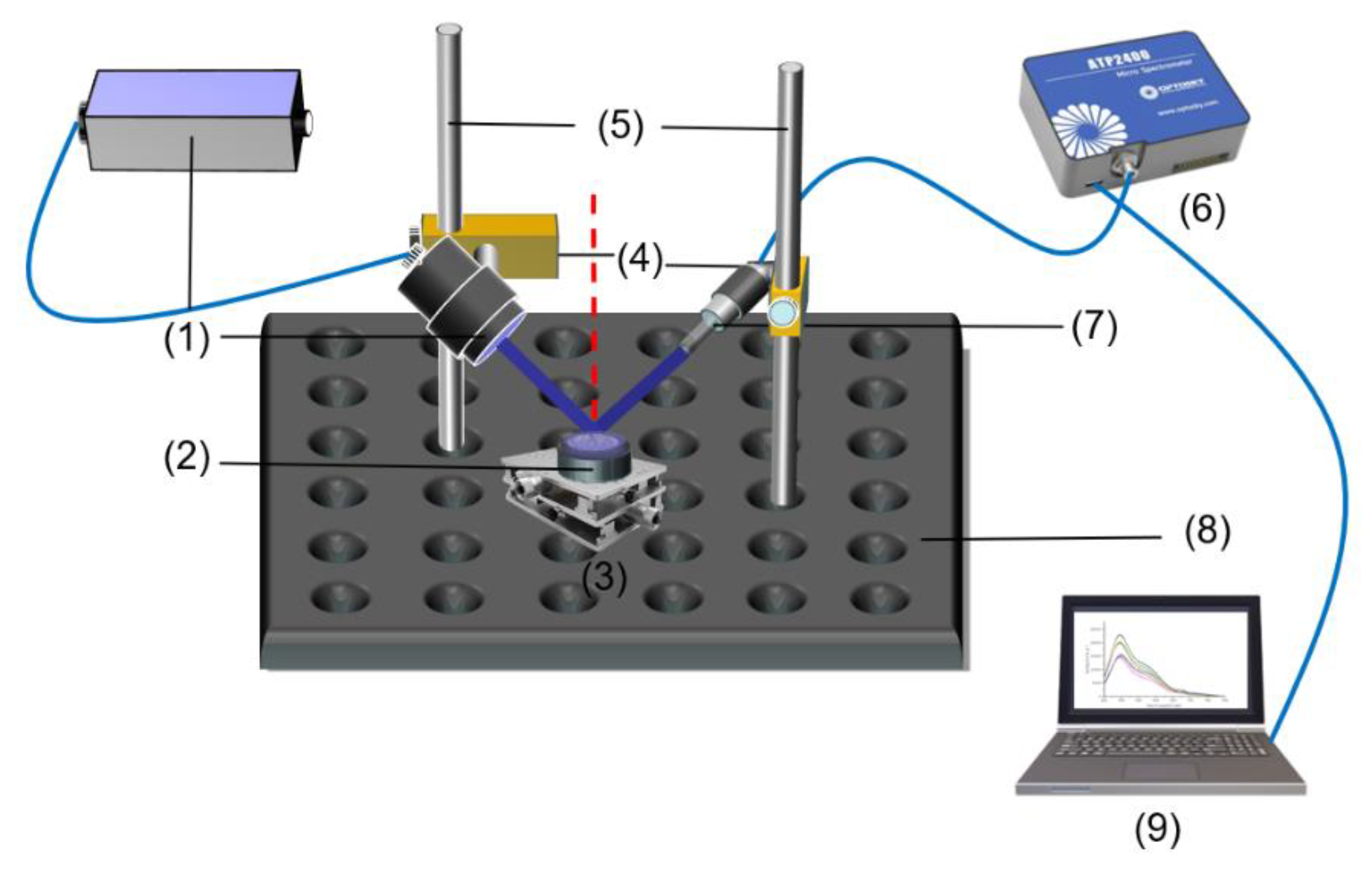
The spectrometer used in this experiment was an ATP2400 spectrometer from Aopu Tiancheng Photoelectric CO., LTD., in Xiamen, China. The spectral detection range of the spectrometer was 350–800 nm; the slit was 50 nm; the resolution was 1.5 nm, and the experimental process adopted an integration time of 20 s. The fluorescence spectral detection light source was a laser light source with an excitation wavelength of 405 nm, which came from Golden Emblem Optoelectronics, branded MTO-LASER. The product power was 50 mW, and the working current was 60 mA. The LED light source with an excitation wavelength of 365 nm was from Zhongshan Yanxi Early Lighting Power Plant, branded as UVGO, with a power of 3 W. The LED light source with an excitation wavelength of 310 nm was from Zhongshan Zigu Lighting Electric Appliance Factory, with beads imported from PW and a power of 3 W. The optical fiber was purchased from Shanghai Wenyi Optoelectronics Co., Ltd. in Shanghai, China. The 2D working platform was a model BRHTXY300 with a load capacity of 70 kg. The optical fiber was purchased from Shanghai Wenyi Optoelectronics Technology Co., Ltd., model number UV600-1.0, with a light transmission range of 200–1100 nm and a core diameter of 600 µm.
Figure 2 shows the fluorescence spectral detection device for subsequent fluorescence spectral detection.
2.3. Data Collection
The total number of rice years to be tested was six years; ten copies of rice and brown rice samples were selected for each year, and about fifty grains of brown rice were selected for each sample. The samples were laid flat in the cuvette, and the detection process was carried out in a dark room. Three different excitation wavelengths (405 nm, 365 nm, and 310 nm) were used for the fluorescence spectroscopy of rice samples from different years. About 100 spectra were collected for each sample, and more than 1000 fluorescence spectra were collected for each storage year sample. After removing the invalid spectra, 1000 spectra with 1586 feature points per spectrum were selected for each year.
2.4. Classification Methods
With the development of big data and intelligent technology, some recently developed stoichiometric variable selection techniques are discussed. Adaptive adjustment algorithms used in the training and testing phases are the foundation of machine learning [
