Abstract
Panicle length (PL) is an important trait closely related to rice yield. More than 200 quantitative trait loci (QTL) for PL have been identified, but only a few can be used for breeding. Dongxiang wild rice contains many excellent genes, and mining favorable PL-related QTL from DXWR is helpful for rice variety improvement. Here, we report a QTL analysis for PL using a recombinant inbred line population consisting of 143 individuals derived from a cross between Dongxiang wild rice and indica cultivar Guangluai 4. A total of four QTL (qPL1-37, qPL4-26, qPL7-25, and qPL8-4) for PL were identified and located on chromosomes 1, 4, 7, and 8. Among them, qPL7-25 showed the largest F-value of 32.32 and 16.80, and the QTL explained 18.66% and 13.06% of the phenotypic variation of Dongxiang wild rice in Hangzhou and Hainan, respectively. QTL mapping was performed using a population of 1800 individuals derived from the crossing of NIL-qPL7-25 and GLA4. qPL7-25 was located between two InDel markers, InDel-24591 and InDel-24710, in a 119 kb region containing 14 predicted genes. Using Sanger sequencing and qRT-PCR analysis, we propose that LOC_Os07g41200 is probably a new allele of the well-known GL7 gene, which affects grain length and appearance quality in rice. These results provide new insights into the use of molecular marker-assisted selection for breeding high-yielding and high-quality rice varieties.
1. Introduction
Food security is a top priority for most countries. Rice is the staple food for more than 60% of the population in China, so improving rice production is crucial to ensuring national food security and is the goal of rice breeding. Rice yield is composed of the effective number of panicles per plant, the number of filled grains per panicle, the seed setting rate, and the kilogram grain weight. As a yield-related trait, panicle length (PL) directly determines the number and length of panicle branches in rice, which in turn affects the number of grains in the panicle [1,2]. Traditional rice breeding has made great strides, but there are also drawbacks of inefficiency, and molecular design breeding technology is beginning to show great potential for application, with mining for favorable alleles being the prerequisite for molecular design breeding.
Common wild rice (Oryza rufipogon Grff.) can provide favorable gene variants for many important agronomic traits in improved cultivated rice varieties [3,4,5,6,7,8,9,10]. Mining and exploiting favorable genes in O. rufipogon could be an effective way to overcome yield stagnation in cultivated rice. Dongxiang wild rice (DXWR) is the most northerly distributed O. rufipogon resource in the world, with many excellent characteristics and rich genetic diversity. Compared with other wild rice, DXWR is a close ancestor of cultivated rice, which is suitable for use as an excellent germplasm resource for cultivating rice genetic improvement. Previous studies have shown that DXWR could provide many favorable genes controlling desirable traits such as high yield [11], cold tolerance [12], drought tolerance [13], and cytoplasmic male sterility [14]. Panicle traits are essential for yield and quality formation; the cloning of favorable panicle architecture genes from DXWR can provide new genetic resources for the high yield and quality breeding of rice, and introducing these favorable genes into the cultivated rice varieties by molecular breeding techniques can accelerate the breeding and use of DXWR.
Panicle traits in rice include the PL, primary branch number (PBN), secondary branch number (SBN), spikelets per panicle (SPP), and grain size. PL is a crucial determinant of panicle architecture, affecting rice yield and quality [15]. Therefore, PL can be used as a criterion for yield improvement breeding. More than 250 quantitative trait loci (QTL) for PL have been mapped and are located across all of the 12 rice chromosomes, but only a few of them have been cloned and used in breeding practice. In addition, some important QTLs for high-yielding traits have also been cloned and applied in rice genetic improvement, such as Grain number 1a (Gn1a), DENSE AND ERECT PANICLE1 (DEP1), Ideal Plant Architecture 1 (IPA1), Grain number, plant height and heading date 7 (Ghd7), Ghd8, grain size 3 (GS3), grain weight 2 (GW2), and ABERRANT PANICLE ORGANIZATION 1 (APO1). The characterization of these genes that supports molecular design breeding and pyramiding beneficial alleles by functional marker-assisted selection is of great help in increasing rice yield [16,17,18].
