An Attention-Based Spatial-Spectral Joint Network for Maize Hyperspectral Images Disease Detection
Abstract
1. Introduction
- (1)
- We proposed a novel hybrid CNN architecture that contains 3D and 2D convolutional layers for detecting pest-infected maize. The 3D layers perform well in extracting spatial-spectral features from redundant information, simultaneously the 2D layers could reduce the complexity of parameters produced by the 3D layers, thereby improving the performance of the architecture.
- (2)
- We incorporated attention mechanisms into the network, aiming to expand the model’s receptive field and enhance its ability to extract significant features. This has enabled the model to maintain its effectiveness even when the amount of necessary training data is reduced.
- (3)
- We tested our model in various field scenarios, and experimental results have demonstrated the effectiveness of the proposed model in this study.
2. Related Works
2.1. Maize Disease Detection
2.2. Attention Mechanism
3. Materials and Methods
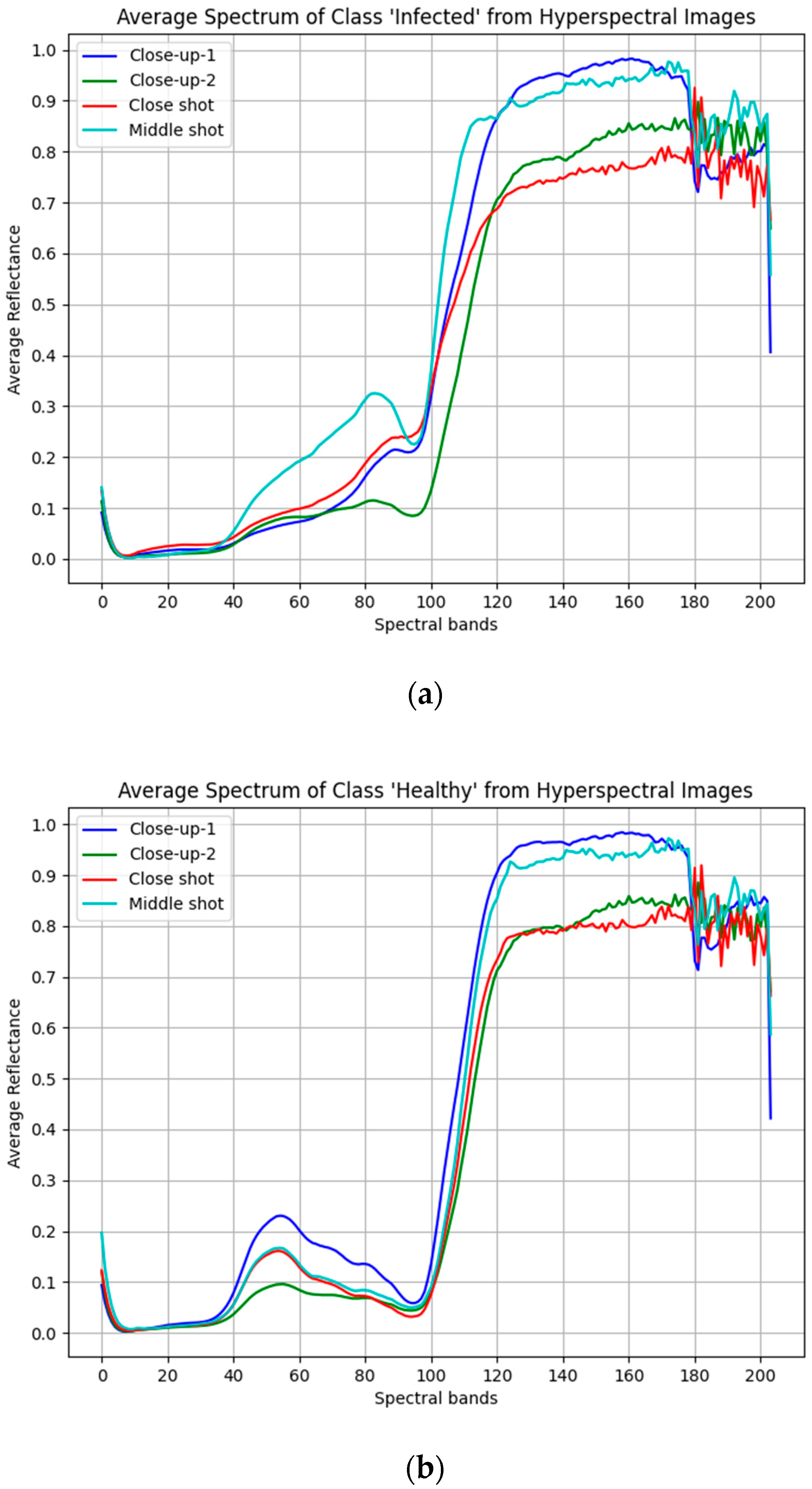
3.1. Study Area and Data Collection
3.2. Hyperspectral Image Preprocessing
3.2.1. Spectral Calibration and Data Labeling
3.2.2. Data Dimensionality Reduction
3.2.3. Creating Data Patches
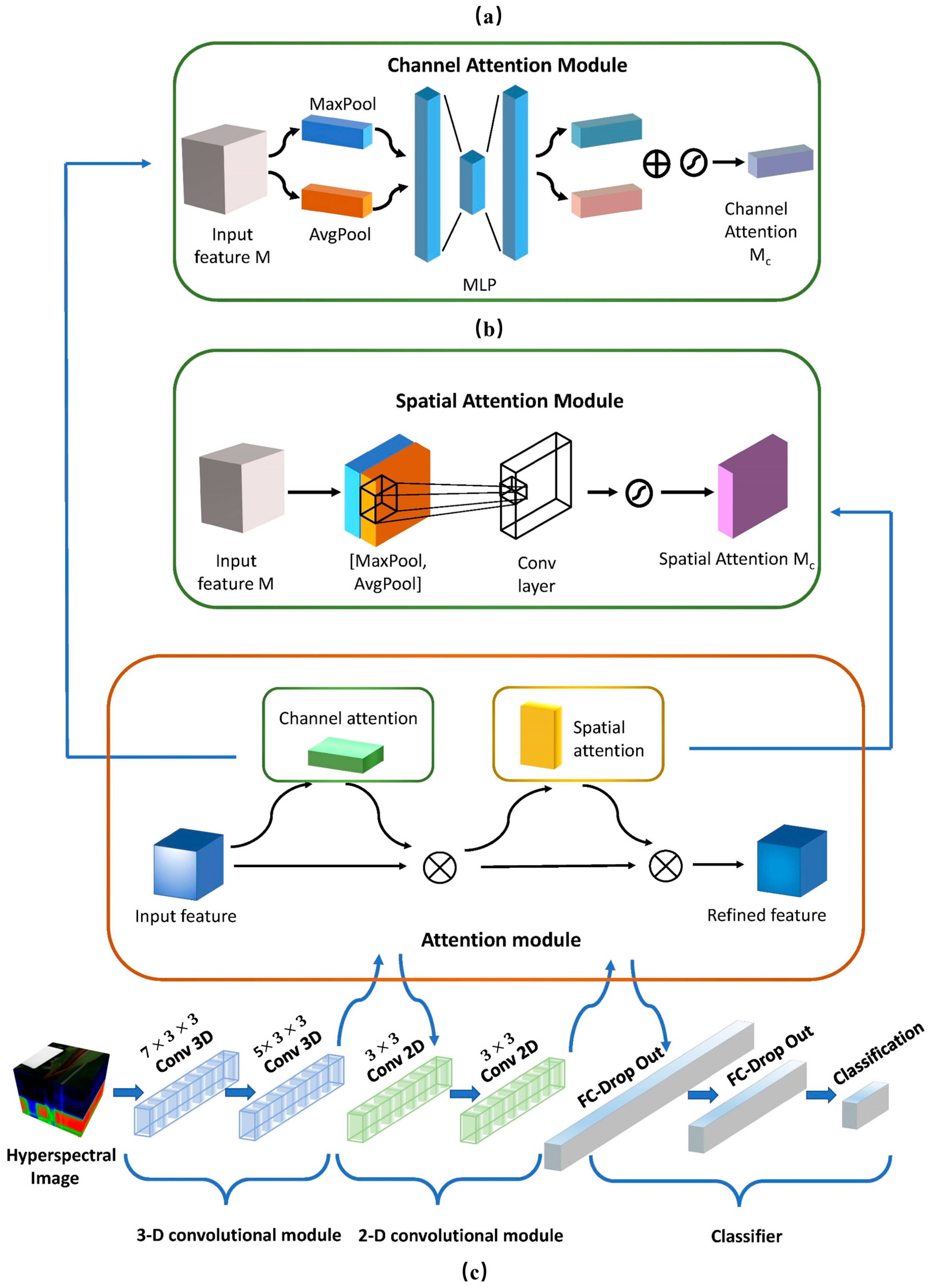
3.3. Proposed Neural Network
4. Experiments and Discussion
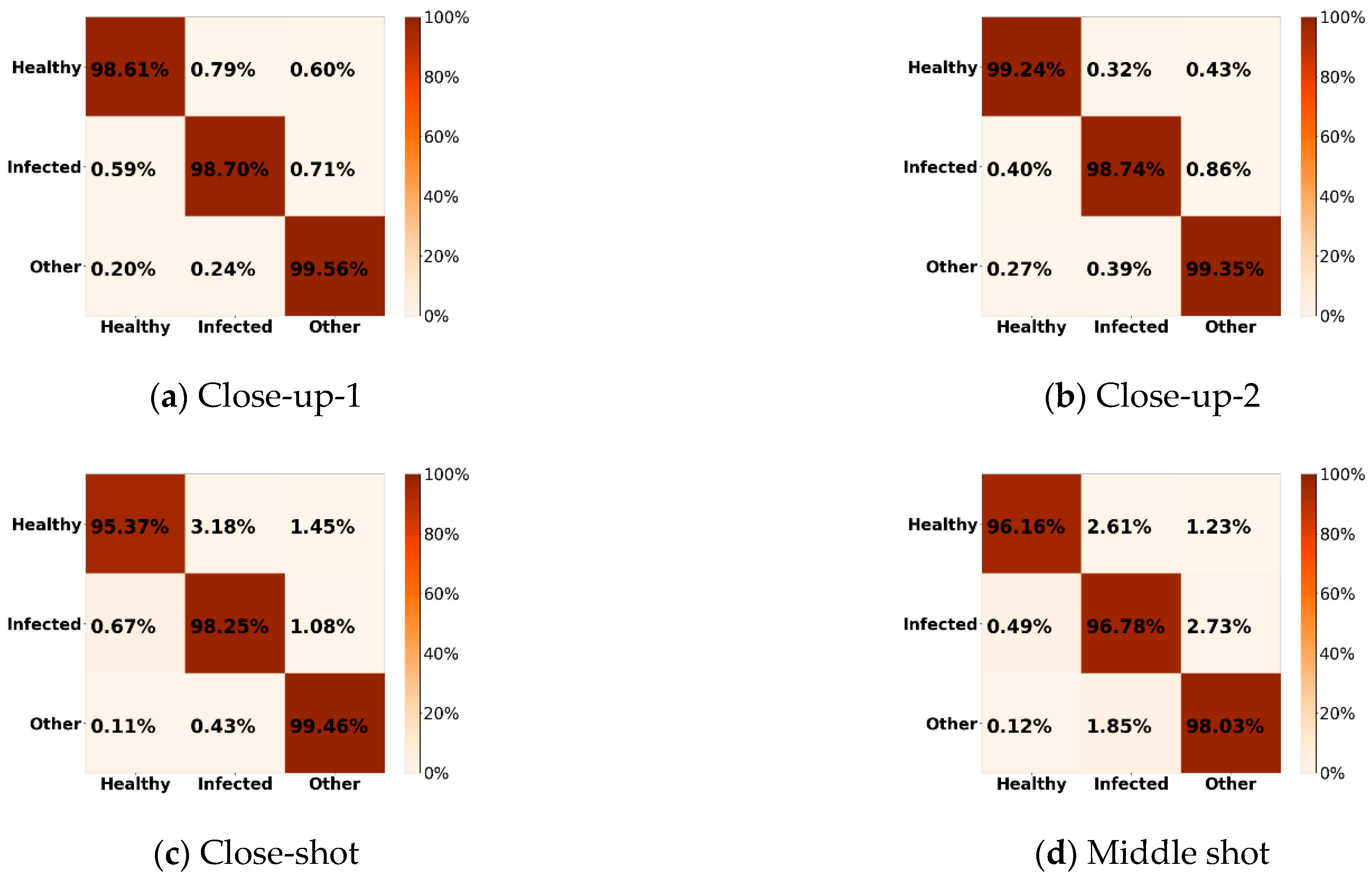
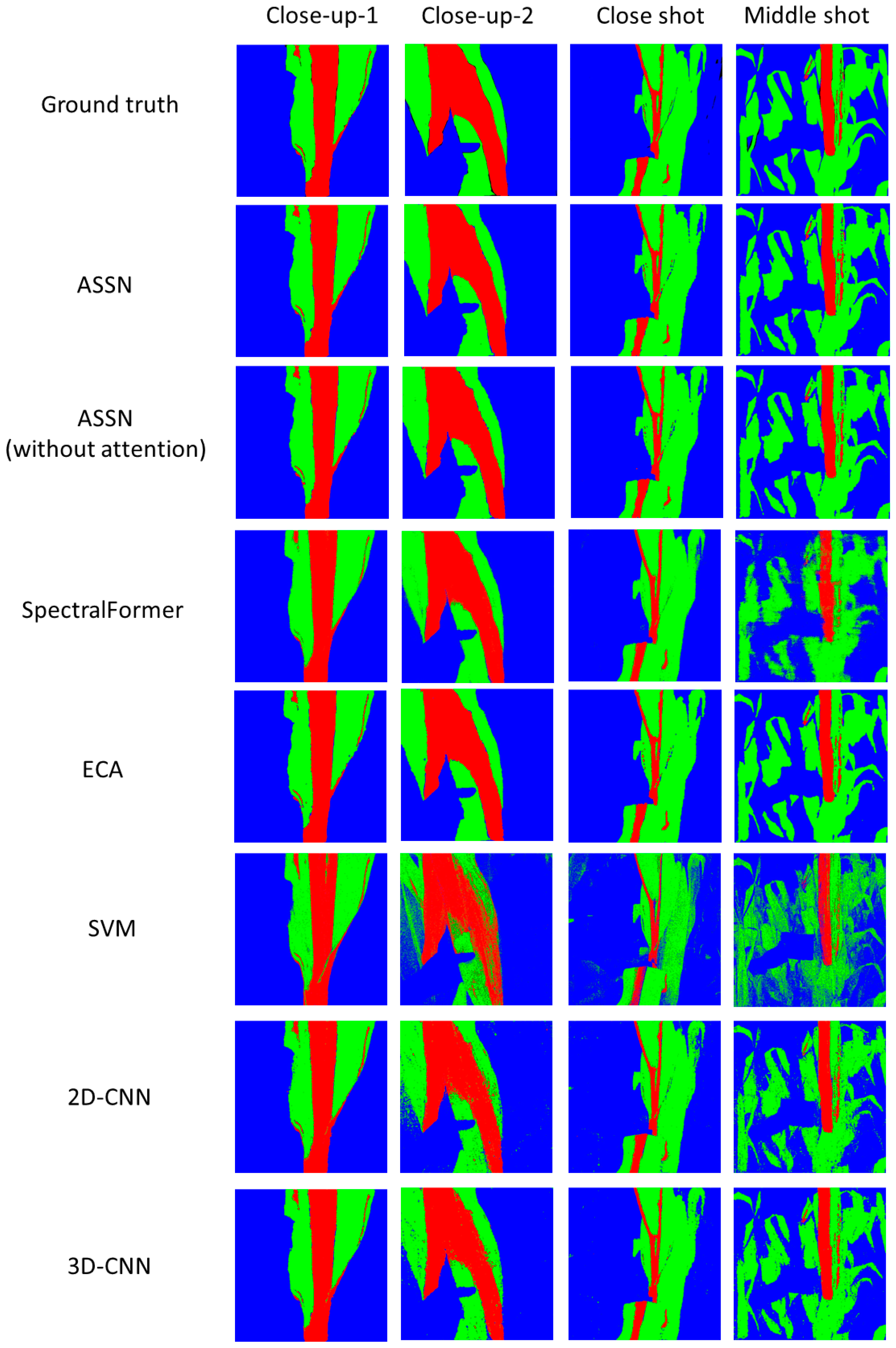
4.1. Experiment with 10% Data as Training Set
4.2. Experiment with 1% Data as Training Set
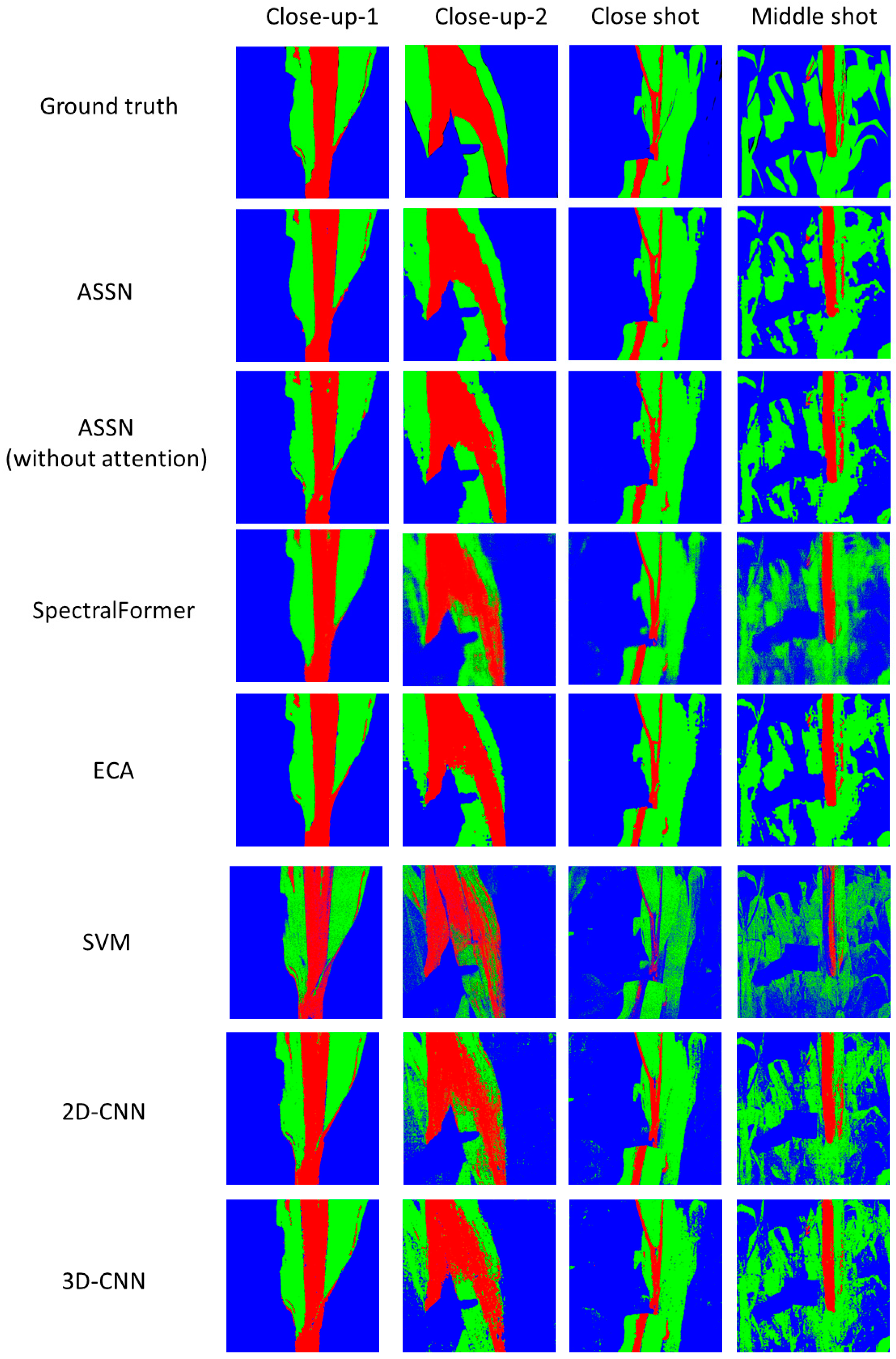
4.3. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Ahila Priyadharshini, R.; Arivazhagan, S.; Arun, M.; Mirnalini, A. Maize leaf disease classification using deep convolutional neural networks. Neural Comput. Appl. 2019, 31, 8887–8895. [Google Scholar] [CrossRef]
- Zhang, X.; Qiao, Y.; Meng, F.; Fan, C.; Zhang, M. Identification of Maize Leaf Diseases Using Improved Deep Convolutional Neural Networks. IEEE Access 2018, 6, 30370–30377. [Google Scholar] [CrossRef]
- Shiferaw, B.; Prasanna, B.M.; Hellin, J.; Bänziger, M. Crops that feed the world 6. Past successes and future challenges to the role played by maize in global food security. Food Secur. 2011, 3, 307–327. [Google Scholar] [CrossRef]