19,
20]. Every machine learning algorithm also has unique properties. This study modeled and analyzed brown rice from several years using the fluorescence spectroscopy approach in conjunction with three classification learners.
2.4.1. SVM
The dimensionality of the input data has no effect on SVM, allowing it to perform well even with highly complex data. Numerous research projects [
21,
22,
23,
24] have effectively used SVM, demonstrating its reliability and effectiveness in the field of food analysis. Overfitting and dimensionality rarely affect SVM, a classification and regression procedure [
25]. SVM uses M-dimensional input data to classify objects in a binary classification problem [
26]. M-dimensional input data reference different wavelengths. To determine the ideal hyperplane for data categorization, we employ a linear SVM. This hyperplane’s equation is as follows:
where
X represents the data matrix; b stands for the coefficient vector, and deviation denotes the bias. We classify it into a different class when the distance to the class boundary (
Y) is less than 0 and larger than 0.del.b. The specific parameters used are as follows: kernel function: linear; kernel scale: automatic; box constraint level: 1; multi-class method: one-to-one; standardized data: yes; and optimizer is not applicable.
2.4.2. KNN
The KNN algorithm generalizes the closest neighbor rule. The KNN algorithm uses the class label of the k samples that have the closest class label to evaluate its generalization offset. In the decision phase, it differs from the nearest neighbor method in that it extends the nearest neighbor to k. This modification allows the KNN algorithm to access and use more data. Compared to previous classification algorithms with various training phases, it skips the learning process [
27]. The relevant parameters are set to the number of neighbors: 10, distance measure: Euclidean, distance weight: equidistant, standardized data: yes, and the optimizer is not applicable.
2.4.3. Wide Neural Networks (WNN)
Wide neural networks are a unique class of neural networks that increase the number or width of neurons in the network layers to improve a model’s performance. In some situations, wide neural networks can outperform deep neural networks, particularly when handling particular kinds of data or tasks.
