Abstract
The soil microbiome plays an important role in maintaining soil health, plant productivity, and soil ecosystem services. Current molecular-based studies have shed light on the fact that the soil microbiome has been quantitatively underestimated. In addition to metagenomic studies, metaproteomics and metatranscriptomic studies that target the functional part of the microbiome are becoming more common. These are important for a better understanding of the functional role of the microbiome and for deciphering plant-microbe interactions. Free-living beneficial bacteria that promote plant growth by colonizing plant roots are called plant growth-promoting rhizobacteria (PGPRs). They exert their beneficial effects in different ways, either by facilitating the uptake of nutrients and synthesizing particular compounds for plants or by preventing and protecting plants from diseases. A better understanding of plant-microbe interactions in both natural and agroecosystems will offer us a biotechnological tool for managing soil fertility and obtaining a high-yield food production system.
1. Introduction
The increased demand for agricultural products worldwide has been a consequence of population growth. Increased production leads to topsoil depletion, reduced organic matter content, and compromised soil ecological function. Soil ecological function is maintained by soil microbes, which confer stability to the soil environment and stability during disturbance.
Other issues related to inadequate land use include groundwater contamination, outbreaks of plant diseases, and air pollution. The need for sustainable and healthy food is also reflected in a population that is sensitive to environmental problems. Therefore, eco-friendly strategies for sustainable agriculture are becoming increasingly popular.
According to De Corato, sustainable agroecosystems are highly resilient, adaptive, and diverse [1]. These issues are all related, with diversity conferring more adaptability, and adaptability being a key component of resilience in agroecosystems. The diversity of the soil microbiota, which plays a crucial role in nutrient recycling and soil formation, is therefore a key issue in sustainable agriculture. The main issue that has to be studied is not taxonomical but the functional diversity of soil bacteria. A better understanding of the role of microbes in agroecosystem functioning in the framework of plant growth and soil fertility is key to sustainable agricultural production.
2. Soil Microbiome
2.1. Spatial Distribution of Soil Microbiome
Microorganisms are one of the most abundant living organisms on Earth, constituting approximately 17% of the global biomass [2]. Soil is the most complex habitat that contains a huge abundance of microbial life, which comprises approximately 4–5 × 1030 microbial cells [3]. The soil microbiome is mainly comprised of soil bacteria, archaea, fungi and viruses. Mendes et al. estimates that 108–109 bacteria, 107–108 viruses and 105–106 fungal cells are in one gram of soil [4]. Soil microbial communities provide ecosystem services as nutrient recycling, carbon sequestration, water retention, plant growth promotion and defense [5,6,7]. The focus on soil microbiota research has become noticeable due to its role in the global carbon cycle and climate change, as well as in sustainable agriculture.
The diversity and abundance of microbes are affected by land-use patterns and soil compartments. Agricultural ecosystems are more homogeneous than natural environments due to lower plant diversity and frequent human disturbance. Two soil compartments can be distinguished based on the strength of their relationship with the plant roots. Bulk soil is defined as the part of the soil that is not or is loosely attached to the root, whereas the attached part of the soil is considered rhizosphere soil. Bacterial community composition also differs considerably between bulk and rhizosphere soils, decreasing diversity from the bulk soil to the roots [8]. In tobacco and Arabidopsis plants, the number of microbes present in the rhizosphere is approximately 10- to 100-fold higher than that in bulk soil [9].
The rhizosphere is considered a biological hotspot where plant–microbe, microbe–microbe, and microbe–plant interactions shape microbial community composition. Plant roots secrete organic compounds that support microbial activity [8]. Rhizosphere soil contains 108–1011 cultivable cells in one gram of soil, which corresponds to approximately 104 microbial species [10]. In addition to plant growth-promoting rhizobacteria, soil also provides habitats for plant pathogenic microorganisms and opportunistic human pathogenic bacteria [4].
The spatial heterogeneity of the soil microbiome is determined on the one hand by environmental factors and on the other hand by populational processes [11]. Environmental factors can be both biotic and abiotic, and soil microbial colonization is influenced by plant root exudates in the rhizosphere and environmental parameters. The bulk soil microbial community is an important factor that shapes the rhizosphere microbiome, being the main reservoir from which soil microorganisms are attracted by chemotaxis to root exudates [9]. The same taxa are therefore present in bulk and rhizosphere soils but differ in their relative abundance [12].
Differences in the microbial community between bulk and rhizosphere soils were studied in maize fields by Li et al. [13]. They observed that the rhizosphere soil microbiota was enriched in Proteobacteria, Bacteroidetes, and Actinobacteria, accounting for 73–80% of total reads versus 46–56% in bulk soil. A decreased abundance was observed for Acidobacteria, Gemmatimonadetes, Chloroflexi, Firmicutes, and Nitrospira in the rhizosphere relative to that in the bulk soil [13]. Fan et al. studied the microbial community of wheat fields with an emphasis on three soil compartments, namely tightly and loosely bound soil and bulk soil [8]. They found that Proteobacteria, Actinobacteria, and Acidobacteria dominated all soils, whereas the abundance of Actinobacteria, Bacteroidetes, Alphaproteobacteria, and Verrucomicrobia was higher and the abundance of Gammaproteobacteria, Chloroflexi, and Deltaproteobacteria was lower in tightly bound soil than in the other soils. The greater relative abundance of Actinobacteria in tightly bound soil was explained by their antibiotic-producing potential, whereas the presence of Alphaproteobacteria was attributed to their fast growth characteristics.
Owing to the complexity of the soil system, in addition to the variation in space (microhabitats), stratification also shapes the microbiome. Rchiad et al. observed that, in a semiarid agroecosystem, although the diversity index of the soil microbiota does not decrease with increasing soil depth, there were differences in the microbial profiles [14]. They detected 43 microbial phyla, of which the distribution of 12 was affected by soil depth. At the phylum/family level, a transition in the microbial community from the top (0–15 cm) to the deepest level (30–60 cm) was observed. The abundances of Verrucomicrobia and Bacteroidetes decreased with soil depth. The abundance of soil functional genes was also affected by depth, and most functional categories were observed either in the top layer or in the deepest level [14].
2.2. Soil Microbiome Taxonomic Diversity: Structure and Function
2.2.1. Study Methods
Microbiological studies, as well as those for the soil microbiome, have traditionally been based on isolation and culturing methods, using different specific growth media to maximize culturable isolates. Owing to the development of molecular genetic methods, we found that culture-based methods missed most of the microbial diversity. Data on the ratio of unculturable bacterial fraction that remains to be explored reflects different opinions: Mendes et al. and Dubey et al. reported it as approximately 90–95%, whereas Yadav et al. reported that it might be from 100 to 1000 fold larger [4,5,15]. Each opinion reflects that the soil and rhizosphere microbiomes are highly underestimated. Most of the bacterial phyla have unculturable representatives whereas Archaea were reported only using culture-independent methods [5,15].
In the late 20th century, culture-independent molecular techniques allowed us to study bacterial genomic material. The first developed methods were based on targeted gene sequencing (16S ribosomal RNA), whereas today we are able to decipher the meta-genome (whole genome sequencing) and even the metatransciptome (mRNA) of a whole environment. Metaproteomics and metatranscriptome analyses target the functional part of the microbiome based on protein extraction and analysis. The last two methods are important for understanding not only the taxonomic profile but also the functional role of the microbiome [16].
2.2.2. Taxonomic Diversity
The rhizosphere microbiome includes different groups of bacteria. They belong to the following phyla: Acidobacteria, Actinobacteria, Ascomycota, Bacteroidetes, Basidiomycota, Deinococcus-Thermus, Euryarchaeota, Firmicutes, and Proteobacteria [15]. The bacterial community diversity and composition differ between different crop plants, with higher differences reported between different plant types, such as legumes, forbs, and grasses. A review of the microbiome distribution of different crops was conducted for six important crop plant species: maize, wheat, rape, cotton, rice, and soybean (Figure 1). Bacterial strains belonging to phyla Proteobacteria, Actinobacteria, Acidobacteria, Gemmatimonadetes and Chloroflexi were observed in all six crop plants [15,17].
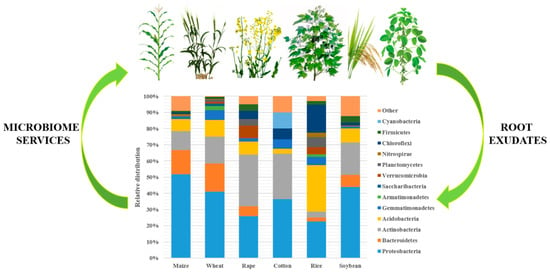
Figure 1.
Relative abundance of different bacterial communities in different crops (based on Li et al. [13], Mahoney et al. [18], Rathore et al. [19], Ullah et al. [20], Edwards et al. [21], and Sugiyama et al. [22]).
Maize rhizosphere was preferentially colonized by Proteobacteria (class Betaproteobacteria and Gammaproteobacteria), Bacteroidetes (class Sphingobacteria) and Actinobacteria, with Massilia, Burkholderia, Ralstonia, Dyella, Chitinophaga and Sphingobium as dominant genera accounting for from 63% to 77% of total bacteria [13]. The core rhizosphere of wheat comprises Proteobacteria, Bacteroidetes, Actinobacteria, Acidobacteria, Gemmatimonadetes, Armatimonadetes, Planctomycetes, Saccharibacteria, Verrucomicrobia, Firmicutes, Nitrospirae, and Chloroflexi. Sphingobacteriaceae (Bacteroidetes) and Gemmatimonadaceae were the most abundant families [18].
The highest similarity was found between the maize and wheat rhizomicrobiomes. The community structures of rape, cotton, rice, and soybeans were more specific. Approximately 99% of the rape microbiota was represented by the phyla Proteobacteria, Bacteroidetes, Actinobacteria, Acidobacteria, Verrucomicrobia, and Chloroflexi. Bacteria belonging to families such as Sphingomonadaceae, Sphingobacteriaceae, Micrococcaceae and Chthoniobacteracaea were among the most abundant groups [19]. The cotton microbiome, studied by Ullah et al., consists mainly of Proteobacteria, Actinobacteria, Gemmatimonadetes, Chloroflexi, Cyanobacteria, and Acidobacteria at the phylum level [20]. Bacterial strains belonging to the Burkholderia, Streptomyces, Rhizobium, Massilia, Pseudonocardia, Shinella, Sphingobium, Agaricicola, Mesorhizobium, Streptomyces, Altererythrobacter, Opitutus, and Sphingopyxis genera were the most abundant in the cotton rhizosphere. The rice microbiome was dominated by Proteobacteria, Chloroflexi, and Acidobacteria phyla, whereas in the case of the soybean microbiome, bacterial strains belonging to Proteobacteria were the dominant phyla, followed by Actinobacteria and Chloroflexi [22].
2.2.3. Factors Affecting Diversity
The main environmental factors affecting the community structure of the rhizosphere microbiome are pH, salinity, moisture, temperature, and nutrient content (C-N content and other nutrients); however, vegetation also has an important role [15,23]. Environmental factors create unique ecological niches that frame specific microbiomes. Numerous studies have shown that environmental stress can alter microbial diversity and shift ecosystem function. Niche-based research has shown that the important processes of microbial communities are influenced by microbial fitness and habitat conditions [5].
The spatial heterogeneity of the soil microbiome was studied in the case of switchgrass vegetation, where the bacterial community structure proved to be patchy, and the abundance of the dominant phyla (Verrucomicrobia) changed 2.5 fold in a 10 cm3 grid [5]. In a study conducted in an alpine environment, environmental parameters explained 41% of the total variation in soil communities, with pH and temperature being the strongest influencing factors [23]. Praeg et al. compared the bulk and rhizosphere soils and found that Ranunculus glacialis roots explained approximately one-third of the variation [23]. Differences among different plant type (legume, grass, or forb) microbiomes were observed by Hannula et al. [24]. They attributed a variation of 4% in bacteria and 11% in fungi to the difference between plant groups and 30% to species-level variations.