- Deutsch, C.A.; Tewksbury, J.J.; Tigchelaar, M.; Battisti, D.S.; Merrill, S.C.; Huey, R.B.; Naylor, R.L. Increase in crop losses to insect pests in a warming climate. Science 2018, 361, 916–919. [Google Scholar] [CrossRef]
- Zbytek, Z.; Dach, J.; Pawłowski, T.; Smurzyńska, A.; Czekała, W.; Janczak, D. Energy and economic potential of maize straw used for biofuels production. In Proceedings of the MATEC Web of Conferences, Amsterdam, The Netherlands, 23–25 March 2016; p. 04008. [Google Scholar]
- Samarappuli, D.; Berti, M.T. Intercropping forage sorghum with maize is a promising alternative to maize silage for biogas production. J. Clean. Prod. 2018, 194, 515–524. [Google Scholar] [CrossRef]
- Mboya, R.M. An investigation of the extent of infestation of stored maize by insect pests in Rungwe District, Tanzania. Food Secur. 2013, 5, 525–531. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, D.; Zeb, A.; Nanehkaran, Y.A. Identification of rice plant diseases using lightweight attention networks. Expert Syst. Appl. 2021, 169, 114514. [Google Scholar] [CrossRef]
- Kusumo, B.S.; Heryana, A.; Mahendra, O.; Pardede, H.F. Machine learning-based for automatic detection of corn-plant diseases using image processing. In Proceedings of the 2018 International Conference on Computer, Control, Informatics and Its Applications (IC3INA), Tangerang, Indonesia, 1–2 November 2018; pp. 93–97. [Google Scholar]
- Ishengoma, F.S.; Rai, I.A.; Said, R.N. Identification of maize leaves infected by fall armyworms using UAV-based imagery and convolutional neural networks. Comput. Electron. Agric. 2021, 184, 106124. [Google Scholar] [CrossRef]
- Li, K.; Xiong, L.; Zhang, D.; Liang, Z.; Xue, Y. The research of disease spots extraction based on evolutionary algorithm. J. Optim. 2017, 2017, 4093973. [Google Scholar] [CrossRef]
- Huang, M.; Xu, G.; Li, J.; Huang, J. A method for segmenting disease lesions of maize leaves in real time using attention YOLACT++. Agriculture 2021, 11, 1216. [Google Scholar] [CrossRef]
- Madhogaria, S.; Schikora, M.; Koch, W.; Cremers, D. Pixel-based classification method for detecting unhealthy regions in leaf images. In Proceedings of the GI-Jahrestagung, Berlin, Germany, 4–7 October 2011; p. 482. [Google Scholar]