Wide neural networks are simple to comprehend and use because of their very straightforward structure. It captures more characteristics and patterns, has fewer gradient issues, is more stable than deep networks, and enhances model representation [
28]. In this paper, its parameters are set as fully connected, number of layers: 1, first layer size: 100, activation function: ReLU, iteration limit: 1000, regularization strength (Lambda): 0, standardized data: yes, and optimizer does not apply.
2.5. Model Evaluation Metrics
Statistics and machine learning use a tabular form known as a confusion matrix to evaluate the performance of categorization algorithms. We visually represent the link between the model’s actual and predicted categories and provide a thorough analysis of the classifier’s performance.
Typically, a confusion matrix is a two-dimensional table with the expected categories represented in columns and the actual categories represented in rows. The confusion matrix of a binary classification problem often contains four entries:
True Positive (TP): the actual case is positive, and the model correctly predicts it as such;
True Negative (TN): the actual case is negative, and the model correctly predicts it as negative;
False Positive (FP): the actual case is negative, but the model incorrectly predicts it as positive (False Positive);
False Negative (FN): an actual positive case, but the model incorrectly predicted it as a negative case (underreporting).
The evaluation parameters of this model, including accuracy, specificity, sensitivity, and precision, can be calculated based on True Positive (
TP), False Positive (
FP), True Negative (
TN), and False Negative (
FN) [
29]. The evaluation parameters of the model were calculated as follows:
The confusion matrix for multi-category situations will be larger, but the general idea remains the same. It aids in the evaluation of the model’s performance by visualizing the predictions between every conceivable pair of categories. The confusion matrix plays a significant role in understanding the model’s classification errors and identifying the frequently confused categories. This study compares the model categorization’s accuracy and recall performance.
2.6. Preprocessing Methods