Mahoney et al. studied the microbiome of wheat cultivars [18]. They found higher differences in Planctomycetes, Acidobacteria, Actinobacteria, and lower differences in Chloroflexi, Fibrobacteres, and Verrucomicrobia in the nine studied wheat cultivars. The bacterial community structures at lower taxonomic ranks (family, genus, and OTU levels) were influenced by different growth stages in Zea mays. At early growth stages bacterial species belonging to Massilia, Flavobacterium, Arenimonas and Ohtaekwangia genera were more abundant, whereas bacterial species belonging to Burkholderia, Ralstonia, Dyella, Chitinophaga, Sphingobium, Bradyrhizobium and Variovorax genera were dominant at later stages [13].
Anthropogenic effects that cause biotic and abiotic stresses and changing climatic conditions modify soil microbial and plant diversity [10,25]. Ullah et al. identified drought-tolerant bacteria in drought-treated cotton plants [20]. Thermophilic bacteria belonging to the phyla Chloroflexi and Gemmatimonadetes were found to be dominant in drought-affected environments [20]. The use of fertilizers, farming, and tillage methods in agroecosystems can induce changes in soil microbiomes. In the case of the switchgrass rhizosphere microbiome, a fertilizer-induced decrease in the relative abundance of the most abundant phylum (Verrucomicrobia) was observed [5].
The use of different soil amendments also reduced bulk soil microbiome diversity and influenced the rhizosphere community in Zea mays. Changes in community structure were caused by a lower abundance of Actinobacteria and Firmicutes and a higher abundance of Proteobacteria, Bacteroidetes, Verrucomicrobia, and Acidobacteria [12]. Fertilizer type and dose also contributed to changes in rhizobial community structure in Z. mays, and differences in the abundance of microbial groups were attributed to their different nutrient contents [26]. Tillage practices modified the root- and shoot-associated bacterial communities in rape plants, whereas farming practices affected the microbial community structure of rice [21].
2.2.4. Ecological Function
In microbial communities, it is important to study species composition and existing ecological functional groups. Different microbes have different roles in the community structure, which might support soil health and plant productivity. Microbial communities are complex dynamic networks with various interactions between microbes, such as resource competition, metabolic dependencies (cross-feeding), spatial organization notably production of biofilms, signaling, horizontal gene transfer, coevolution, and viral looting [27]. Usually, a higher diversity of microbes increases the quantity of metabolites, secondary metabolites, phytohormones, biocontrol substances, and other beneficial substances, thereby contributing to soil structure and fertility, root system architecture and nutrient foraging, plant nutrition and hormonal balance, plant stress tolerance, agricultural productivity, and resilience to climate, land use, and agronomic practices [10]. Wei et al. studied the role of the initial microbiome in the occurrence and phytopathogeny of Rhizoctonia solanum [28]. They observed higher abundances of Alphaproteobacteria, Firmicutes, and Cyanobacteria in soils associated with healthy tomato plants, whereas diseased plants were found in soils rich in Acidobacteria, Actinobacteria, and Verrucomicrobia [28].
In a study realized in a semiarid region regarding the functional metagenome analysis of different soil layers, twenty-eight functional categories of subsystem level 1 were detected. Differences between the soil layers were observed in 20 functional categories. In the top layer (0–15 cm), which is rich in organic matter, functional genes related to DNA and RNA metabolism, and nutrient metabolism (N, S, carbohydrate) were more abundant than in the deeper layers. The 15–30 cm layer proved to be overlapping, showing a transition in the microbial community and function. In the deeper soil layer (30–60 cm), where the organic matter content is lower and mineral content is higher, functional genes related to P metabolism, nucleosides and nucleotides, amino acids and derivatives were observed in abundance. Understanding the changes in the microbial community and function between soil layers could be important for the usefulness of intercropping and further development of sustainable agricultural practices [14].
The microbial diversity of soil is important for maintaining the function of soil ecosystem services. Microbes with plant growth promotion and protection potentials are important for sustainable agricultural practices. A better understanding of plant–microbe interactions in both natural and agroecosystems will offer us a biotechnological tool for managing soil fertility and obtaining a high-yield food production system [29].
2.3. Beneficial Plant–Microbe Interactions
2.3.1. Biostimulant Microbes
Microorganisms and plants live in nature in association, but the microorganisms can be free-living, attached, or enter symbiosis with the host plants. There are different interactions such as commensalism, mutualism or parasitism. During evolution, plants interacted with a broad range of plant growth-promoting rhizobacteria (PGPR). Owing to recent advances in metagenomics, massive genome-sequencing strategies, and new identification techniques, bacterial rhizobiome mapping is rapidly progressing. These findings revealed novel bacterial species and their mechanisms involved in biocontrol and plant growth promotion [30].
Microorganisms living in soil can indirectly promote plant growth (Figure 2), especially by fixation of atmospheric N2, production of siderophores, plant growth hormones (cytokinins, auxins, and gibberellins), volatile compounds, and solubilization of nutrients and minerals (phosphorus, potassium, zinc, etc.). Additionally, some PGPR species play a role in plant stress resistance. They can help the host plant overcome drought and saline stresses and can increase the plant’s capacity to sequester heavy metals or other toxic elements [13,26,31].
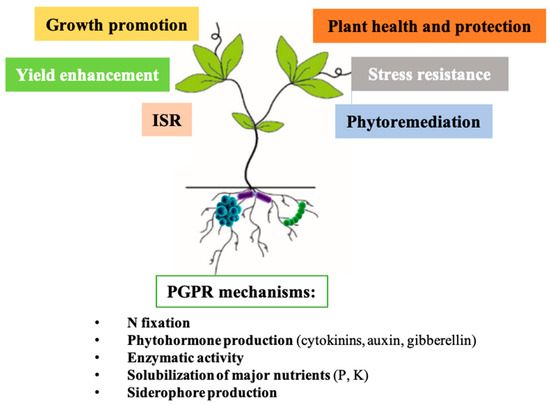
Figure 2.
Plant growth-promoting mechanisms by soil microbes. PGPRs play an important role in plant growth promotion, stress resistance, plant health and protection, phytoremediation, and ISR.
PGPRS are agriculturally important bacteria because they contribute to plant growth by increasing germination rate, biomass, chlorophyll, and nitrogen content. These can improve the leaf area, shoot and root length, hydraulic activity, and crop yield. They can also confer plant tolerance to biotic (pathogen bacteria, fungi, yeast, pests, insects etc.) and abiotic stresses (drought, flood, heavy metals, salinity, temperature etc.) [15]. Fungi that promote plant growth belong to the following genera: Aspergillus, Penicillium, Rhizoctonia, Talaromyces, and Trichoderma, among which the most promising plant growth-promoting fungi are Trichoderma sp. (T. viride and T. harzianum) and Penicillium chrysogenum [32].
Biological Nitrogen Fixation
Nitrogen from soil is available to plants in inorganic forms (nitrate, ammonium) and organic ones (urea, amino acids, small peptides). Organic forms can be used only in special environments [33]. The Earth’s atmosphere is rich in elemental dinitrogen (N2) but this is biologically unavailable for plants. Fowler et al. calculated the amount of biologically available nitrogen convertible from the total amount of dinitrogen gas 4 × 109 Tg N to be 473 Tg N [34,35].
Atmospheric nitrogen is reduced to ammonia (NH3) gas, and this reduction can be made artificially by the Haber–Bosch procedure or occurs naturally as thunderstorms and biological nitrogen fixation (BNF), which accounts for 66% of the total fixed N2 [36]. After photosynthesis, BNF is the second most important process on Earth, due to its significant role in agroecosystem sustainability [34,35].
The nitrogenase activity of nitrogen-fixing microorganisms is responsible for BNF, whereas atmospheric N2 is reduced to ammonia. Bacteria with BNF capacity are categorized into three groups: free-living, associated, and symbiotic bacteria. Free-living N2 fixing bacteria belong to different genera such as Gluconacetobacter, Azospirillum, and Azotobacter spp., but their contribution is negligible compared with the total BNF. The highest proportion of BNF is due to symbiotic nitrogen-fixing bacteria called rhizobia. In addition, other PGPRS with nitrogenase complexes, called diazotrophs, fix N2 in non-leguminous plants (including cereals). The nitrogenase complex is a two-component metalloenzyme. The first component is dinitrogenase reductase, a homodimeric iron protein that acts as a reductase. They have a high reducing capacity and are responsible for providing electrons. The second is dinitrogenase, a heterotetrameric Mo-Fe protein that utilizes the electrons provided to reduce N2 to NH3 [35].
Rhizobium bacteria are symbiotic bacteria linked to leguminous plants. The non-symbiotic or free-living type N2-fixing bacteria are cyanobacteria (blue-green algae, Anabaena, Nostoc) and other species belonging to different genera, such as Azotobacter, Beijerinckia, and Clostridium. Associative nitrogen-fixing bacteria, such as Azospirillum sp. (maize, rice, and wheat), Klebsiella sp., Azotobacter sp. and Alcaligenes sp. live around roots in the rhizosphere and they have the role to stream the fixed nitrogen to the plant. Endophytic nitrogen-fixing microorganisms are linked to cereals, grasses, sugarcane, and Azoarcus sp. (Kallar grass, sorghum, rice), Herbaspirillum sp. (rice, sorghum, sugarcane), Gluconacetobacter diazotrophicus (sorghum, sugarcane) and Burkholderia sp. (rice) [33,36].
Phytohormone Production
Phytohormones are organic compounds that influence physiological processes in plants even at very low concentrations. The ability of soil microbiota to produce phytohormones is a potential source of phytohormones. Plant growth hormones, such as auxins (indole-3-acetic acid), gibberellins, abscisic acids, ethylene, and cytokinins, are biosynthesized by microorganisms. For synthesis, a considerable amount of metabolic energy and nutrients are required [35,37]. Ethylene, jasmonic acid, and salicylic acid are stress-related regulators of plant immunity that are involved in the creation of a central signaling backbone that coordinates defense responses against phytopathogens [38].
Phytohormones play a significant role in plant growth during cell division and enlargement, seed germination, root formation, and stem elongation. Phytohormones produced by bacteria are released into the plant body and have a positive effect on plant growth and development. Several reports have shown that bacteria can produce 60 times more plant growth regulator substances than plants can. Bacteria belonging to the Rhizobium, Sinorhizobium, Bradyrhizobium, Azospirillum, Bacillus, Paenibacillus, and Pseudomonas genera have the capacity to produce phytohormones (auxins, ABA, cytokinins, and gibberellins), thereby improving plant growth and productivity under natural conditions [27,35].
All plant-associated microbes produce auxins, but not all PGP microbes have the ability to produce gibberellin. This capacity is related to root-associated microbes. Auxins, mostly indole-3-acetic acid (IAA), are synthesized by 80% of rhizosphere bacteria. Tryptophan is the main precursor for IAA biosynthesis in bacteria. Bacteria that promote IAA synthesis can take up tryptophan present in root exudates. There are five different tryptophan-dependent and tryptophan-independent pathways, as in Azospirillum brasilense, in which the biosynthetic intermediates are unknown [38].
Among bacterial phytohormones, IAA, which promotes root elongation and lateral root development, is the most studied. These plant hormones are highly effective under stressful conditions. Some plants are unable to produce enough auxins to cope with stress effects; therefore, bacterial auxins can alleviate stress conditions in plants [39]. Bacterial strains with IAA production capacity include Pseudomonas fluorescens, Pseudomonas syringae, Agrobacterium tumefaciens, Pantoea agglomerans, Azospirillum brasilense, Bacillus cereus, Bacillus amyloliquefaciens, Rhizobium sp., and Bradyrhizobium sp. [38]. According to Shahzad et al., inoculation with Micrococcus yunnanensis RWL-2, Pantoea dispersa RWL-3, Micrococcus luteus RWL-3, and Staphylococcus epidermidis RWL-7 in rice plants resulted in a significant increase in dry biomass, shoot and root length, chlorophyll, and protein content [40,41]. IAA-producing fungi include Aspergillus, Mortierella, Talaromyces, Fusarium, Penicillium, and Trichoderma spp. Penicillium janczewskii, which produces IAA, and inhibits Rhizoctonia solani, a phytopathogen that causes stem rot [32].