- Thomas, S.; Kuska, M.T.; Bohnenkamp, D.; Brugger, A.; Alisaac, E.; Wahabzada, M.; Behmann, J.; Mahlein, A.-K. Benefits of hyperspectral imaging for plant disease detection and plant protection: A technical perspective. J. Plant Dis. Prot. 2018, 125, 5–20. [Google Scholar] [CrossRef]
- Rayhana, R.; Ma, Z.; Liu, Z.; Xiao, G.; Ruan, Y.; Sangha, J.S. A Review on Plant Disease Detection Using Hyperspectral Imaging. IEEE Trans. AgriFood Electron. 2023, 1, 108–134. [Google Scholar] [CrossRef]
- Yan, T.; Xu, W.; Lin, J.; Duan, L.; Gao, P.; Zhang, C.; Lv, X. Combining multi-dimensional convolutional neural network (CNN) with visualization method for detection of aphis gossypii glover infection in cotton leaves using hyperspectral imaging. Front. Plant Sci. 2021, 12, 604510. [Google Scholar] [CrossRef]
- Li, L.; Zhang, S.; Wang, B. Plant Disease Detection and Classification by Deep Learning—A Review. IEEE Access 2021, 9, 56683–56698. [Google Scholar] [CrossRef]
- Yu, R.; Luo, Y.; Li, H.; Yang, L.; Huang, H.; Yu, L.; Ren, L. Three-dimensional convolutional neural network model for early detection of pine wilt disease using UAV-based hyperspectral images. Remote Sens. 2021, 13, 4065. [Google Scholar] [CrossRef]
- Zhang, X.; Han, L.; Dong, Y.; Shi, Y.; Huang, W.; Han, L.; González-Moreno, P.; Ma, H.; Ye, H.; Sobeih, T. A deep learning-based approach for automated yellow rust disease detection from high-resolution hyperspectral UAV images. Remote Sens. 2019, 11, 1554. [Google Scholar] [CrossRef]
- Ma, H.; Huang, W.; Dong, Y.; Liu, L.; Guo, A. Using UAV-based hyperspectral imagery to detect winter wheat fusarium head blight. Remote Sens. 2021, 13, 3024. [Google Scholar] [CrossRef]
- Shahi, T.B.; Xu, C.-Y.; Neupane, A.; Fresser, D.; O’Connor, D.; Wright, G.; Guo, W. A cooperative scheme for late leaf spot estimation in peanut using UAV multispectral images. PLoS ONE 2023, 18, e0282486. [Google Scholar] [CrossRef]
- Behmann, J.; Acebron, K.; Emin, D.; Bennertz, S.; Matsubara, S.; Thomas, S.; Bohnenkamp, D.; Kuska, M.T.; Jussila, J.; Salo, H. Specim IQ: Evaluation of a new, miniaturized handheld hyperspectral camera and its application for plant phenotyping and disease detection. Sensors 2018, 18, 441. [Google Scholar] [CrossRef]