Several spectral preprocessing methods were used to improve the characterization of the spectra, including Multiplicative Scattering Correction (MSC), Standard Normal Variable (SNV), and Savitzky–Golay smoothing (SG). The MSC method is used to calculate the average spectrum or reference spectrum from a set of spectral data. Each individual spectrum is adjusted by a multiplication factor based on the reference spectrum. Each spectrum using the SNV method is adjusted by subtracting the mean of the spectrum so that the data are centered around zero. The average value of the data is converted to 0, and the standard deviation is converted to 1. Savitzky–Golay smoothing (SG) is based on polynomial fitting and is commonly used to eliminate spectral noise. Different algorithms are used to recognize the storage time of rice, including SVM, KNN, WNN.
3. Results
3.1. Fluorescence Spectral Analysis of Brown Rice
With the change in storage time, the structure and content of some chemical components in brown rice will change, resulting in a certain difference in the fluorescence spectrum. One thousand fluorescence spectra datasets of each year were selected and averaged for spectral plotting. The characteristic information of the fluorescence spectra of rice was obtained by raw spectral analysis. Rice contains nutrients such as starch, protein, and riboflavin, which exhibit different vibrational spectral information under excitation at different excitation wavelengths due to their different chemical compositions, contents, and structures. As shown in
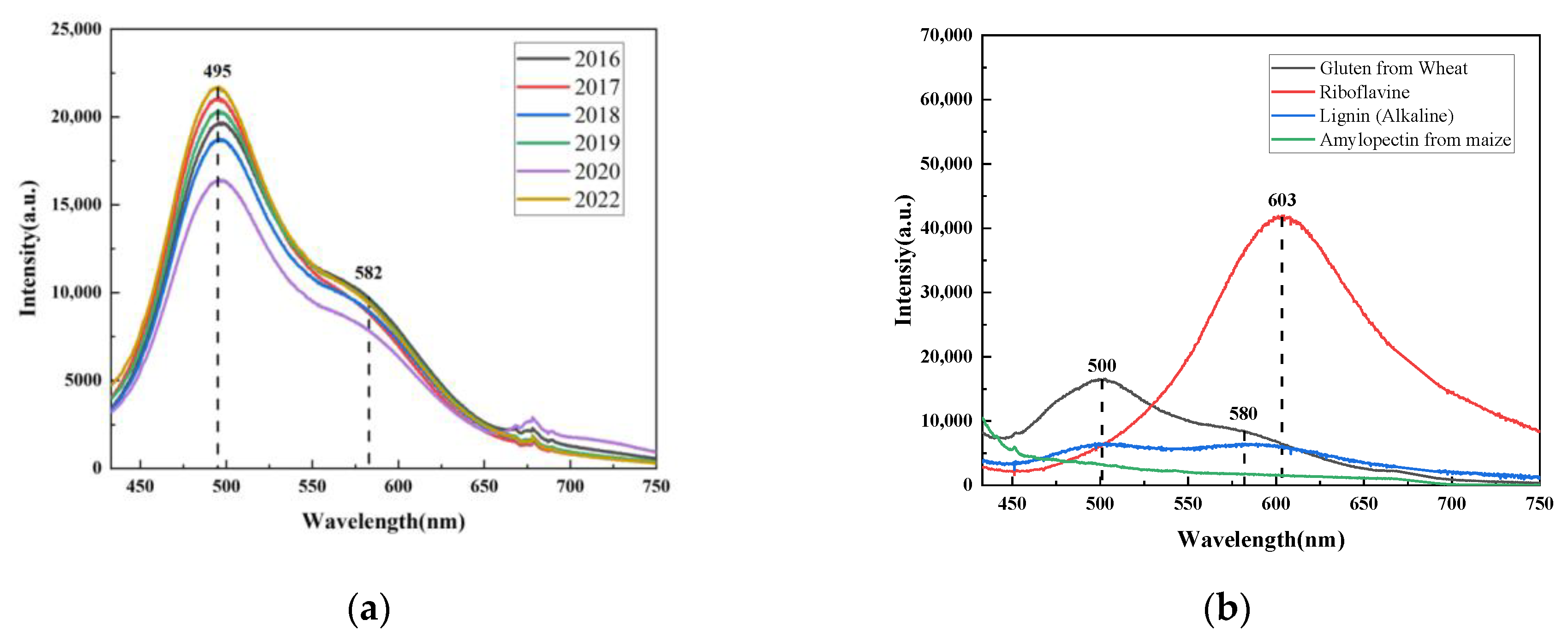
Figure 3, the fluorescence spectrum of rice is mainly concentrated in the range of 450–750 nm.
It can be seen in
Figure 3a that brown rice’s fluorescence spectrum showed two main peaks around 495 nm and 580 nm when excited by a 405 nm light source. The peak at 495 nm was more noticeable. There was also a slight peak shift with the season, which could indicate that the structure and content of some nutrients varied. The fluorescence peak at 495 nm in
Figure 3b might represent a combination of alkaline lignin and glutenin. At 580 nm, the fluorescence peak is a shoulder peak. A combination of riboflavin, basic lignin, and glutenin may be present in this fluorescence peak. There is no noticeable fluorescence peak on the branched starch under the 405 nm excitation light source.