Abscisic acid (ABA) is a stress-related hormone that plays a key role in photoperiodic induction of flowering, contributing to plant growth and development. It is involved in plant responses to different environmental stresses such as cold, salinity, and desiccation [39]. Several plant-associated bacteria can produce ABA, thereby increasing phytohormone levels in plants. ABA is an important factor in modulating plant defenses, so plant mutants with altered ABA biosynthesis or that are ABA-insensitive are more resistant to pathogens than wild-type plants [38]. ABA-producing endophytic bacteria include Achromobacter xylosoxidans, Brevibacterium halotolerans, Bacillus licheniformis, Bacillus pumilus, and Lysinibacillus fusiformis [42].
The gibberellin (GA) phytohormone plays a major role in leaf expansion and stem elongation in plants. When GA is applied exogenously, it can promote parthenocarpy in fruits, bolting plants, breaking tuber dormancy, and increasing fruit size and the number of buds. Several soil microorganisms have been reported to produce gibberellin, with positive or negative effects on nodulation and plant growth. These microorganisms can induce nodule organogenesis and inhibit nodulation during the infection stage [39]. The first described bacterium with GA production ability was Rhizobium phaseoli, which produces GA1 and GA4. Biologically active GA1 and GA3 are produced by Azospirillum lipoferum, Acetobacter diazotrophicus, and Herbaspirillum seropedicae [42]. GA-producing fungi such as Cladosporium sp. were reported by Adedayo and Babalola, and were found to improve tomato plant growth due to GA production as well as playing an important role in pea plant colonization [32].
Cytokinins (CK) play a role in many stages of plant development by stimulating plant cell division, root development, and root hair formation, activating dormant buds, and inducing the germination of plant seeds. These plant hormones affect apical dominance and regulate nodulation and nitrogen fixation. Some pathogenic and beneficial microbes produce cytokinin phytohormones. It has been reported that PGPRs from Pseudomonas and Bacillus genera produce cytokinin, especially zeatin [15,35]. Pseudomonas fluorescens and Rhizobium spp. are cytokinin-producing bacteria [43].
Ethylene (ET) is a plant stress hormone. Under stress conditions, higher amounts of ethylene can negatively affect plant growth. Ethylene production can be modulated by bacterial strains possessing 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase activity. PGPRS acts as a sink for the ET precursor, 1-aminocyclopropane-1-carboxylic acid (ACC), consequently reducing ET levels in roots and simultaneously increasing root length and plant growth. ACC exuded by roots and seeds can be taken up by rhizobacteria, and due to activity of ACC deaminase (ACCd) is split into ammonia and α-ketobutyrate. ACCd activity has been detected in many PGPRs, and the genes encoding ACCd are widespread within these bacteria. PGPRs with ACCd activity enhance plant tolerance to biotic and abiotic stresses by reducing ET production in colonized roots and promoting plant growth [27,35]. It has been reported that Actinobacteria alleviate stress in plants by reducing ethylene levels in roots by secreting ACCd enzymes [44]. Patil et al. revealed that bacterial strain Bradyrhizobium elkanii is an inhibitor in ethylene synthesis and alleviated the negative effect of stress-induced ethylene production on nodulation [45].
The phytohormones produced and secreted by microbes are used as growth regulators in crop production. These provide benefits to the host plant by facilitating root system expansion, enhancing the absorption of water and nutrients, and improving plant survival [15].
Enzymatic Activity
1-aminocyclopropane-1-carboxylate deaminase (ACC-deaminase, ACCd) enzyme plays an important role in plant hormone and ethylene regulation. ACC deaminase is found in numerous microbial species, including Gram-negative and Gram-positive bacteria and fungi [46,47].
Solubilization of Major Nutrients
Soil microorganisms play a major role in nutrient cycling. The crop residues incorporated in the soil represent the carbon, energy, and nutrient sources of microorganisms. Rhizobia can solubilize nutrients such as phosphorus, iron, potassium, and zinc, thus increasing their availability to plants [46].
Among the macronutrients, phosphorus (P) is essential for plant growth and development. P is abundantly available in the soil in organic (phytin) and inorganic forms (P minerals such as apatite and secondary P minerals such as Al, Fe, and Ca phosphates). P is a major growth-limiting nutrient despite being present in soil in abundancy in insoluble form. The soluble level of P in soil determines the P accessibility to plants [38,44].
PGP microbes are a biological rescue system because they are capable of solubilizing insoluble inorganic P in soil, increasing its availability to plants in the form of orthophosphate. The major mechanism of P solubilization involves the production of organic acids. As a result, insoluble P is transformed into its soluble form. The produced organic acids decrease the soil pH or chelate mineral ions, resulting in phosphate solubilization [38]. The organic acids most frequently produced by Gram-negative PGPRs are gluconic acid and 2-ketogluconic acid. Other organic acids produced by phosphate-solubilizing PGPRs are lactic, isobutyric, isovaleric, acetic, glycolic, oxalic, succinic, and malonic acids [38]. PGP microbes with phosphate solubilization capacity belong to different genera such as Arthrobacter, Azotobacter, Bacillus, Beijerinckia, Burkholderia, Citrobacter, Enterobacter, Erwinia, Flavobacterium, Halolamina, Microbacterium, Pantoea, Paenibacillus, Pseudomonas, Rhizobium, Rhodococcus, Serratia and Thiobacillus [15,38,40].
PGPRs can release from soil organic and inorganic phosphorus by producing several enzymes, such as phytases, phosphatases, phosphanatases and lyases [38]. In this process, the microbes also produce organic acids (gluconate, acetate, ketogluconate, oxalate, lactate, tartarate, succinate, citrate, and glycolate), but this depends on the type of carbon source used as substrate. The highest amount of solubilized P was observed in in vitro conditions when glucose, sucrose or galactose was used as the sole carbon source [15].
Potassium (K) is considered the third major macronutrient for plant growth and crop yields. More than 90% of the potassium that exists in soil is in the form of insoluble rocks and silicate minerals. The soluble form of K is present in soil in low concentration. One of the major constraints in crop production is the potassium deficiency due to imbalanced fertilizer application. Lack of potassium causes poorly developed roots, small seeds, lower crop yields and slow growth. An alternative indigenous source of potassium for plants is the potassium solubilized by soil microorganisms. The potassium-solubilizing microbes (KSMs) have an important role in transforming insoluble K into available form for uptake by plants through production and secretion of different organic acids. Many bacteria such as Acidithiobacillus ferrooxidans, Bacillus mucilaginosus, Bacillus edaphicus, Bacillus circulans, Burkholderia sp., Paenibacillus sp. and Pseudomonas sp. possess the capacity to solubilize K into available form. These bacteria contribute to the enhancement of K+ availability in agricultural soils [48].
KSMs excrete low molecular organic acids (citric, tartaric, oxalic, succininc, syringic, malic, coumaric, and ferulic acids), thus modifying potassium uptake. The organic acids dissolve K+ from the minerals, thereby lowering the pH and forming metal–organic complexes with Si4+, thus bringing the potassium into solution. Through weathering, biofilms, polymers, capsular polysaccharides, and low molecular weight ligands produced by soil microbiota can mobilize K [33]. The application of KSMs as biofertilizers in agriculture can reduce the use of agrochemicals and support environmental sustainability [48].
Adedayo and Babalola reported that Ceriporia lacerata, Aspergillus awamori, and Penicillium digitatum were able to solubilize phosphate owing to the activity of organic acids and phytase [32]. Trichoderma sp. improved the absorption of several minerals and nutrients (K, N, and P), whereas T. viride fungi increased the nitrogen content of the soil [32].
Solubilization of Iron with Siderophore Production
The bioavailability of iron as an essential micronutrient is limited by the soil. Siderophores produced by soil bacteria play a key role in plant iron nutrition. These compounds are low-molecular-weight chelators with a high affinity for iron (III), the most common form of iron in nature. The iron solubilization mechanism relies on the formation of a stable siderophore-Fe+3 complex that can be absorbed by plants [38,48]. To date, more than 500 siderophores have been identified. Plant growth-promoting Pseudomonas fluorescens produces pyoverdine among other siderophores. Microorganisms can produce other siderophoric compounds such as catechol, hydroxamate, carboxylate, and phenolate, which contribute to plant protection against pathogens. Bacterial strains with iron chelation properties belong to Azotobacter, Bacillus, Enterobacter, Arthrobacter, Nocardia, and Streptomyces [46].
The direct beneficial effect of siderophores is the improvement in the iron nutritional status of the plant, contributing to plant growth promotion. It has been hypothesized that bacterial siderophores chelate Fe+3 from the soil, making it accessible to phytosiderophores, but the exact mechanism is unknown. It has been shown that plants can incorporate Fe+3-pyoverdine complexes resulting in an increase in the iron content of plant tissues. The indirect beneficial role of bacterial siderophores in plant growth promotion is their capacity to reduce the availability of iron to phytopathogens [38].
Siderophore synthesis is influenced by several environmental factors such as pH, the level of iron, the presence of other trace elements, and an adequate supply of carbon, phosphorus, and nitrogen sources [48]. Siderophores transport iron into bacterial cells by means of a system involving ferric-specific ligands (siderophores) and their corresponding membrane receptors, which are chelating agents in bacteria. Studies have shown that siderophores suppress pathogens, and this mechanism consists of the fact that siderophores sequester a limited supply of iron (III) in the rhizosphere, thereby limiting the availability of iron for pathogens and suppressing their growth [46].
2.3.2. Biocontrol Activity of Microbes
The overuse of chemicals in agriculture, such as pesticides, insecticides, herbicides, and fertilizers, negatively affects consumer health, biomagnification of chemicals, and economic loss [49,50]. Biological control organisms are defined as living organisms other than disease-resistant host plants that suppress the activity of plant pathogens in the soil environment [51].
Microbial control agents can exert their plant-protecting characteristics based on their mode of interaction with pathogens through direct and indirect mechanisms (Figure 3). Indirect mechanisms are those that do not require interaction with pathogens, such as microbial-induced systemic resistance (ISR), nutrients, and space competition. The production of antibiotics and lytic enzymes to inhibit pathogen taxa is considered a direct mechanism [52]. Bacterial isolates with biocontrol potential produce a broad spectrum of bioactive metabolites such as antibiotics, siderophores, volatiles, and growth-promoting substances. These bacteria can compete aggressively with other microorganisms and can easily adapt to environmental stress [51]. Trichoderma spp. produce biological antifungal volatile organic compounds (VOCs), such as 6-pentyl-2H-pyran-2-one (6-PP) [32].
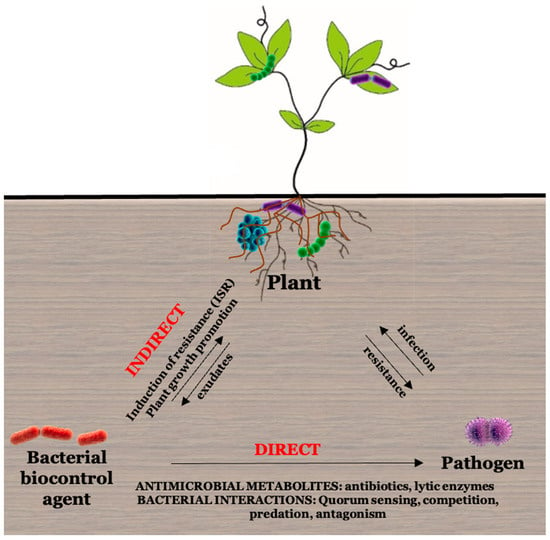
Figure 3.
Indirect and direct mechanisms of biocontrol agents. Indirect methods for biocontrol agents include induced systemic resistance and plant growth-promoting mechanisms. Biocontrol agents directly protect plants through antimicrobial metabolites and bacterial interactions. Arrows indicate the direction of the relationship.
Accordingly, the mechanisms of biocontrol by PGPRS include competition, antibiosis, hydrolytic enzymes, siderophore production, and induced systemic resistance. Species belonging to different genera such as Bacillus, Serratia, Enterobacter, Pseudomonas, Burkholderia, Herbaspirillum, Staphylococcus, Ochrobactrum, Streptomyces, and Stenotrophomonas are known as antagonistic species against plant pathogens and are also used as biopesticides for the control of bacterial and fungal diseases, whereas species belonging to genera Bacillus, Pseudomonas, and Serratia have been reported to induce plant systemic resistance [46,53,54,55,56,57].