Rice PL is a quantitative trait controlled by multiple genes with low heritability, and some genes regulating other panicle traits may also affect PL. Based on the functions of the cloned genes, these genes regulating PL can be classified into the following categories: (1) Several genes/QTL regulate PL by modulating the hormone metabolism, such as ELONGATED UPPERMOST INTERNODE1 (EUI1), LONELY GUY (LOG), DEP1, Large Panicle (LP)/Erect Panicle 3 (EP3), Oryza sativa PIN-FORMED 5b (OsPIN5b), HOMEOBOX12 (HOX12), Oryza sativa GROWTH-REGULATING FACTOR4 (OsGRF4)/PT2, Short Panicle (SP3), and semidwarf-1 (sd1) [19,20,21,22,23,24,25,26,27]. (2) Some genes control PL by affecting the heading date, such as Ghd7, Ghd8, and Heading date quantitative trait locus7.2 (qHD7.2) [28,29,30]. (3) Other genes are responsible for PL by regulating the cell wall components and nutrients required for growth, including APO1, LARGE2, DEP3, Oryza sativa Curled leaf and Dwarf mutant1 (OsCD1), and Oryza sativa Arginine (OsARG) [31,32,33,34,35,36,37]. In addition, other genes such as SP1, IPA1, DEP2, Aberrant spikelet and panicle1 (ASP1), SHORT GRAIN1 (SG1), and TAWAWA1 are involved in the regulation of PL development [38,39,40,41,42,43,44]. Grain length on chromosome 7 (GL7) is a major QTL controlling grain length and grain width in rice, and Wang et al. (2015) found that tandem duplication of a 17.1-kb segment at the GL7 locus led to the upregulation of GL7 and downregulation of its nearby negative regulator, resulting in increased grain length and improved grain appearance quality [45]. Slender grain on chromosome 7 (SLG7) was allelic to GL7 and GW7 [45,46], highly expressed alleles of SLG7/GL7/GW7 produced slender grains with low chalkiness, and the PL of near-isogenic line (NIL)-SLG7 was 14.2% longer than that of its background parent, 9311 [47]. APO1 is an important panicle architecture gene that can directly interact with APO2 and positively regulate the PBN and SPP [48]. The weak allele STRONG CULM2 (SCM2) carrying APO1 can increase culm strength, which is useful for breeding applications [32,49]. The above results suggest that several factors, such as plant hormones, cell wall components, and heading date, are involved in the genetic regulation of PL.
In this study, a high-density single-nucleotide polymorphism (SNP) linkage map using specific locus-amplified fragment (SLAF) markers was constructed in the recombinant inbred line (RIL) population consisting of 143 individuals derived from a cross between DXWR and indica cultivar Guangluai 4 (GLA4) to detect the QTL associated with PL. The major QTL qPL7-25 was stably detected on chromosome 7 in Hangzhou (HZ) and Hainan (HN) and was restricted to a 119 kb region between InDel-24591 and InDel-24710. Our results revealed that qPL7-25 was probably a new allele of GL7, which can be used to improve the breeding of elite rice varieties.
2. Materials and Methods
2.1. Plant Materials
An RIL population was developed from a single seed offspring of a backcross (BC1F1) of DXWR as the donor and GLA4 as the recurrent parent. This population consisted of 143 lines backcrossed with GLA4 as the parent for one generation and then self-crossed for 12 generations. The parental lines DXWR and GLA4, together with the RIL population, were planted as a plot in the experimental field of the China National Rice Research Institute in Hangzhou (119.54′ E, 30.04′ N, May to October 2014) and Hainan (110.01′ E, 18.30′ N, December 2014 to May 2015). Each plot consisted of four rows separated by 20 cm, with each row consisting of ten plants, each separated from its neighbor by 20 cm. All experimental plots had uniform fertility and medium fertilizer application, and field management was carried out according to general field cultivation techniques.