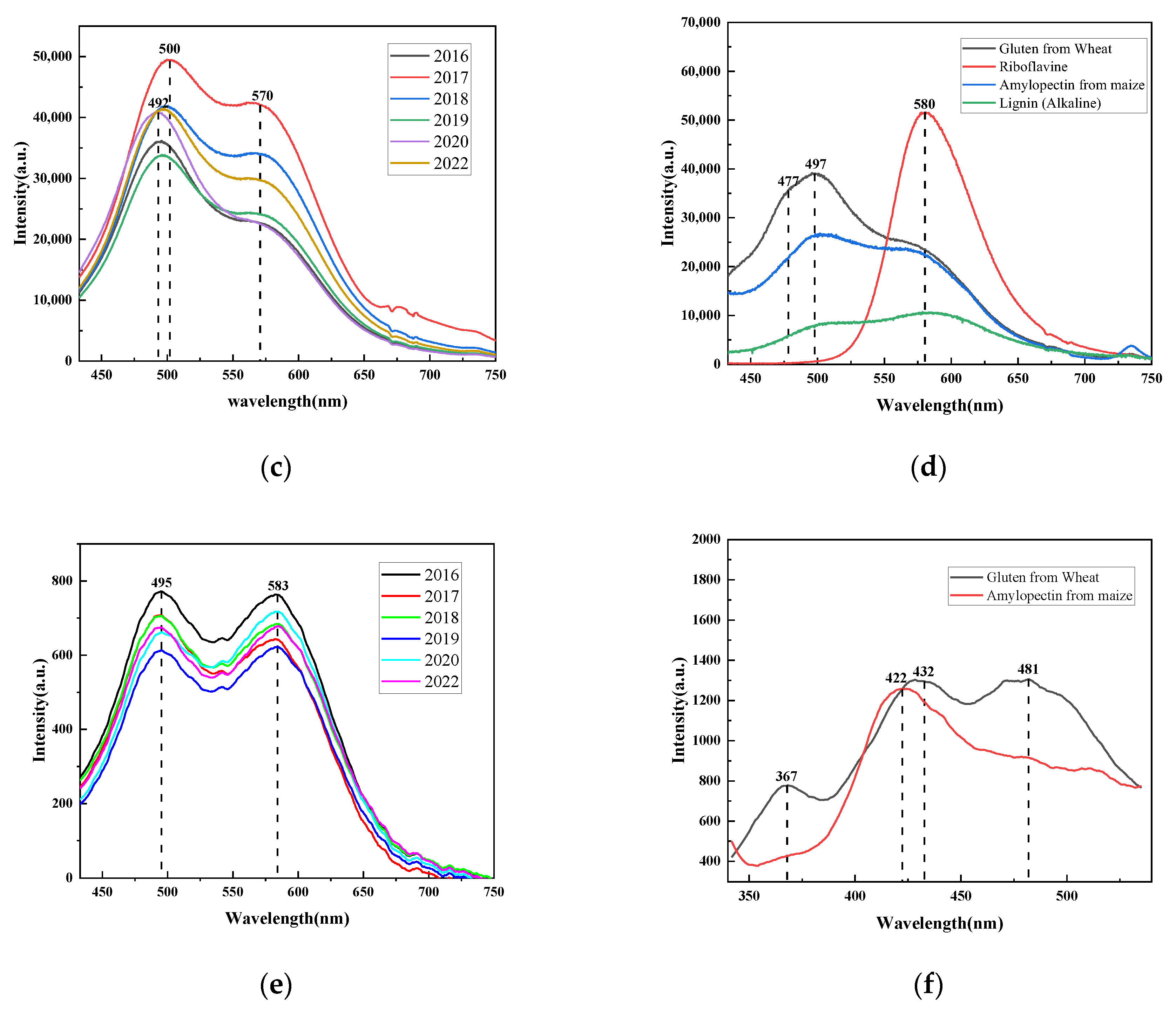
Figure 3c shows the fluorescence spectra of brown rice from different years. It had two clear fluorescence peaks at around 495 nm and 575 nm when it was exposed to a 365 nm excitation light. As the years passed, there was a noticeable shift in the peak location, and the fluorescence peak was more visible at 495 nm.
Figure 3d shows that this fluorescence peak might be a combination of basic lignin and gluten, both of which can have some degree of fluorescence peak in this wavelength range. We found the gluten peak more clearly and strongly in the 365 nm light source than in the 405 nm light source. Additionally, we can simultaneously see the fluorescence peaks of branched-chain starch, which are invisible to the 405 nm light source, and enriched fluorescence spectral features. There is a small fluorescence shoulder peak around 435 nm, which is undetectable by the 405 nm excitation light, thus also providing richer spectral features for machine learning. According to
Figure 3d, it can be seen that it may be the fluorescence peak of gluten. The fluorescence peak at 575 nm is a shoulder peak, which may be a mixture of gluten, branched-chain starch, basic lignin, and riboflavin compared with
Figure 3d.