Biocontrol agents produce and release many metabolites (lipopeptides, bacteriocins, antibiotics, biosurfactants, cell wall-degrading enzymes, and microbial volatile compounds), which reduce pathogen growth and metabolic activity. In contrast, BCAs affect pathogen virulence and compete for nutrients and space by producing antimicrobial compounds [58].
Antibiotics
The production of antibiotics by various microorganisms is a biological control mechanism. A microbially synthetized antibiotic can inhibit the metabolic activities of pathogenic agents. The mechanisms involved in pathogen inhibition include inhibition of cell wall and protein synthesis, and deformation of cellular membranes. The biochemical nature of these metabolites largely determines their modes of action. Antibiotics also play a pivotal role in the induced systemic resistance (ISR) mechanism in plants [59,60].
Several microorganisms belonging to different genera have been characterized for their antibiotic production capacity, including Agrobacterium sp., Pseudomonas sp., Bacillus sp., Pantoea sp., Serratia sp., Stenotrophomonas sp., Streptomyces sp., and Trichoderma sp. [46,52]. Ram et al. listed various bacterial species and strains that are authorized for producing agroinoculants such as Agrobacterium tumefaciens, Agrobacterium radiobacter strain 84, Bacillus subtilis, Streptomyces lydicus, Burkholderia cepacia, Erwinia amylovora, Alcaligenes sp., Serratia marcescens GPS 5, and Streptomyces griseoviridis [60].
Antibiotics synthetized by biocontrol strains include: polyketides, heterocyclic nitrogenous compounds, phenazine, phenylpyrroles, cyclic lipopeptides, aminopolyols, and volatile antibiotics [51,59,61]. Several reports have been published on the production of antibiotics by various biocontrol bacterial strains to target pathogens [60,61,62,63]. Ongena and Jacques investigated the production of iturin, fengycin, and surfactin by Bacillus sp. [64]. Raaijmakers and Mazzola investigated the production of antibiotic metabolites (2, 4-diacetylphloroglucinol, pyrrolnitrin, phenazine) in Pseudomonas sp. [65].
Adedayo and Babalola reported that T. harzianum had an inhibitory effect on the growth of phytopathogen fungi: F. oxysporum, Phythium sp. and R. solani [32]. Gliocladium virens produces an antibiotic (gliovirin) that inhibits the growth of Pythium ultimum and F. oxysporum [32].
Interference of Quorum Sensing with Virulence
Quorum sensing is a cell–cell communication process in bacteria that involves extracellular signaling molecules (autoinducers) and serves to share information about cell density. Because many processes are advantageous only in this group, when the bacterial population increases, gene expression is altered. Processes such as biofilm formation, antibiotic production, and virulence factor secretion are controlled by QS. Quorum sensing (QS) is important for expressing bacterial pathogenicity in plants. QS is required for the colonization and expression of virulence factors in plant pathogenic strains, such as Pseudomonas syringae, Pantoea stewartii, Erwinia chrysanthemi, and Burkholderia glumae [66,67,68,69].
QS inhibitory activity in the raw extracts of plants or microbes has been used in the food packaging industry and in the treatment of multi-drug-resistant bacteria [70]. Interference with pathogen QS, termed quorum quenching, can be used as a sustainable biocontrol strategy [70,71,72]. QS interference can be accomplished in three ways, namely preventing the signal molecule biosynthesis, degrading the signal molecules, and blocking the signal corpuscle receptors [72]. BCAs can also interfere with QS against pathogens due to their enzymatic degradation capacity, by inhibiting the production of molecules (lactonases, pectinases, chitinases) which initiate infections [54,73].
Acinetobacter sp., Bacillus sp., Burkholderia sp., Lysinibacillus sp., Serratia sp., Pseudomonas sp., Rhodococcus sp., Strepsporangium sp., Streptomyces sp., Enterobacter sp., and Myroides sp. were reported as AHL-degrading bacteria [71,74,75]. AHL-degrading Lysinibacillus has been shown to reduce potato soft rot disease caused by Pectobacterium carotovorum subsp. carotovorum [76,77].
Lytic Enzymes
A potential mechanism of action against pathogens is the production of lytic enzymes. PGPRs inhibit the growth of fungal pathogens (Fusarium oxysporum, Sclerotinia sclerotiorum, and Botrytis cinerea) and other soil-borne pathogens through the excretion of enzymes such as chitinases, hydrolases, proteases, and glucanases [46,48].
The production and secretion of cell wall lytic enzymes (CWLEs) are the most important effects of BCAs in restraining the growth of soil-borne pathogens [78]. The mode of action of CWLEs is to disrupt the structural integrity of the target pathogen cell wall. β-1,4-N-acetyl glucoseamine and chitin are the principal components of fungal pathogenic cell walls. Numerous CWLEs, such as β-1,3-glucanase and chitinase, have been shown to play a role in most BCAs. CWLEs are generally involved in the lysis of the cell wall and thus neutralize the inhibitory action of pathogens [79]. PGPRS inhibit widely known phytopathogens such as Phytophthora capsici and Rhizoctonia solani [80,81].
Induced Systemic Resistance (ISR)
Disease control by various beneficial bacterial strains involves the induction of systemic resistance. Different microbial metabolites and biocontrol agents can generate an immune response in the host plant, resulting in systemic disease resistance [46]. Plants recognize microbial compounds (flagellin, lipopolysaccharide, exopolysaccharide, and chitin oligosaccharides) produced by beneficial microorganisms. Different bacterial species are effective against fungal, bacterial, and viral infections through ISR, including Bacillus amyloliquefaciens, B. atrophaeus, B. cereus, B. megaterium, B. subtilis, Paenibacillus alvei, Pseudomonas fluorescens, Pseudomonas aeruginosa, and Streptomyces pactum [82].
In ISR, some plant hormones such as salicylic acid, jasmonic acid, and ethylene are involved. The host plant defense response against phytopathogens occurs through the jasmonic acid and ethylene signaling pathways. Manifestation of ISR is related to cell wall integrity and biochemical and physiological changes. Several bacterial strains with ISR-eliciting traits have been reported to be effective against a broad spectrum of fungal pathogens. These bacteria include Pseudomonas aeruginosa, Pseudomonas fluorescens, Bacillus amyloliquefaciens, B. subtilis, B. pasteurii, B. cereus, B. pumilus, B. mycoides, and B. sphaericus [73,80].
Treatment of cotton plants with P. chrysogenum induced ISR against Verticillium dahlia. P. simplicissimum, T. harzianum, T. viride and Acremonium sclerotigenum were effective in inducing ISR in crop plants such as cucumber and tomato [32].
2.4. Plant-Beneficial Function Encoding Gene Clusters and Mobile Genetic Elements
Horizontal gene transfer (HGT) is an event in which bacteria incorporate advantageous genes into their genomes. Horizontally transmitted genes are crucial for bacterial adaptation to changing environments or to plant-microbe interactions. They are often grouped into genomic islands and gene clusters [83]. Up to 20% of the bacterial genome is disseminated during horizontal gene transfer events [84]. The rhizosphere is considered to be one of the hotspots of microbial gene transfer whereas the microbiome is a rich reservoir of genetic functionality [85,86].
Many function genes in soil bacteria are encoded by plasmids that act as mobile genetic elements (MGEs). Plasmids are most commonly considered antibiotic resistance gene carriers; however, they are also important carriers of heavy metal detoxification genes, N fixation genes, and other plant growth stimulation genes. The pSym plasmid found in Rhizobium sp., in addition to nodulation and atmospheric nitrogen fixation genes, is involved in phytohormone synthesis and transport of root exudate compounds. This conjugative plasmid is commonly transferred to the soil or rhizosphere community, mainly after sensing certain plant compounds such as flavonoids [86,87]. An approximately 150 kb plasmid was observed in an endophytic plant growth-promoting Enterobacter sp. (pENT638-1), which has a role in host colonization [85]. In the case of Enterobacteriaceae and Pseudomonadaceae, the most abundant and highly efficiently transferred is the IncP plasmid [85]. IncP plasmids have clustered restriction sites that represent an integration site for mobile genetic elements (MGEs) carrying antibiotic and heavy metal resistance determinant genes [87].
HGT is a common strategy for changing adaptation-related genes, such as those related to antibiotic resistance and heavy metal resistance among soil bacteria. Lekired et al. performed an extensive analysis of mobile genetic elements (MGEs) in Pantoea eucrina OB49 and found a plethora of MGEs (transposons, insertion sequences, a putative integrative and conjugative element) related to antibiotics (beta-lactamase class) and a broad range of heavy metal tolerance [83].
In the rhizosphere, microbes sense surrounding environmental stimuli and modulate the process of gene exchange; therefore, appropriate environmental conditions can promote HGT events. Higher temperature values (up to 35 °C), loamy soil, use of organic fertilizer, and higher soil toxicity have been found to increase the frequency of plasmid transfer between bacteria [86].
Plant growth-promoting rhizobacteria possess more than one beneficial function as a result of gene accumulation in the rhizosphere and soil environment governed by selection mechanisms. The major function genes related to plant-beneficial function are as follows: (i). nitrogen fixation-contributing nifHDK genes (encoding nitrogenase), (ii). Mineral phosphate solubilization pqqBCDEFG genes (encoding pyrroloquinoline quinone), (iii). inhibition of ethylene biosynthesis acdS gene (encoding 1-aminocyclopropane-1-carboxylate), (iv). IAA-producing ipdC/ppdC genes (encoding indole-3-pyruvate decarboxylase/phenylpyruvate decarboxylase), (v). antimicrobial compound synthesis hcnABC/phlACBD (hydrogen cyanide/2,4-diacetylphloroglucinol) genes, and (vi). induced systemic resistance conferring budAB/budC (acetoine/2,3-butanediol) genes, which was reported previously and studied in Proteobacteria by Bruto et al. [88].
Co-occurrence of budAB with ipdC, the nifHDK operon, clustered pqqBCDE genes, and hcnABC with phlABCD gene clusters was observed in 25 PGPR genomes [88]. Bruto et al. also studied the genome plasticity of the Proteobacteria and identified acquisition and loss events of the plant-beneficial function encoding genes [88]. They observed clade specificity in phloroglucinol-producing (phlACBD) and IAA-producing (ipdC/ppdC) genes. Among the genes conferring induced systemic resistance, budAB was clade-specific, whereas budC was observed several times in other taxa. Although the genes responsible for mineral phosphate solubilization were detected in the LCA of Pseudomonas, they were acquired 15 times by other taxa. The highest number of acquisition events were observed in nitrogen fixation-contributing genes (nifHDK) and the ACC deaminase (acdS) gene [88]. Occurrence of function genes related to plant-beneficial function in other root-adapted, non-PGPR bacteria was also observed [88].
Cross-kingdom MGE transfers also promote environmental adaptation. Due to the co-existence of bacteria, fungi, and plants in the soil environment and rhizosphere, MGE transfers occur among all parties. The network structure of fungal mycelia and root exudates enable the transfer of MGEs among bacteria, and MGE transfers occur between fungi and bacteria [86].
2.5. Synergistic Microbial Processes
In many cases, plant inoculation with bacterial consortia proved to be more efficient than inoculation with a single bacterial strain. Current research is focused on deciphering how synergistic microbial processes work and how different plant beneficial traits influence each other. Synergistic processes between ACC deaminase and IAA production and N2 fixation [89,90,91,92,93,94,95,96], ACC deaminase and IAA production and stress tolerance [93,97], and phosphate solubilization and ACC deaminase [98] were reported.
The role of ACC deaminase and IAA in BNF fixation process is complex; they can enhance nodule formation, improve the competitiveness of rhizobia for nodulation, suppress nodule senescence, and upregulate genes associated with legume–rhizobia symbiosis [96].
The role of ACC deaminase in nodule formation was studied using either knockout or overproducing strains for the ACC deaminase-producing gene [96]. When Mesorhizobium loti ACC deaminase-overproducing mutant strain was tested for the efficiency of plant colonization and nodulation, it was found to be more efficient than the wild type [89]. This relationship is relatively complex, whereas in Mesorhizobium loti, the acdS gene was found in the symbiotic island and its expression was regulated by the N2 fixation regulator NifA2 [99]. In senescent nodules, increased gene expression of PsACS2 encoding ACC synthase (an enzyme involved in ethylene synthesis) and increased transcription have been observed [90]. Therefore, ACC deaminase-producing PGP bacteria can enhance N2 fixation by extending the lifespan of functional nodules. Nascimento et al. investigated the physiological function of ACC deaminase in a transformed Mesorhizobium sp. expressing ACC deaminase and its parental type strain without this activity, and they observed nitrogenase activity in 31-day-old nodules [94]. Nascimento et al. also observed that ACC deaminase-producing Pseudomonas fluorescens YsS6 strain increased the nodulation ability of both alpha- and beta-rhizobia compared to its ACC deaminase mutant strain [95]. Thus, free-living ACC deaminase-producing bacteria play an important role in facilitating rhizobia nodulation.