- Cen, Y.; Huang, Y.; Hu, S.; Zhang, L.; Zhang, J. Early detection of bacterial wilt in tomato with portable hyperspectral spectrometer. Remote Sens. 2022, 14, 2882. [Google Scholar] [CrossRef]
- Nguyen, C.; Sagan, V.; Maimaitiyiming, M.; Maimaitijiang, M.; Bhadra, S.; Kwasniewski, M.T. Early detection of plant viral disease using hyperspectral imaging and deep learning. Sensors 2021, 21, 742. [Google Scholar] [CrossRef]
- Wang, L.; Jin, J.; Song, Z.; Wang, J.; Zhang, L.; Rehman, T.U.; Ma, D.; Carpenter, N.R.; Tuinstra, M.R. LeafSpec: An accurate and portable hyperspectral corn leaf imager. Comput. Electron. Agric. 2020, 169, 105209. [Google Scholar] [CrossRef]
- Pan, T.-t.; Chyngyz, E.; Sun, D.-W.; Paliwal, J.; Pu, H. Pathogenetic process monitoring and early detection of pear black spot disease caused by Alternaria alternata using hyperspectral imaging. Postharvest Biol. Technol. 2019, 154, 96–104. [Google Scholar] [CrossRef]
- Fazari, A.; Pellicer-Valero, O.J.; Gómez-Sanchıs, J.; Bernardi, B.; Cubero, S.; Benalia, S.; Zimbalatti, G.; Blasco, J. Application of deep convolutional neural networks for the detection of anthracnose in olives using VIS/NIR hyperspectral images. Comput. Electron. Agric. 2021, 187, 106252. [Google Scholar] [CrossRef]
- Pérez-Roncal, C.; Arazuri, S.; Lopez-Molina, C.; Jarén, C.; Santesteban, L.G.; López-Maestresalas, A. Exploring the potential of hyperspectral imaging to detect Esca disease complex in asymptomatic grapevine leaves. Comput. Electron. Agric. 2022, 196, 106863. [Google Scholar] [CrossRef]
- Wang, L.; Liu, J.; Shao, J.; Yang, F.; Gao, J. Remote sensing index selection of leaf blight disease in spring maize based on hyperspectral data. Trans. Chin. Soc. Agric. Eng. 2017, 33, 170–177. [Google Scholar]
- Fu, J.; Liu, J.; Zhao, R.; Chen, Z.; Qiao, Y.; Li, D. Maize disease detection based on spectral recovery from RGB images. Front. Plant Sci. 2022, 13, 1056842. [Google Scholar] [CrossRef]
- Xu, J.; Miao, T.; Zhou, Y.; Xiao, Y.; Deng, H.; Song, P.; Song, K. Classification of maize leaf diseases based on hyperspectral imaging technology. J. Opt. Technol. 2020, 87, 212–217. [Google Scholar] [CrossRef]