2.2. Measurement of Traits
To find the relationships between PL and other agronomic traits, ten and five plants from two parents and 143 lines, respectively, were harvested at the maturity stage to measure the plant height (PH), PL, PBN, SBN, and SPP. PH was measured as the distance from the ground to the tip of the main panicle. PL was measured as the distance from the neck node to the tip of the main panicle using a ruler. More than 100 fully filled grains were used to measure seed length, width and length/width ratio using a seed phenotyping system (Wan Sheng, Hangzhou, China). The 1000-grain weight was measured by weighing 1000 full-filled grains with an electronic balance with three replicates. The mean values of all the characters measured were used for the analysis.
2.3. Linkage Map Construction and QTL Analysis
The SLAF sample preparation, sequencing, sequence comparison, polymorphic analysis, and associated marker identification were processed as previously reported [50]. Briefly, two restriction enzymes (Hae III and Hpy 166II) were selected for their uniform distribution and prevalence in the simulations of fragment alignments to the NPB reference genome. Arabidopsis thaliana was used as a control genome to verify the accuracy of the restriction digestion protocol. Fragments of 200–350 bp were isolated to be used as the SLAF tags. The fragments were sequenced on the Illumina HiSeq 2500 system. The SLAFs were grouped into 12 linkage groups with their positions in the reference genome. The modified logarithm of odds scores (MLOD) value between two adjacent markers was determined [51], and SLAFs with MLOD values less than 5 were filtered out. QTL analysis was performed using the QTL Network 2.0 software based on a mixed linear model with all the genotype data from the RIL mapping population. The contribution rates and additive effects of each QTL to the PL were calculated, and the detected QTL were named according to the method proposed by McCouch et al. [52]. If the additive effect value was positive, then the allele was from the DXWR parent, and if the additive effect value was negative, then the allele was from the GLA4 parent.
2.4. Construction of NIL
A VB431 line of RILs with the DXWR genotype was selected for backcrossing with the recurrent parent GLA4. The F1 generation was then backcrossed with GLA4, combined with the PL phenotype of the progeny, and two InDel markers (InDel-24091 and InDel-25971) on either side of qPL7-25 were used for marker-assisted selection (MAS) of each generation. After three consecutive backcrosses and one self-cross, a BC3F2 population containing 100 individuals with a GLA4 genetic background was isolated and used for primary mapping of qPL7-25 (Figure S2A). The NIL-qPL7-25 contains 1.88 Mb of the qPL7-25 locus from a fragment of DXWR located on chromosome 7 between the InDel markers InDel-24091 and InDel-25971 (Figure S2B). Rice variety P13 with the GL7 gene was used as the male, the restoring line Huazhan as the female, and the F1 generation was then backcrossed with recurrent parent Huazhan. A pair of dominant functional markers (NGL7-F: TGACACGCCACAGTCCAAGACGAGCAGT, NGL7-R: AAGGGAGTTGAGAGTAGAAAAAA) was used for MAS of each generation. After four consecutive backcrosses and one self-cross, a BC4F2 population was obtained. An NIL carrying a homozygous allele of P13 in the target QTL region between markers NGL7-F and NGL7-R, designated NIL-Huazhan-GL7p13, was also developed on a Huazhan background.
2.5. QTL Mapping and Statistical Analysis
QTL analysis was performed with the QTLNetwork2.0 software using the mixed linear models [53]. The VB431 line was crossed with the GLA4 variety to develop a mapping population. The F2 population was constructed by self-crossing of the F1 hybrid. A primary linkage of the QTLs for PL was obtained using 100 recessive plants from the F2 population. According to the resequencing and alignment results of GLA4 and DXWR, the InDel loci were identified on chromosome 7. Primers were designed using Vector NTI 11.5 biological software with the following design parameters: GC % 40–55%, base length 19–30 bp, and Tm value 55–60 °C. Seven new InDel markers were developed for fine mapping of the qPL7-25 locus (Table S1), and the qPL7-25 locus was mapped to the interval between InDel-24591 and InDel-24710 using a population of 1800 individuals derived from the crossing of NIL-qPL7-25 and GLA4. All statistical analyses were performed using Student’s t-test (*, p < 0.05; **, p < 0.01).