IAA is a biomolecule that can affect legume–rhizobia interactions because nodule initiation and development require high levels of IAA to regulate cell division and nodule primordium formation. IAA also alters the expression of genes associated with rhizobia and nodule initiation in plant cells [96]. Camerini et al. demonstrated a close relationship between IAA and nodulation when a lower nodule formation capacity was observed in IAA-negative Rhizobium leguminosarum bv. viciae compared to the parent strain [91]. Moreover, short-chain fatty acid production (caproic acid) of IAA-producing nodule-enhancing rhizobacteria facilitated rhizobia colonization when used as a co-inoculum [96]. The IAA-overproducing Ensifer meliloti RD64-induced 45-day-old nodules had an extended nitrogen-fixing zone and a reduction in the senescent zone compared with plants nodulated with the wild-type strain [92]. In the same IAA-overproducing bacterial strain, Defez et al. observed an increase that reached maximum induction of the fixNOQP1,2 operon gene in the nodules after 40 days of inoculation [93].
Crosstalk between ACC deaminase and IAA-producing bacteria exists; ACC deaminase acts as a sink for ACC, whereas IAA may facilitate plant growth or activate the transcription of the enzyme ACC synthase. If ethylene synthesis occurs, IAA signal transduction is limited [97].
Orozco-Mosqueda et al. and Defez et al. studied the role of ACC deaminase and IAA in the stress response [93,100]. In Pseudomonas sp. UW4 ACC deaminase (acdS) and trehalose (treS) mutant strains (mutated at one or two traits), synergism between the two traits was observed when used as a bioinoculant on salt-stressed tomato plants, since single gene mutants have a better effect than double ones [100]. Stress tolerance-associated gene expression upregulation was observed in IAA overproducing E. meliloti RD64 compared to its wild derivative [93].
Alemneh et al. reported the presence of diverse ACC deaminase producing bacteria that showed phosphate solubilizing capacity and proved that ACC deaminase mediated P solubilization [98]. They also observed improved nodulation by increasing the P nutrition of chickpeas when inoculated with ACC expressing Burkholderia sp. 12F.
Synergism occurs not only between bacteria, but also between bacterial ACC deaminase and arbuscular mycorrhizal fungi [97]; therefore, metabolomic studies of the whole soil microbiome are very useful in understanding plant–soil–microbe interactions.
2.6. Innovations in Carrier Materials for Bioinoculants
Carrier materials for bioinoculants must be chemically stable, nontoxic, low-cost, and able to provide a protective niche for microorganisms to ensure the viability of cells during storage and controlled release [101,102]. Many types of bioinoculant carriers have been studied in the recent decades. They can be classified as solid, liquid, organic, or inorganic. Additives that nutritionally support microorganisms are used in these bioformulations [101,102].
Peat, biochar, bagasse, cork compost, attapulgite, sepiolite, perlite, and amorphous silica were used as media for the solid bioformulations. They provide support for beneficial microbes, in contrast with liquid bioformulations that are more sensitive to prolonged storage. Immobilized formulations or encapsulation is an emerging technology with significant advantages over the above-mentioned formulations [101,102]. Microbial cells are immobilized by adhesion/biofilm formation on solid supports or entrapment, thereby conferring a protective environment for bacterial cells [103]. Various polymer matrices were tested to study their effect on microbial cell viability during encapsulation and storage [104,105,106,107].
The use of environmentally friendly biopolymer matrices is well suited to sustainable agriculture. Microbial cells are encapsulated using various techniques such as ionic gelation, emulsification, and spray drying [102]. Additives are used to improve the stability, encapsulation efficiency, and mechanical properties of the carrier polymer, as well as fillers to improve microbial survival [106,108].
Alginates are the most widely studied microbial carriers, mainly for Azospirillum sp. and Pseudomonas sp. [102]. Alginate bead-entrapped A. brasilense showed better viability during prolonged storage [104]. Calcium alginate microspheres have been used for Trichoderma viride spore encapsulation and provided a supportive environment for growth [109]. Panichikkal et al. used alginate beads supplemented with salicylic acid and zinc oxide nanoparticles to immobilize Pseudomonas sp. DN18 which proved to be a promising bioformulation for Oryza sativa [108]. The drawbacks of alginate beads such as low mechanical strength, poor appearance, and high porosity have also been reported [105].
Starch presents important characteristics as a potential biopolymeric carrier, as it is nontoxic, shows high solubility, and confers the controlled release of PGPB [102]. Marcelino et al. used an innovative biodegradable foam containing as major component starch combined with low-cost industrial by-products such as sugarcane bagasse, glycerol, rock phosphate, crystal sugar, powdered skim milk and yeast extract for Azospirillum brasilense Ab-V5 immobilization [106]. Microbial cell viability was maintained for up to 120 d at room temperature [106]. Starch beads combined with chitosan were tested for release of A. brasilense and P. fluorescens. After one year of storage, the recovery of bacteria was of the order of 108–109 CFU/g for both bacterial strains [107]. Alginate–chitosan nanoparticles supplemented with starch were used for encapsulation of B. licheniformis. The bioformulation was tested on chili plants, and plant beneficial traits were observed [110].
Nanofibers have also been used for the immobilization of bacterial co-cultures based on Pantoea agglomerans and Burkholderia caribensis, and on testing, showed beneficial effects on soybean [111].
The above-mentioned studies using immobilized formulations are based mainly on laboratory-scale experiments and rarely on field experiments. Therefore, future research should address how the production of bioformulations can be scaled up, how efficient they are in the field, and what their long-term effects on soil and microbiome are.
2.7. Engineering Microbiome
Many plant growth-promoting microbes and microbial consortia have been studied and proposed as potential bioinoculants. Various carriers have been tested to maximize their colonization and persistence in harsh soil environments. Nevertheless, limitations of natural bioinoculant use have been reported due to the complexity of soil–microbe–plant interconnectedness.
A better understanding of the rhizosphere biochemical and molecular specificity that governs plant–microbe interactions is required to be used in rhizosphere microbiome engineering [112]. Rhizosphere microbiome engineering has gained much attention in advanced agricultural research [113,114].
Microbiome engineering uses a microbe-focused approach that is based on constructing synthetic communities called SynComs. These communities can be constructed using bottom-up strategies. The bottom-up approach involves the identification of keystone microbial taxa (e.g., Agrobacterium, Pseudomonas, Enterobacter) and the use of a combination of microbial isolates. SynComs complexity is important in terms of their effectiveness and stability in a changing environment, and functional species can be substituted because of their stable metabolic network [113].
Other possibilities rely on the genetic engineering of PGPRs, when PGP gene clusters isolated from rhizobacteria are introduced on broad-host-range plasmids or phage transduction. Integrative and conjugative plasmid systems and chassis-independent recombinase-assisted genome engineering are more advantageous than conjugative plasmids, which are rapidly lost from bacterial populations [114]. A new strategy for PGP gene cluster transmission is based on the introduction of foreign genes in situ through a conjugal donor strain, which has led to the emergence of microbiome engineering [114].
Plant-secreted secondary metabolites such as carbohydrates, amino acids, and flavonoids are key drivers for the colonization of the rhizosphere [112] and play a role in microbial gene expression [114]. Therefore, plant-derived signals can be used as controllers of the expression of the introduced genes. Ryu et al. identified such legume-derived flavonoid-inducible expression systems in rhizobia and used them to N-fixation in Pseudomonas protogens Pf-5 [115].
The plant-microbe relationship specificity and their species-specific signal make it possible to use them in a very targeted way [114]. Rhizopine was recognized as a specific signal to select rhizobia (Rhizobium and Sinorhizobium) in legumes and was used to obtain transgenic Medicago and barley plants. Rhizopine signaling allows the establishment of a control-engineered PGPR [116].
Owing to recent advances in this field, at least two directions for improving plant growth promoting biopreparates can be defined. Both synthetic microbiome/PGPR genetic engineering and plant genome engineering to recruit the desired microbiome can revolutionize the field of sustainable agriculture.
2.8. Farm-Derived Products in Sustainable Agricultural Practices
Reduced use or lack of use of external inputs such as agrochemicals and the use instead of farm-derived organic inputs are common practices in sustainable agriculture. The main aim of these practices is to increase the soil organic carbon level, which is important from both environmental and agricultural points of view.
Farm-derived organic amendments, such as compost, compost tea, and manure, are used more frequently for the substitution of agrochemicals. Organic wastes can be successfully converted through anaerobic decomposition to form organic fertilizers [117]. The reduced use of chemical fertilizers and higher use of farm amendments mitigates greenhouse gas (GHG) emissions [118]. In a meta-analysis by Wei et al., an amount of 0.203 MgCO2 eq/ha was calculated in the case of full substitution with organic fertilizer (manure, compost, or commercial organic fertilizer) [119]. A considerable carbon sink resulted when partial substitution of mineral fermentation with organic amendments was used [118,119]. The impact of the use of fermented liquid amendments was evaluated in comparison with mineral fertilization on lettuce growth and soil quality at the microcosm and field scales [117]. Urra et al. reported that the commercial and farm-made fermented liquid organic amendments used in experiments beside sustaining crop yield had ameliorative effect on soil quality [117].
Sujatha et al. proposed the conversion of weed biomass to obtain an environment-friendly and climate-friendly amendment [120]. Substances such as urea, cow dung, a microbial consortium (Bacillus subtilis), and a farm-derived liquid organic formulation (jeevamrutham) were used as activators. The most efficient activator was the farm-derived organic formulation, which converted the weed biomass to a carbon and humic substance-rich amendment [120].
Sustainable agricultural practices, such as recycling farm-derived products, are in accordance with global strategies to reduce greenhouse gas emissions by diminishing agricultural waste and agrochemical-related emissions.
3. Concluding Remarks and Future Perspectives
Owing to stratification and various microhabitats, the complexity of the soil system supports the formation of a diverse microbial community. Analyzing microbial community data is currently the biggest challenge. Beyond taxonomy, understanding the functional groups of bacterial taxa and the dynamics of the bacterial community structure are important issues for better understanding soil ecosystem functioning. Because the rhizosphere is considered one of the hotspots of microbial gene transfer and the microbiome is a rich reservoir of genetic functionality, PGPR genetic engineering can be an important tool in the field of sustainable agriculture. How is the soil ecological function affected by environmental changes and how can this function be maintained under sustainable agricultural practices? How these are shaped by plant and microorganism interactions remains to be elucidated. Another tool for sustainable agriculture is based on the specificity of plant-microbe communication and relies on plant genome engineering. Therefore, the key to sustainable agriculture relies on an enhanced comprehension of the complexity of soil–microbe–plant systems and their impact on short- or long-term sustainability. In light of these facts, future efforts and research topics should focus on the following: (i). exploitation of the potential of genetically engineered microbes and plants for sustainable agriculture; and (ii). long-term effects on the soil and microbiome of the immobilized bioformulations; and (iii). clean and sustainable practices based on farm-derived product conversion for soil amendments.
Author Contributions
Conceptualization, G.M. and É.L.; investigation, É.-B.V., A.B., G.M. and É.L.; writing—original draft preparation, É.-B.V., A.B., G.M. and É.L.; writing—review and editing, G.M., and É.L.; visualization, É.-B.V. and G.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors acknowledge the University of Pécs, Faculty of Sciences, Institute of Chemistry, Chemical Doctoral School and Collegium Talentum Programme of Hungary for the PhD scholarship accorded to É.-B.V and A.B.
Conflicts of Interest
The authors declare no conflict of interest.