- Paliwal, J.; Joshi, S. An overview of deep learning models for foliar disease detection in maize crop. J. Artif. Intell. Syst. 2022, 4, 1–21. [Google Scholar] [CrossRef]
- Aravind, K.; Raja, P.; Mukesh, K.; Aniirudh, R.; Ashiwin, R.; Szczepanski, C. Disease classification in maize crop using bag of features and multiclass support vector machine. In Proceedings of the 2018 2nd International Conference on Inventive Systems and control (ICISC), Coimbatore, India, 19–20 January 2018; pp. 1191–1196. [Google Scholar]
- Kilaru, R.; Raju, K.M. Prediction of maize leaf disease detection to improve crop yield using machine learning based models. In Proceedings of the 2021 4th International Conference on Recent Trends in Computer Science and Technology (ICRTCST), Jamshedpur, India, 11–12 February 2022; pp. 212–217. [Google Scholar]
- Masood, M.; Nawaz, M.; Nazir, T.; Javed, A.; Alkanhel, R.; Elmannai, H.; Dhahbi, S.; Bourouis, S. MaizeNet: A deep learning approach for effective recognition of maize plant leaf diseases. IEEE Access 2023, 11, 52862–52876. [Google Scholar] [CrossRef]
- Kundu, N.; Rani, G.; Dhaka, V.S.; Gupta, K.; Nayaka, S.C.; Vocaturo, E.; Zumpano, E. Disease detection, severity prediction, and crop loss estimation in MaizeCrop using deep learning. Artif. Intell. Agric. 2022, 6, 276–291. [Google Scholar] [CrossRef]
- Lv, M.; Zhou, G.; He, M.; Chen, A.; Zhang, W.; Hu, Y. Maize Leaf Disease Identification Based on Feature Enhancement and DMS-Robust Alexnet. IEEE Access 2020, 8, 57952–57966. [Google Scholar] [CrossRef]
- He, J.; Liu, T.; Li, L.; Hu, Y.; Zhou, G. MFaster r-CNN for maize leaf diseases detection based on machine vision. Arab. J. Sci. Eng. 2023, 48, 1437–1449. [Google Scholar] [CrossRef]
- Zhang, Y.; Wa, S.; Liu, Y.; Zhou, X.; Sun, P.; Ma, Q. High-accuracy detection of maize leaf diseases CNN based on multi-pathway activation function module. Remote Sens. 2021, 13, 4218. [Google Scholar] [CrossRef]
- Luo, L.; Chang, Q.; Wang, Q.; Huang, Y. Identification and severity monitoring of maize dwarf mosaic virus infection based on hyperspectral measurements. Remote Sens. 2021, 13, 4560. [Google Scholar] [CrossRef]
- Adam, E.; Deng, H.; Odindi, J.; Abdel-Rahman, E.M.; Mutanga, O. Detecting the Early Stage of Phaeosphaeria Leaf Spot Infestations in Maize Crop Using In Situ Hyperspectral Data and Guided Regularized Random Forest Algorithm. J. Spectrosc. 2017, 2017, 6961387. [Google Scholar] [CrossRef]