2.6. RNA Extraction and QRT-PCR Analysis
Total RNA was isolated from 3 cm young panicles of DXWR and GLA4 plants according to the manufacturer’s instructions of the AxyPerp Multisource Total RNA miniprep kit (Corning Life Sciences (WuJiang) Co., Ltd, Suzhou, China). DNAase-treated RNA (1 μg) was reverse transcribed using a ReverTra Ace qPCR RT master mix (FSQ-301, Toyobo, Osaka, Japan). qRT-PCR was performed using GL7 gene-specific primers, as described by Wang et al. [45], using the SYBR Green Real-Time-PCR master mix (QPK-201, Toyobo) and the Bio-Rad (Hercules, CA, USA) CFX96 Real Time-PCR system. Ubiquitin was used as an internal standard, and the results were calculated using the 2−ΔΔCT method [54].
3. Results
3.1. Phenotypic Data of Panicle Traits of Two Parents and RIL Population
The phenotypes of the DXWR and GLA4 parents were relatively different. The PH of GLA4 in HZ and HN was 54.09% and 64.66% of that of DXWR, respectively. The PL of DXWR was 23.7 cm and 22.1 cm, while that of GLA4 was 21.2 cm and 20.0 cm in HZ and HN, respectively, and was significantly shorter than that of DXWR (Table 1). The grain length and grain length/width ratio of DXWR were significantly greater than those of GLA4, but the grain width and 1000-grain weight were significantly lower than those of GLA4 (Figure S1). In addition, the PBN and SBN of DXWR were significantly higher than those of GLA4, but there was no significant difference in the SPP between the parents (Figure 1, Table 1). The skewness and kurtosis values of the PL in HZ and HN were both less than 1, indicating that the PL was normally distributed and was a polygene-controlled quantitative trait. The data obtained for the PL in the RIL population conformed to a continuous normal distribution with a wide range (Figure 2). In addition, we identified significant positive correlations between PL pairs, the PBN, SBN, and SPP in both HZ and HN (Table 2).

Table 1.
Data collection of panicle traits of DXWR, GLA4, and their RIL population.

Figure 1.
Plant and panicle of GLA4 and DXWR at the mature stage. (A) Outward appearance comparison of the plants, scale bar = 20 cm. (B) Outward appearance comparison of the panicles, scale bar = 3 cm.
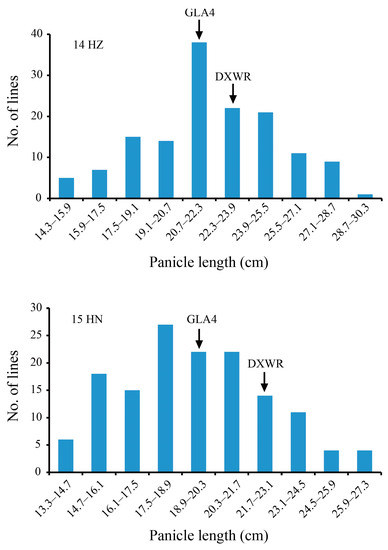
Figure 2.
Frequency distribution of panicle length in recombinant inbred line populations in Hangzhou and Hainan. DXWR, Dongxiang wild rice; GLA4, Guangluai 4; HZ, Hangzhou; HN, Hainan.

Table 2.
Correlation analysis of the panicle traits in Hangzhou and Hainan recombinant inbred lines.