References
- De Corato, U. Towards New Soil Management Strategies for Improving Soil Quality and Ecosystem Services in Sustainable Agriculture: Editorial Overview. Sustainability 2020, 12, 9398. [Google Scholar] [CrossRef]
- Bar-On, Y.M.; Phillips, R.; Milo, R. The biomass distribution on Earth. Proc. Natl. Acad. Sci. USA 2018, 115, 6506–6511. [Google Scholar] [CrossRef] [PubMed]
- Dubey, D.-A.; Malla, M.; Khan, F.; Chowdhary, K.; Yadav, S.; Kumar, A.; Sharma, S.; Khare, P.K.; Khan, M.; Khan, M. Soil microbiome: A key player for conservation of soil health under changing climate. Biodivers. Conserv. 2019, 28, 2405–2429. [Google Scholar] [CrossRef]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, S.L.; Gibbons, S.M.; Owens, S.M.; Hampton-Marcell, J.; Johnston, E.R.; Jastrow, J.D.; Gilbert, J.A.; Meyer, F.; Antonopoulos, D.A. Spatial scale drives patterns in soil bacterial diversity. Environ. Microbiol. 2016, 18, 2039–2051. [Google Scholar] [CrossRef] [PubMed]
- Jansson, J.K.; Hofmockel, K.S. Soil microbiomes and climate change. Nat. Rev. Microbiol. 2020, 18, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Thakur, M.P.; Geisen, S. Trophic Regulations of the Soil Microbiome. Trends Microbiol. 2019, 27, 771–780. [Google Scholar] [CrossRef]
- Fan, K.; Cardona, C.; Li, Y.; Shi, Y.; Xiang, X.; Shen, C.; Wang, H.; Jack, G.; Chu, H. Rhizosphere-associated bacterial network structure and spatial distribution differ significantly from bulk soil in wheat crop fields. Soil Biol. Biochem. 2017, 113, 275–284. [Google Scholar] [CrossRef]
- Bakker, P.; Berendsen, R.; Doornbos, R.; Wintermans, P.; Pieterse, C. The rhizosphere revisited: Root microbiomics. Front. Plant Sci. 2013, 4, 165. [Google Scholar] [CrossRef]
- Saleem, M.; Hu, J.; Jousset, A. More than the Sum of Its Parts: Microbiome Biodiversity as a Driver of Plant Growth and Soil Health. Annu. Rev. Ecol. Evol. Syst. 2019, 50, 145–168. [Google Scholar] [CrossRef]
- Ettema, C.H.; Wardle, D.A. Spatial soil ecology. Trends Ecol. Evol. 2002, 17, 177–183. [Google Scholar] [CrossRef]
- Bakker, M.G.; Chaparro, J.M.; Manter, D.K.; Vivanco, J.M. Impacts of bulk soil microbial community structure on rhizosphere microbiomes of Zea mays. Plant Soil 2015, 392, 115–126. [Google Scholar] [CrossRef]
- Li, X.; Rui, J.; Mao, Y.; Yannarell, A.; Mackie, R. Dynamics of the bacterial community structure in the rhizosphere of a maize cultivar. Soil Biol. Biochem. 2014, 68, 392–401. [Google Scholar] [CrossRef]
- Rchiad, Z.; Dai, M.; Hamel, C.; Bainard, L.D.; Cade-Menun, B.J.; Terrat, Y.; St-Arnaud, M.; Hijri, M. Soil Depth Significantly Shifted Microbial Community Structures and Functions in a Semiarid Prairie Agroecosystem. Front. Microbiol. 2022, 13, 815890. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.N.; Kumar, V.; Dhaliwal, H.; Prasad, R.; Saxena, A. Microbiome in Crops: Diversity, Distribution, and Potential Role in Crop Improvement. In New and Future Developments in Microbial Biotechnology and Bioengineering: Crop Improvement through Microbial Biotechnology; Elsevier: Amsterdam, The Netherlands, 2018; pp. 305–332. ISBN 978-0-444-63987-5. [Google Scholar]
- Dubey, R.K.; Tripathi, V.; Prabha, R.; Chaurasia, R.; Singh, D.P.; Rao, C.S.; El-Keblawy, A.; Abhilash, P.C. (Eds.) Metatranscriptomics and Metaproteomics for Microbial Communities Profiling. In Unravelling the Soil Microbiome: Perspectives for Environmental Sustainability; Springer Briefs in Environmental Science; Springer International Publishing: Cham, Switzerland, 2020; pp. 51–60. ISBN 978-3-030-15516-2. [Google Scholar]
- Tan, L.; Qu, M.; Zhu, Y.; Peng, C.; Wang, J.; Gao, D.; Chen, C. ZINC TRANSPORTER5 and ZINC TRANSPORTER9 Function Synergistically in Zinc/Cadmium Uptake. Plant Physiol. 2020, 183, 1235–1249. [Google Scholar] [CrossRef] [PubMed]
- Mahoney, A.K.; Yin, C.; Hulbert, S.H. Community Structure, Species Variation, and Potential Functions of Rhizosphere-Associated Bacteria of Different Winter Wheat (Triticum aestivum) Cultivars. Front. Plant Sci. 2017, 8, 132. [Google Scholar] [CrossRef]
- Rathore, R.; Dowling, D.N.; Forristal, P.D.; Spink, J.; Cotter, P.D.; Bulgarelli, D.; Germaine, K.J. Crop Establishment Practices Are a Driver of the Plant Microbiota in Winter Oilseed Rape (Brassica napus). Front. Microbiol. 2017, 8, 1489. [Google Scholar] [CrossRef]
- Ullah, A.; Akbar, A.; Luo, Q.; Khan, A.H.; Manghwar, H.; Shaban, M.; Yang, X. Microbiome Diversity in Cotton Rhizosphere Under Normal and Drought Conditions. Microb. Ecol. 2019, 77, 429–439. [Google Scholar] [CrossRef]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef]
- Sugiyama, A.; Ueda, Y.; Zushi, T.; Takase, H.; Yazaki, K. Changes in the Bacterial Community of Soybean Rhizospheres during Growth in the Field. PLoS ONE 2014, 9, e100709. [Google Scholar] [CrossRef]
- Praeg, N.; Pauli, H.; Illmer, P. Microbial Diversity in Bulk and Rhizosphere Soil of Ranunculus glacialis Along a High-Alpine Altitudinal Gradient. Front. Microbiol. 2019, 10, 1429. Available online: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01429 (accessed on 24 August 2023). [CrossRef] [PubMed]
- Hannula, S.E.; Ma, H.; Pérez-Jaramillo, J.E.; Pineda, A.; Bezemer, T.M. Structure and ecological function of the soil microbiome affecting plant–soil feedbacks in the presence of a soil-borne pathogen. Environ. Microbiol. 2020, 22, 660–676. [Google Scholar] [CrossRef] [PubMed]
- Lakshmanan, V.; Selvaraj, G.; Bais, H.P. Functional soil microbiome: Belowground solutions to an aboveground problem. Plant Physiol. 2014, 166, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Aira, M.; Gómez-Brandón, M.; Lazcano, C.; Bååth, E. Plant genotype strongly modifies the structure and growth of maize rhizosphere microbial communities. Soil Biol. Biochem. 2010, 42, 2276–2281. [Google Scholar] [CrossRef]
- Geller, A.M.; Levy, A. “What I cannot create, I do not understand”: Elucidating microbe-microbe interactions to facilitate plant microbiome engineering. Curr. Opin. Microbiol. 2023, 72, 102283. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Gu, Y.; Friman, V.-P.; Kowalchuk, G.A.; Xu, Y.; Shen, Q.; Jousset, A. Initial soil microbiome composition and functioning predetermine future plant health. Sci. Adv. 2019, 5, eaaw0759. [Google Scholar] [CrossRef] [PubMed]
- Suman, J.; Rakshit, A.; Ogireddy, S.D.; Singh, S.; Gupta, C.; Chandrakala, J. Microbiome as a Key Player in Sustainable Agriculture and Human Health. Front. Soil Sci. 2022, 2, 821589. [Google Scholar] [CrossRef]
- Khan, A.A.H. Plant-Bacterial Association and Their Role as Growth Promoters and Biocontrol Agents. In Plant Growth Promoting Rhizobacteria for Sustainable Stress Management; Springer: Cham, Switzerland, 2019; pp. 389–419. ISBN 9789811369858. [Google Scholar]
- Husna; Hussain, A.; Shah, M.; Hamayun, M.; Iqbal, A.; Qadir, M.; Alataway, A.; Dewidar, A.Z.; Elansary, H.O.; Lee, I.-J. Phytohormones producing rhizobacteria alleviate heavy metals stress in soybean through multilayered response. Microbiol. Res. 2023, 266, 127237. [Google Scholar] [CrossRef]
- Adedayo, A.A.; Babalola, O.O. Fungi That Promote Plant Growth in the Rhizosphere Boost Crop Growth. J. Fungi 2023, 9, 239. [Google Scholar] [CrossRef]
- Eva, L.; Gyongyver, M. Is PGPR an Alternative for NPK Fertilizers in Sustainable Agriculture? In Microbial Interventions in Agriculture and Environment; Springer: Cham, Switzerland, 2019; pp. 51–62. ISBN 9789811383908. [Google Scholar]
- Fowler, D.; Steadman, C.E.; Stevenson, D.; Coyle, M.; Rees, R.M.; Skiba, U.M.; Sutton, M.A.; Cape, J.N.; Dore, A.J.; Vieno, M.; et al. Effects of global change during the 21st century on the nitrogen cycle. Atmos. Chem. Phys. 2015, 15, 13849–13893. [Google Scholar] [CrossRef]
- Ladha, J.K.; Peoples, M.B.; Reddy, P.M.; Biswas, J.C.; Bennett, A.; Jat, M.L.; Krupnik, T.J. Biological nitrogen fixation and prospects for ecological intensification in cereal-based cropping systems. Field Crops Res. 2022, 283, 108541. [Google Scholar] [CrossRef] [PubMed]
- Turan, M.; Topcuoğlu, B.; Kıtır, N.; Alkaya, Ü.; Erçelik, F.; Nikerel, E.; Güneş, A.; Turan, M.; Topcuoğlu, B.; Kıtır, N.; et al. Plant Growth Promoting Rhizobacteria’s (PGPRS) Enzyme Dynamics in Soil Remediation. In Soil Contamination—Current Consequences and Further Solutions; IntechOpen: London, UK, 2016; ISBN 978-953-51-2816-8. [Google Scholar]
- de Bruijn, F.J.; Hungria, M. Biological Nitrogen Fixation. In Good Microbes in Medicine, Food Production, Biotechnology, Bioremediation, and Agriculture; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2022; pp. 466–475. ISBN 978-1-119-76262-1. [Google Scholar]
- Matilla, M.; Krell, T. Plant Growth Promotion and Biocontrol Mediated by Plant-Associated Bacteria. In Plant Microbiome: Stress Response; Springer: Cham, Switzerland, 2018; pp. 45–80. ISBN 978-981-10-5513-3. [Google Scholar]
- Ahmad, M.; Nadeem, S.M.; Zahir, Z.A. Plant-Microbiome Interactions in Agroecosystem: An Application. In Microbiome in Plant Health and Disease; Springer: Singapore, 2019; pp. 251–291. [Google Scholar] [CrossRef]
- Chaudhary, P.; Agri, U.; Chaudhary, A.; Kumar, A.; Kumar, G. Endophytes and their potential in biotic stress management and crop production. Front. Microbiol. 2022, 13, 933017. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, R.; Waqas, M.; Khan, A.L.; Al-Hosni, K.; Kang, S.-M.; Seo, C.-W.; Lee, I.-J. Indoleacetic acid production and plant growth promoting potential of bacterial endophytes isolated from rice (Oryza sativa L.) seeds. Acta Biol. Hung. 2017, 68, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Salazar-Cerezo, S.; Martínez-Montiel, N.; García-Sánchez, J.; Pérez-Y-Terrón, R.; Martínez-Contreras, R.D. Gibberellin biosynthesis and metabolism: A convergent route for plants, fungi and bacteria. Microbiol. Res. 2018, 208, 85–98. [Google Scholar] [CrossRef] [PubMed]