- Zhang, N.; Yang, G.; Pan, Y.; Yang, X.; Chen, L.; Zhao, C. A Review of Advanced Technologies and Development for Hyperspectral-Based Plant Disease Detection in the Past Three Decades. Remote Sens. 2020, 12, 3188. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 5998–6008. [Google Scholar]
- Wang, X.; Girshick, R.; Gupta, A.; He, K. Non-local Neural Networks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018. [Google Scholar]
- Li, L.; Xu, M.; Wang, X.; Jiang, L.; Liu, H. Attention Based Glaucoma Detection: A Large-Scale Database and CNN Model. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 10563–10572. [Google Scholar]
- Zhang, H.; Goodfellow, I.; Metaxas, D.; Odena, A. Self-attention generative adversarial networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 10–15 June 2019; pp. 7354–7363. [Google Scholar]
- Hu, H.; Li, Q.; Zhao, Y.; Zhang, Y. Parallel deep learning algorithms with hybrid attention mechanism for image segmentation of lung tumors. IEEE Trans. Ind. Inform. 2020, 17, 2880–2889. [Google Scholar] [CrossRef]
- Chen, B.; Deng, W. Hybrid-attention based decoupled metric learning for zero-shot image retrieval. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 2750–2759. [Google Scholar]
- Zhao, W.; Chen, X.; Chen, J.; Qu, Y. Sample generation with self-attention generative adversarial adaptation network (SaGAAN) for hyperspectral image classification. Remote Sens. 2020, 12, 843. [Google Scholar] [CrossRef]
- Woo, S.; Park, J.; Lee, J.-Y.; Kweon, I.S. Cbam: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Jiang, H.; Zhang, C.; He, Y.; Chen, X.; Liu, F.; Liu, Y. Wavelength Selection for Detection of Slight Bruises on Pears Based on Hyperspectral Imaging. Appl. Sci. 2016, 6, 450. [Google Scholar] [CrossRef]
- Kara, S.; Dirgenali, F. A system to diagnose atherosclerosis via wavelet transforms, principal component analysis and artificial neural networks. Expert Syst. Appl. 2007, 32, 632–640. [Google Scholar] [CrossRef]
- Makantasis, K.; Karantzalos, K.; Doulamis, A.; Doulamis, N. Deep supervised learning for hyperspectral data classification through convolutional neural networks. In Proceedings of the 2015 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Milan, Italy, 26–31 July 2015. [Google Scholar]