There are two distinct fluorescence absorption peaks at 495 and 580 nm. The fluorescence spectra exhibit greater differences, and the second peak shoulder becomes more conspicuous compared to the fluorescence spectra of the 365 nm and 405 nm excitation lights. Compared with
Figure 3f, the fluorescence peak around 495 nm may be a mixture of glutenin and branched-chain starch.
3.2. Raw Data Analysis
The rice samples were divided into a training set and a test set in the ratio of 7:3. The training set includes 700 rice spectra for each of the six rice samples, and the test set includes 300 rice spectra for each of the six rice samples. In order to preserve the original spectral features and analyze and compare the recognition accuracy, rice fluorescence spectra were classified and recognized using different classification and recognition methods.
Since differences in spectral intensities may arise due to operational problems in the detection process, the raw spectra are normalized to eliminate inaccuracies in the classification results due to excessive differences in spectral intensities. The normalized spectra are also referred to as raw spectral data (data) in the following. All classification accuracies, precision, and recall in this paper are averaged after the results of five operations.
As shown in
Table 1, the original fluorescence spectra of brown rice were classified and recognized using three classification learners, in which the fluorescence spectral data under a 405 nm excitation light source combined with the wide neural network WNN could reach an average classification accuracy of 94.8%. The fluorescence spectral data under the 365 nm excitation light source combined with the wide neural network WNN reached an average classification accuracy of 93.7%. The average classification accuracy of fluorescence spectral data under 310 nm excitation light source combined with KNN reaches 99.2%. In summary, it can be seen that when the original fluorescence spectral data are classified and recognized, the wide neural network WNN classification recognizer is the best choice under 405 nm and 365 nm light sources, and the classification accuracy can reach more than 90%. The recall and precision rates also reach more than 90%. The KNN classification recognizer under 310 nm excitation light source has the highest classification accuracy of 99.2%, and the recall and precision rates are also over 99%. This means that the raw spectra combined with the classification learner can accurately distinguish the storage time of rice.
3.3. Analysis of Spectral Preprocessing Results
The classification recognition accuracy under two light sources, 405 nm and 365 nm, still needs to be improved, even though the raw fluorescence spectral data + KNN under the 310 nm light source have reached 99.2%. We preprocess the raw spectra using multiplicative scattering correction (MSC), standard normal variance (SNV), and Savitzky–Golay smoothing (SG) to mitigate the effects of scattering and other interferences.
After three preprocessing analyses of the spectrum data under each type of excitation light source and modeling analysis using the same classification modeling approach,
Table 2 displays the validation results. With an average classification accuracy of 95.9% and a recall and precision rate of 95.9%, the SG + WNN classification modeling approach has the highest classification accuracy under 405 nm excitation. With an average recall rate of 94.8%, an average precision rate of 94.7%, and an average classification accuracy of 94.8% at 365 nm excitation, the SG + WNN classification modeling approach has the highest classification accuracy. The SG + KNN classification accuracy can reach 99.3% under 310 nm excitation, and it can also achieve 99.3% recall and precision rates.
A data fusion rice vintages differentiate model was creatively established based on three types of excitation light (405 nm, 365 nm, and 310 nm), which are raw data layer fusion, SG preprocessing data layer fusion, SNV preprocessing data layer fusion, and MSC preprocessing data layer fusion, respectively, in order to further improve the accuracy of the identification model. SVM, KNN, and WNN were used to classify and identify the pooled data.
3.4. Data Fusion Technology
Three light sources (405 nm, 365 nm, and 310 nm) of fluorescence raw spectral data fusion, preprocessing data fusion, together with three chemometric analysis algorithms (SVM, KNN, and WNN) were utilized to identify rice in various years in order to further increase the recognition accuracy.