- de Garcia Salamone, I.E.; Hynes, R.K.; Nelson, L.M. Role of Cytokinins in Plant Growth Promotion by Rhizosphere Bacteria. In PGPR: Biocontrol and Biofertilization; Siddiqui, Z.A., Ed.; Springer: Dordrecht, The Netherlands, 2006; pp. 173–195. ISBN 978-1-4020-4152-5. [Google Scholar]
- Sakure, S.; Bhosale, S. Actinobacteria for Biotic Stress Management. In Plant Growth Promoting Rhizobacteria for Sustainable Stress Management; Springer: Cham, Switzerland, 2019; pp. 363–378. ISBN 9789811369858. [Google Scholar]
- Patil, A.; Patil, S.; Sayyed, R. Interaction of Rhizobacteria with Soil Microorganisms: An Agro-Beneficiary Aspect. In Plant Growth Promoting Rhizobacteria for Sustainable Stress Management; Springer: Cham, Switzerland, 2019; pp. 241–259. ISBN 9789811369858. [Google Scholar]
- Ramadan, E.; Abdelhafez, A.; Enas, A.; Saber, F. Plant growth promoting rhizobacteria and their potential for biocontrol of phytopathogens. Afr. J. Microbiol. Res. 2016, 10, 486–504. [Google Scholar] [CrossRef]
- Zhang, S.; Gan, Y.; Xu, B. Mechanisms of the IAA and ACC-deaminase producing strain of Trichoderma longibrachiatum T6 in enhancing wheat seedling tolerance to NaCl stress. BMC Plant Biol. 2019, 19, 22. [Google Scholar] [CrossRef] [PubMed]
- Mathur, A.; Koul, A.; Hattewar, J. Plant Growth-Promoting Rhizobacteria (PGPRs): Significant Revolutionary Tools for Achieving Long-Term Sustainability and Combating the Biotic Stress Caused by the Attack of Pathogens Affecting Crops in Agriculture. In Plant Growth Promoting Rhizobacteria for Sustainable Stress Management; Springer: Cham, Switzerland, 2019; pp. 379–388. ISBN 9789811369858. [Google Scholar]
- Tandon, S.; Vats, S. Microbial Biosynthesis of Cadmium Sulfide (CDS) Nanoparticles and their Characterization. Eur. J. Pharm. Med. Res. 2016, 3, 545–550. [Google Scholar]
- Vats, S.; Bhargava, P. Alternate Energy: Fuel for “Modi’s India” and “Smart cities”. Int. J. Curr. Res. 2017, 9, 49090–49097. [Google Scholar]
- Saxena, P.; Srivastava, J.; Pandey, S.; Srivastava, S.; Maurya, N.; Chand, K.; Mishra, S.; Asthana, G.; Bhargava, P.; Kumar, R.; et al. Plants for Biocontrol and Biological Control of Plant Pathogens. In Plant Biotic Interactions; Springer: Cham, Switzerland, 2019; pp. 147–179. ISBN 978-3-030-26656-1. [Google Scholar]
- Köhl, J.; Kolnaar, R.; Ravensberg, W.J. Mode of Action of Microbial Biological Control Agents Against Plant Diseases: Relevance Beyond Efficacy. Front. Plant Sci. 2019, 10, 845. [Google Scholar] [CrossRef]
- Abbamondi, G.R.; Tommonaro, G.; Weyens, N.; Thijs, S.; Sillen, W.; Gkorezis, P.; Iodice, C.; de Melo Rangel, W.; Nicolaus, B.; Vangronsveld, J. Plant growth-promoting effects of rhizospheric and endophytic bacteria associated with different tomato cultivars and new tomato hybrids. Chem. Biol. Technol. Agric. 2016, 3, 1. [Google Scholar] [CrossRef]
- Brígido, C.; Singh, S.; Menéndez, E.; Tavares, M.J.; Glick, B.R.; Félix, M.d.R.; Oliveira, S.; Carvalho, M. Diversity and Functionality of Culturable Endophytic Bacterial Communities in Chickpea Plants. Plants 2019, 8, 42. [Google Scholar] [CrossRef] [PubMed]
- Egamberdieva, D. Alleviation of salt stress by plant growth regulators and IAA producing bacteria in wheat. Acta Physiol. Plant. 2009, 31, 861–864. [Google Scholar] [CrossRef]
- Tabatabaei, S.; Ehsanzadeh, P.; Etesami, H.; Alikhani, H.; Glick, B. Indole-3-acetic acid (IAA) producing Pseudomonas isolates inhibit seed germination and alpha-amylase activity in durum wheat (Triticum turgidum L.). Span. J. Agric. Res. 2016, 14, e0802. [Google Scholar] [CrossRef]
- Shaharoona, B.; Arshad, M.; Zahir, Z. Effect of plant growth promoting rhizobacteria containing ACC-deaminase on maize (Zea mays L.) grown under axenic conditions and on nodulation in mungbean (Vigna radiata L.). Lett. Appl. Microbiol. 2006, 42, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Bonaterra, A.; Badosa, E.; Daranas, N.; Francés, J.; Roselló, G.; Montesinos, E. Bacteria as Biological Control Agents of Plant Diseases. Microorganisms 2022, 10, 1759. [Google Scholar] [CrossRef] [PubMed]
- Arseneault, T.; Filion, M. Biocontrol through antibiosis: Exploring the role played by subinhibitory concentrations of antibiotics in soil and their impact on plant pathogens. Can. J. Plant Pathol. 2017, 39, 267–274. [Google Scholar] [CrossRef]
- Ram, R.M.; Keswani, C.; Bisen, K.; Tripathi, R.; Singh, S.P.; Singh, H.B. Biocontrol Technology; Elsevier: Amsterdam, The Netherlands, 2018; pp. 177–190. [Google Scholar]
- Glick, B.R. Biocontrol of Bacteria and Fungi. In Beneficial Plant-Bacterial Interactions; Glick, B.R., Ed.; Springer International Publishing: Cham, Switzerland, 2020; pp. 181–230. ISBN 978-3-030-44368-9. [Google Scholar]
- Kenawy, A.; Joe Dailin, D.; Abo-Zaid, G.; Malek, R.A.; Ambehabati, K.; Zakaria, K.; Sayyed, R.; El Enshasy, H. Biosynthesis of Antibiotics by PGPR and Their Roles in Biocontrol of Plant Diseases. In Plant Growth Promoting Rhizobacteria for Sustainable Stress Management; Springer: Cham, Switzerland, 2019; pp. 1–35. ISBN 9789811369858. [Google Scholar]
- Cesa, C.; Baez, A.; Quintero-Hernandez, V.; De la cruz-Enriquez, J.; Castañeda, D.; Muñoz-Rojas, J. The importance of antimicrobial compounds produced by beneficial bacteria on the biocontrol of phytopathogens. Acta Biol. Colomb. 2020, 25, 140–154. [Google Scholar] [CrossRef]
- Ongena, M.; Jacques, P. Bacillus lipopeptides: Versatile weapons for plant disease biocontrol. Trends Microbiol. 2008, 16, 115–125. [Google Scholar] [CrossRef]
- Raaijmakers, J.M.; Mazzola, M. Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu. Rev. Phytopathol. 2012, 50, 403–424. [Google Scholar] [CrossRef]
- Quiñones, B.; Dulla, G.; Lindow, S.E. Quorum sensing regulates exopolysaccharide production, motility, and virulence in Pseudomonas syringae. Mol. Plant Microbe Interact. 2005, 18, 682–693. [Google Scholar] [CrossRef]
- Koutsoudis, M.D.; Tsaltas, D.; Minogue, T.D.; von Bodman, S.B. Quorum-sensing regulation governs bacterial adhesion, biofilm development, and host colonization in Pantoea stewartii subspecies stewartii. Proc. Natl. Acad. Sci. USA 2006, 103, 5983–5988. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.B.B.M.; Zhang, H.-B.; Xu, J.-L.; Liu, Q.; Jiang, Z.; Zhang, L.-H. The acyl-homoserine lactone-type quorum-sensing system modulates cell motility and virulence of Erwinia chrysanthemi pv. zeae. J. Bacteriol. 2008, 190, 1045–1053. [Google Scholar] [CrossRef]
- Kang, B.R.; Anderson, A.J.; Kim, Y.C. Hydrogen Cyanide Produced by Pseudomonas chlororaphis O6 Exhibits Nematicidal Activity against Meloidogyne hapla. Plant Pathol. J. 2018, 34, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Mookherjee, A.; Singh, S.; Maiti, M.K. Quorum sensing inhibitors: Can endophytes be prospective sources? Arch. Microbiol. 2018, 200, 355–369. [Google Scholar] [CrossRef] [PubMed]
- Noorashikin, M.N.; Li, L.Y.; Karim, M.; Daud, H.M.; Natrah, F.M.I. Screening and identification of quorum sensing degraders from live feed Artemia. J. Environ. Biol. 2016, 37, 811–816. [Google Scholar] [PubMed]
- Torabi Delshad, S.; Soltanian, S.; Sharifiyazdi, H.; Bossier, P. Effect of quorum quenching bacteria on growth, virulence factors and biofilm formation of Yersinia ruckeri in vitro and an in vivo evaluation of their probiotic effect in rainbow trout. J. Fish. Dis. 2018, 41, 1429–1438. [Google Scholar] [CrossRef] [PubMed]
- Panpatte, D.; Jhala, Y.; Vyas, R. Signaling pathway of induced systemic resistance. In Molecular Aspects of Plant Beneficial Microbes in Agriculture; Elsevier: Amsterdam, The Netherlands, 2020; pp. 133–141. ISBN 978-0-12-818469-1. [Google Scholar]
- Chan, K.-G.; Atkinson, S.; Mathee, K.; Sam, C.-K.; Chhabra, S.R.; Cámara, M.; Koh, C.-L.; Williams, P. Characterization of N-acylhomoserine lactone-degrading bacteria associated with the Zingiber officinale (ginger) rhizosphere: Co-existence of quorum quenching and quorum sensing in Acinetobacter and Burkholderia. BMC Microbiol. 2011, 11, 51. [Google Scholar] [CrossRef]
- Ma, A.; Lv, D.; Zhuang, X.; Zhuang, G. Quorum Quenching in Culturable Phyllosphere Bacteria from Tobacco. Int. J. Mol. Sci. 2013, 14, 14607–14619. [Google Scholar] [CrossRef]
- Garge, S.S.; Nerurkar, A.S. Attenuation of Quorum Sensing Regulated Virulence of Pectobacterium carotovorum subsp. carotovorum through an AHL Lactonase Produced by Lysinibacillus sp. Gs50. PLoS ONE 2016, 11, e0167344. [Google Scholar] [CrossRef]
- Jose, P.; Ramasamy, K.; Kwon, S.-W.; Veeranan, J.; Murugaiyan, S.; Gopal, N.O.; Kumutha, K.; Anandham, R. Interference in quorum sensing and virulence of the phytopathogen Pseudomonas syringae pv. passiflorae by Bacillus and Variovorax species. BioControl 2019, 64, 423–433. [Google Scholar] [CrossRef]
- Singh, V.K.; Singh, A.K.; Kumar, A. Disease management of tomato through PGPB: Current trends and future perspective. 3 Biotech 2017, 7, 255. [Google Scholar] [CrossRef]
- Goswami, D.; Thakker, J.N.; Dhandhukia, P.C. Portraying mechanics of plant growth promoting rhizobacteria (PGPR): A review. Cogent Food Agric. 2016, 2, 1127500. [Google Scholar] [CrossRef]
- Kannojia, P.; Choudhary, K.K.; Srivastava, A.K.; Singh, A.K. PGPR Bioelicitors: Induced Systemic Resistance (ISR) and Proteomic Perspective on Biocontrol. In PGPR Amelioration in Sustainable Agriculture; Elsevier: Amsterdam, The Netherlands, 2019; ISBN 978-0-12-816019-0. [Google Scholar]
- Islam, S.; Akanda, A.M.; Prova, A.; Islam, M.T.; Hossain, M.M. Isolation and Identification of Plant Growth Promoting Rhizobacteria from Cucumber Rhizosphere and Their Effect on Plant Growth Promotion and Disease Suppression. Front. Microbiol. 2016, 6, 1360. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Gui, Y.; Li, Z.; Jiang, C.; Guo, J.; Niu, D. Induced Systemic Resistance for Improving Plant Immunity by Beneficial Microbes. Plants 2022, 11, 386. [Google Scholar] [CrossRef] [PubMed]