- Li, Y.; Zhang, H.; Shen, Q. Spectral–Spatial Classification of Hyperspectral Imagery with 3D Convolutional Neural Network. Remote Sens. 2017, 9, 67. [Google Scholar] [CrossRef]
- Hong, D.; Han, Z.; Yao, J.; Gao, L.; Zhang, B.; Plaza, A.; Chanussot, J. SpectralFormer: Rethinking hyperspectral image classification with transformers. IEEE Trans. Geosci. Remote Sens. 2021, 60, 5518615. [Google Scholar] [CrossRef]
- Wang, Q.; Wu, B.; Zhu, P.; Li, P.; Zuo, W.; Hu, Q. ECA-Net: Efficient channel attention for deep convolutional neural networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 11534–11542. [Google Scholar]




| Different Scenarios | Infected | Healthy | Others | Total |
|---|---|---|---|---|
| Close-up-1 | 40,624 | 54,387 | 166,036 | 261,047 |
| Close-up-2 | 61,999 | 56,034 | 141,815 | 259,848 |
| Close shot | 13,419 | 76,228 | 171,632 | 261,279 |
| Middle shot | 16,520 | 106,573 | 137,589 | 260,682 |
| Methods | OA (%) | AA (%) | Kappa (%) |
|---|---|---|---|
| SVM | 97.90 | 96.64 | 96.01 |
| 2D-CNN | 98.49 | 97.92 | 97.13 |
| 3D-CNN SpectralFormer | 98.89 98.40 | 98.34 97.78 | 97.90 96.97 |
| ECA | 99.18 | 98.85 | 98.45 |
| ASSN (without attention) | 99.19 | 98.92 | 98.47 |
| ASSN | 99.24 | 98.95 | 98.55 |
| Methods | OA (%) | AA (%) | Kappa (%) |
|---|---|---|---|
| SVM | 88.16 | 83.96 | 80.00 |
| 2D-CNN | 95.50 | 94.59 | 92.48 |
| 3D-CNN | 97.74 | 97.22 | 96.22 |
| SpectralFormer | 98.30 | 98.09 | 97.15 |
| ECA | 99.11 | 98.97 | 98.51 |
| ASSN (without attention) | 99.11 | 98.99 | 98.51 |
| ASSN | 99.19 | 99.11 | 98.64 |
| Methods | OA (%) | AA (%) | Kappa (%) |
|---|---|---|---|
| SVM | 92.58 | 84.77 | 84.22 |
| 2D-CNN | 97.64 | 94.98 | 95.06 |
| 3D-CNN | 98.37 | 96.71 | 96.63 |
| SpectralFormer | 97.90 | 95.62 | 95.64 |
| ECA | 98.87 | 97.37 | 97.65 |
| ASSN (without attention) | 98.88 | 97.24 | 97.68 |
| ASSN | 98.90 | 97.69 | 97.70 |
| Methods | OA (%) | AA (%) | Kappa (%) |
|---|---|---|---|
| SVM | 78.81 | 77.62 | 61.15 |
| 2D-CNN | 92.34 | 92.48 | 86.12 |
| 3D-CNN | 95.17 | 95.21 | 91.22 |
| SpectralFormer | 88.98 | 86.25 | 79.88 |
| ECA | 97.30 | 96.56 | 95.09 |