The data fusion model validation results are displayed in
Table 3. With the exception of the SNV preprocessed data fusion + WNN accuracies, which are decreased, the results demonstrate that all three preprocessed data fusion classification accuracies are improved over the original spectrum data classification accuracies when compared to the preprocessed data classification accuracies. This demonstrates how well the spectral data fusion approach may raise classification accuracy. With 100% classification accuracy, precision, and recall rate, the three-excitation light source spectral raw data fusion + WNN rice vintage identification model is the best among them.
4. Discussion
According to
Figure 3, we can characterize different nutrient compositions using different excitation lights, each of which has its own unique fluorescence spectra. The fluorescence spectra of brown rice from different years showed similarities when exposed to the same excitation light, but noted variations in specific wavelength regions. These variations indicate alterations in the composition and structure of specific chemical constituents in brown rice that has been stored for varying durations. In this paper’s fluorescence spectroscopy of rice brown rice, we employ three distinct excitation lights to enhance the fluorescence spectral information, enable classification modeling, and gather additional compositional data from the tested samples.
By integrating various classification learners, brown rice was categorized. As shown in
Table 1, when combined with raw spectra and SVM, KNN, and WNN classification, the accuracy of raw data + KNN classification under 310 nm excitation light can reach 99.2%, with a recall and precision rate of 99.2% and 98.7%, respectively. The spectra were preprocessed using SG, SNV, and MSC in order to further increase the classification accuracy. The classification results are shown in
Table 2. Using the same classification technique, SG + WNN’s classification accuracy under 310 nm excitation light was 99.3%, with 99.3% recall and precision. Not much has changed when further using spectral fusion, the data layer fusion of spectral data, and then using the same classification algorithm. The results are shown in
Table 3. The raw data fusion + WNN classification accuracy of the three excitation lights can reach 100%, and its recall and precision rate also reach 100%. However, after SG, SNV, and MSC data processing, the model’s accuracy dropped instead. This could be due to the loss of useful information from pure data processing, as well as weak fluorescence spectrum signals of minor changes in the concentration of key components. The validation results of the original spectra, preprocessed spectra, and fused spectra following classification using three similar classification modeling techniques are optimally compared in
Table 4. It clearly shows the difference in classification results under various methods. It is shown that the original fluorescence spectral data layer fusion method has excellent potential for identifying rice storage years. A fast and accurate identification of different years of rice can be achieved.
5. Conclusions and Prospect
5.1. Conclusions
In this study, three excitation lights were used to analyze the fluorescence spectra of brown rice in different storage years to evaluate the changes in the content of key components with storage time. Brown rice was categorized using classification models, achieving 99.2% accuracy with raw spectra + KNN classification at 310 nm excitation, a 99.2% recall rate, and a 98.7% accuracy rate. Preprocessing with SG, SNV, and MSC improved classification accuracy, with SG + WNN yielding 99.3% accuracy, recall, and precision at the same excitation. Spectral fusion enables raw data fusion + WNN to achieve 100% accuracy, precision, and recall. The results showed that rapid and precise fluorescence spectroscopic tests offer fresh possibilities for rice quality detection. These assays, when combined with classification modeling frameworks and techniques, may provide new perspectives for evaluating rice storage vintages.
5.2. Prospects
Spectral detection technology has broad prospects in the identification of agricultural products. Spectral detection technology can quickly and non-destructively analyze the chemical characteristics of agricultural products and improve the accuracy of identification. With the development of data analysis and machine learning, combined with big data, a perfect year identification database can be established to enhance consumers’ trust. At the same time, the miniaturized equipment makes on-site detection feasible and is expected to replace traditional detection methods.
However, this method still has some limitations. Insufficient sample size or insufficient representativeness of samples may affect the generalization ability and practical application effect of the model. More samples are collected as much as possible, especially samples covering different varieties, storage conditions, and years, in order to improve the robustness of the model.