- Lekired, A.; Cherif-Silini, H.; Silini, A.; Ben Yahia, H.; Ouzari, H.-I. Comparative genomics reveals the acquisition of mobile genetic elements by the plant growth-promoting Pantoea eucrina OB49 in polluted environments. Genomics 2023, 115, 110579. [Google Scholar] [CrossRef] [PubMed]
- Maheshwari, M.; Abulreesh, H.H.; Khan, M.S.; Ahmad, I.; Pichtel, J. Horizontal Gene Transfer in Soil and the Rhizosphere: Impact on Ecological Fitness of Bacteria. In Agriculturally Important Microbes for Sustainable Agriculture: Volume I: Plant-Soil-Microbe Nexus; Meena, V.S., Mishra, P.K., Bisht, J.K., Pattanayak, A., Eds.; Springer: Singapore, 2017; pp. 111–130. ISBN 978-981-10-5589-8. [Google Scholar]
- Sánchez-Salazar, A.M.; Taparia, T.; Olesen, A.K.; Acuña, J.J.; Sørensen, S.J.; Jorquera, M.A. An overview of plasmid transfer in the plant microbiome. Plasmid 2023, 127, 102695. [Google Scholar] [CrossRef] [PubMed]
- Ku, Y.-S.; Wang, Z.; Duan, S.; Lam, H.-M. Rhizospheric Communication through Mobile Genetic Element Transfers for the Regulation of Microbe–Plant Interactions. Biology 2021, 10, 477. [Google Scholar] [CrossRef]
- Popowska, M.; Krawczyk-Balska, A. Broad-host-range IncP-1 plasmids and their resistance potential. Front. Microbiol. 2013, 4, 44. [Google Scholar] [CrossRef]
- Bruto, M.; Prigent-Combaret, C.; Muller, D.; Moënne-Loccoz, Y. Analysis of genes contributing to plant-beneficial functions in plant growth-promoting rhizobacteria and related Proteobacteria. Sci. Rep. 2014, 4, 6261. [Google Scholar] [CrossRef]
- Ma, W.; Charles, T.C.; Glick, B.R. Expression of an exogenous 1-aminocyclopropane-1-carboxylate deaminase gene in Sinorhizobium meliloti increases its ability to nodulate alfalfa. Appl. Environ. Microbiol. 2004, 70, 5891–5897. [Google Scholar] [CrossRef]
- Serova, T.A.; Tikhonovich, I.A.; Tsyganov, V.E. Analysis of nodule senescence in pea (Pisum sativum L.) using laser microdissection, real-time PCR, and ACC immunolocalization. J. Plant Physiol. 2017, 212, 29–44. [Google Scholar] [CrossRef] [PubMed]
- Camerini, S.; Senatore, B.; Lonardo, E.; Imperlini, E.; Bianco, C.; Moschetti, G.; Rotino, G.L.; Campion, B.; Defez, R. Introduction of a novel pathway for IAA biosynthesis to rhizobia alters vetch root nodule development. Arch. Microbiol. 2008, 190, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Defez, R.; Esposito, R.; Angelini, C.; Bianco, C. Overproduction of Indole-3-Acetic Acid in Free-Living Rhizobia Induces Transcriptional Changes Resembling Those Occurring in Nodule Bacteroids. Mol. Plant Microbe Interact. 2016, 29, 484–495. [Google Scholar] [CrossRef] [PubMed]
- Defez, R.; Andreozzi, A.; Romano, S.; Pocsfalvi, G.; Fiume, I.; Esposito, R.; Angelini, C.; Bianco, C. Bacterial IAA-Delivery into Medicago Root Nodules Triggers a Balanced Stimulation of C and N Metabolism Leading to a Biomass Increase. Microorganisms 2019, 7, 403. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, F.X.; Brígido, C.; Glick, B.R.; Oliveira, S.; Alho, L. Mesorhizobium ciceri LMS-1 expressing an exogenous 1-aminocyclopropane-1-carboxylate (ACC) deaminase increases its nodulation abilities and chickpea plant resistance to soil constraints. Lett. Appl. Microbiol. 2012, 55, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, F.X.; Tavares, M.J.; Franck, J.; Ali, S.; Glick, B.R.; Rossi, M.J. ACC deaminase plays a major role in Pseudomonas fluorescens YsS6 ability to promote the nodulation of Alpha- and Betaproteobacteria rhizobial strains. Arch. Microbiol. 2019, 201, 817–822. [Google Scholar] [CrossRef]
- Alemneh, A.A.; Zhou, Y.; Ryder, M.H.; Denton, M.D. Mechanisms in plant growth-promoting rhizobacteria that enhance legume-rhizobial symbioses. J. Appl. Microbiol. 2020, 129, 1133–1156. [Google Scholar] [CrossRef]
- Orozco-Mosqueda, M.d.C.; Glick, B.R.; Santoyo, G. ACC deaminase in plant growth-promoting bacteria (PGPB): An efficient mechanism to counter salt stress in crops. Microbiol. Res. 2020, 235, 126439. [Google Scholar] [CrossRef]
- Alemneh, A.A.; Zhou, Y.; Ryder, M.H.; Denton, M.D. Is phosphate solubilizing ability in plant growth-promoting rhizobacteria isolated from chickpea linked to their ability to produce ACC deaminase? J. Appl. Microbiol. 2021, 131, 2416–2432. [Google Scholar] [CrossRef]
- Nukui, N.; Minamisawa, K.; Ayabe, S.-I.; Aoki, T. Expression of the 1-aminocyclopropane-1-carboxylic acid deaminase gene requires symbiotic nitrogen-fixing regulator gene nifA2 in Mesorhizobium loti MAFF303099. Appl. Environ. Microbiol. 2006, 72, 4964–4969. [Google Scholar] [CrossRef]
- Orozco-Mosqueda, M.d.C.; Duan, J.; DiBernardo, M.; Zetter, E.; Campos-García, J.; Glick, B.R.; Santoyo, G. The Production of ACC Deaminase and Trehalose by the Plant Growth Promoting Bacterium Pseudomonas sp. UW4 Synergistically Protect Tomato Plants Against Salt Stress. Front. Microbiol. 2019, 10, 1392. [Google Scholar] [CrossRef] [PubMed]
- Balla, A.; Silini, A.; Cherif-Silini, H.; Chenari Bouket, A.; Alenezi, F.N.; Belbahri, L. Recent Advances in Encapsulation Techniques of Plant Growth-Promoting Microorganisms and Their Prospects in the Sustainable Agriculture. Appl. Sci. 2022, 12, 9020. [Google Scholar] [CrossRef]
- Pereira, J.F.; Oliveira, A.L.M.; Sartori, D.; Yamashita, F.; Mali, S. Perspectives on the Use of Biopolymeric Matrices as Carriers for Plant-Growth Promoting Bacteria in Agricultural Systems. Microorganisms 2023, 11, 467. [Google Scholar] [CrossRef] [PubMed]
- Lobo, C.B.; Juárez Tomás, M.S.; Viruel, E.; Ferrero, M.A.; Lucca, M.E. Development of low-cost formulations of plant growth-promoting bacteria to be used as inoculants in beneficial agricultural technologies. Microbiol. Res. 2019, 219, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Zago, S.; Fonseca dos Santos, M.; Konrad, D.; Fiorini, A.; Rosado, F.; Missio, R.; Vendruscolo, E. Shelf Life of Azospirillum brasilense in Alginate Beads Enriched with Trehalose and Humic Acid. J. Agric. Sci. 2019, 11, 269. [Google Scholar] [CrossRef]
- Rohman, S.; Kaewtatip, K.; Kantachote, D.; Tantirungkij, M. Encapsulation of Rhodopseudomonas palustris KTSSR54 using beads from alginate/starch blends. J. Appl. Polym. Sci. 2020, 138, 50084. [Google Scholar] [CrossRef]
- Marcelino, P.R.F.; Milani, K.M.L.; Mali, S.; Santos, O.J.A.P.D.; de Oliveira, A.L.M. Formulations of polymeric biodegradable low-cost foam by melt extrusion to deliver plant growth-promoting bacteria in agricultural systems. Appl. Microbiol. Biotechnol. 2016, 100, 7323–7338. [Google Scholar] [CrossRef]
- Perez, J.J.; Francois, N.J.; Maroniche, G.A.; Borrajo, M.P.; Pereyra, M.A.; Creus, C.M. A novel, green, low-cost chitosan-starch hydrogel as potential delivery system for plant growth-promoting bacteria. Carbohydr. Polym. 2018, 202, 409–417. [Google Scholar] [CrossRef]
- Panichikkal, J.; Prathap, G.; Nair, R.A.; Krishnankutty, R.E. Evaluation of plant probiotic performance of Pseudomonas sp. encapsulated in alginate supplemented with salicylic acid and zinc oxide nanoparticles. Int. J. Biol. Macromol. 2021, 166, 138–143. [Google Scholar] [CrossRef]
- Jurić, S.; Đermić, E.; Topolovec-Pintarić, S.; Bedek, M.; Vinceković, M. Physicochemical properties and release characteristics of calcium alginate microspheres loaded with Trichoderma viride spores. J. Integr. Agric. 2019, 18, 2534–2548. [Google Scholar] [CrossRef]
- Panichikkal, J.; Puthiyattil, N.; Raveendran, A.; Nair, R.; Krishnankutty, R. Application of Encapsulated Bacillus licheniformis Supplemented with Chitosan Nanoparticles and Rice Starch for the Control of Sclerotium rolfsii in Capsicum annuum (L.) Seedlings. Curr. Microbiol. 2021, 78, 911–919. [Google Scholar] [CrossRef] [PubMed]
- De Gregorio, P.; Michavila, G.; Muller, L.; Borges, C.; Pomares, M.; Sá, E.; Pereira, C.; Vincent, P. Beneficial rhizobacteria immobilized in nanofibers for potential application as soybean seed bioinoculants. PLoS ONE 2017, 12, e0176930. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Dubey, A. Rhizosphere microbiome: Engineering bacterial competitiveness for enhancing crop production. J. Adv. Res. 2020, 24, 337–352. [Google Scholar] [CrossRef]
- Bano, S.; Wu, X.; Zhang, X. Towards sustainable agriculture: Rhizosphere microbiome engineering. Appl. Microbiol. Biotechnol. 2021, 105, 7141–7160. [Google Scholar] [CrossRef] [PubMed]
- Haskett, T.L.; Tkacz, A.; Poole, P.S. Engineering rhizobacteria for sustainable agriculture. ISME J. 2021, 15, 949–964. [Google Scholar] [CrossRef] [PubMed]
- Ryu, M.-H.; Zhang, J.; Toth, T.; Khokhani, D.; Geddes, B.A.; Mus, F.; Garcia-Costas, A.; Peters, J.W.; Poole, P.S.; Ané, J.-M.; et al. Control of nitrogen fixation in bacteria that associate with cereals. Nat. Microbiol. 2020, 5, 314–330. [Google Scholar] [CrossRef] [PubMed]
- Wexler, M.; Gordon, D.; Murphy, P.J. The distribution of inositol rhizopine genes in Rhizobium populations. Soil Biol. Biochem. 1995, 27, 531–537. [Google Scholar] [CrossRef]
- Urra, J.; Alkorta, I.; Mijangos, I.; Garbisu, C. Commercial and farm fermented liquid organic amendments to improve soil quality and lettuce yield. J. Environ. Manag. 2020, 264, 110422. [Google Scholar] [CrossRef]
- Rehberger, E.; West, P.C.; Spillane, C.; McKeown, P.C. What climate and environmental benefits of regenerative agriculture practices? an evidence review. Environ. Res. Commun. 2023, 5, 052001. [Google Scholar] [CrossRef]
- Wei, Z.; Ying, H.; Guo, X.; Zhuang, M.; Cui, Z.; Zhang, F. Substitution of Mineral Fertilizer with Organic Fertilizer in Maize Systems: A Meta-Analysis of Reduced Nitrogen and Carbon Emissions. Agronomy 2020, 10, 1149. [Google Scholar] [CrossRef]
- Sujatha, M.P.; Lathika, C.; Smitha, J.K. Sustainable and efficient utilization of weed biomass for carbon farming and productivity enhancement: A simple, rapid and ecofriendly approach in the context of climate change scenario. Environ. Chall. 2021, 4, 100150. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).