| ASSN (without attention) | 97.27 | 96.65 | 95.05 |
| ASSN | 97.40 | 96.99 | 95.27 |
| Methods | OA (%) | AA (%) | Kappa (%) |
|---|---|---|---|
| SVM | 94.36 | 89.98 | 88.97 |
| 2D-CNN | 97.07 | 95.61 | 94.44 |
| 3D-CNN | 97.98 | 97.04 | 96.16 |
| SpectralFormer | 97.56 | 96.25 | 95.37 |
| ECA | 98.52 | 97.89 | 97.19 |
| ASSN (without attention) | 97.82 | 96.54 | 95.86 |
| ASSN | 98.29 | 97.54 | 96.75 |
| Methods | OA (%) | AA (%) | Kappa (%) |
|---|---|---|---|
| SVM | 80.65 | 72.34 | 65.90 |
| 2D-CNN | 89.35 | 86.12 | 82.08 |
| 3D-CNN | 92.58 | 90.55 | 87.55 |
| SpectralFormer | 91.18 | 88.56 | 85.16 |
| ECA | 97.09 | 96.54 | 95.13 |
| ASSN (without attention) | 97.13 | 96.57 | 95.19 |
| ASSN | 97.48 | 97.13 | 95.78 |
| Methods | OA (%) | AA (%) | Kappa (%) |
|---|---|---|---|
| SVM | 87.00 | 66.26 | 70.70 |
| 2D-CNN | 95.36 | 92.12 | 90.29 |
| 3D-CNN | 96.03 | 92.80 | 91.70 |
| SpectralFormer | 94.56 | 88.30 | 88.50 |
| ECA | 97.58 | 93.60 | 94.94 |
| ASSN (without attention) | 97.52 | 93.99 | 94.83 |
| ASSN | 97.84 | 94.93 | 95.50 |
| Methods | OA (%) | AA (%) | Kappa (%) |
|---|---|---|---|
| SVM | 70.06 | 59.52 | 43.30 |
| 2D-CNN | 81.72 | 83.04 | 66.68 |
| 3D-CNN | 85.50 | 85.27 | 73.47 |
| SpectralFormer | 78.84 | 78.48 | 61.25 |
| ECA | 91.70 | 91.14 | 84.90 |
| ASSN (without attention) | 91.20 | 91.39 | 84.03 |
| ASSN | 92.18 | 91.59 | 85.76 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Liu, F.; Fu, J. An Attention-Based Spatial-Spectral Joint Network for Maize Hyperspectral Images Disease Detection. Agriculture 2024, 14, 1951. https://doi.org/10.3390/agriculture14111951
Liu J, Liu F, Fu J. An Attention-Based Spatial-Spectral Joint Network for Maize Hyperspectral Images Disease Detection. Agriculture. 2024; 14(11):1951. https://doi.org/10.3390/agriculture14111951
Chicago/Turabian StyleLiu, Jindai, Fengshuang Liu, and Jun Fu. 2024. "An Attention-Based Spatial-Spectral Joint Network for Maize Hyperspectral Images Disease Detection" Agriculture 14, no. 11: 1951. https://doi.org/10.3390/agriculture14111951
APA StyleLiu, J., Liu, F., & Fu, J. (2024). An Attention-Based Spatial-Spectral Joint Network for Maize Hyperspectral Images Disease Detection. Agriculture, 14(11), 1951. https://doi.org/10.3390/agriculture14111951