3.2. QTL Analysis of PL
The 368 polymorphic SNP markers were evenly distributed among the 12 chromosomes and were used to construct a molecular linkage map with the SLAF markers. QTL analysis was performed using QTLNetwork2.0 software based on a mixed linear model, and p = 0.005 was used as the statistical detection threshold. A total of four QTL were detected for the PL and were distributed on chromosomes 1, 4, 7, and 8. Among them, qPL1-37, qPL4-26, and qPL7-25 were detected repeatedly in both HZ and HN, but qPL8-4 was detected only in HN (Figure 3, Table 3). qPL1-37 was located in the marker M37-M38 on chromosome 1, with a physical distance of 1.0 Mb and an F-value of 17.13 and 9.42, and the QTL explained 8.99% and 7.31% of the phenotypic variation in HZ and HN, respectively. qPL4-26 was located in the marker M142-M143 on chromosome 4 with a physical distance of 1.0 Mb and an F-value of 18.77 and 10.28, and the QTL explained 12.92% and 7.48% of the phenotypic variation in HZ and HN, respectively. qPL7-25 was located in the marker M234-M235 on chromosome 7 with a physical distance of 1.0 Mb and an F-value of 32.32 and 16.80, and the QTL explained 18.66% and 13.06% of the phenotypic variation in HZ and HN, respectively. qPL8-4 was located in the marker M242-M243 on chromosome 8 with a physical distance of 1.2 Mb and an F-value of 10.25, and the QTL explained 6.17% of the phenotypic variation in HN. The enhanced alleles qPL1-37, qPL7-25, and qPL8-4 were from the DXWR parent (Table 3). As the PBN and SBN of DXWR were significantly different from GLA4, we also identified other QTL related to the panicle traits, and three QTL (qPBN1-8, qPBN2-4, and qPBN7-25) for the PBN and five QTL (qSBN1-2, qSBN1-5, qSBN1-6, qSBN5-22, and qSBN7-20) for the SBN were also detected. qPBN1-8 and qPBN2-4 were repeatedly detected in both HZ and HN, but other QTL were only detected in HZ or HN. Notably, qPBN7-25 was mapped to the same interval as qPL7-25 on chromosome 7 (Figure 3, Table 3).
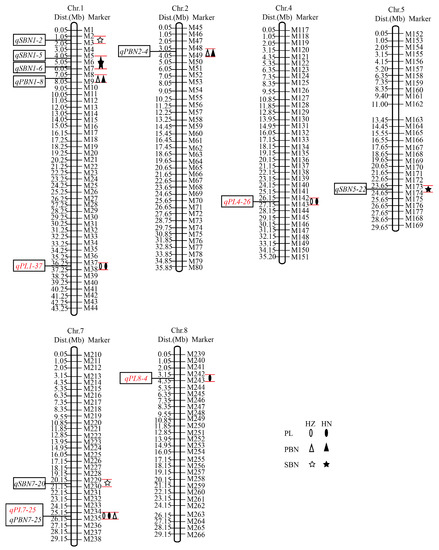
Figure 3.
Location of the quantitative trait loci for panicle traits on the single-nucleotide polymorphism map. The number indicates the physical distance (Mb) along each chromosome. Abbreviations: PL, panicle length; PBN, primary branch number; SBN, secondary branch number; HZ, Hangzhou; HN, Hainan.

Table 3.
Quantitative trait loci for panicle length were detected in recombinant inbred line populations in Hangzhou and Hainan.
3.3. Fine Mapping of qPL7-25
As qPL7-25 showed the greatest potential to increase the PL, we focused on determining its underlying gene using a map-based cloning strategy. The VB431 line from the RILs with the DXWR genotype in the qPL7-25 region was selected to backcross with the recurrent GLA4 parent to construct the near-isogenic line (NIL-qPL7-25). The phenotypic characteristics were measured in the F2 population, which included 100 individuals derived from a BC3F1 line with a GLA4 genetic background (Figure S2A). By combining the genotype and phenotype of individuals, the QTL was mapped primarily between the two insertion-deletion (InDel) markers, InDel-24091 and InDel-25971, at 1.88 Mb intervals (Figure S2B). To delineate the genomic region of the qPL7-25, InDel markers were designed from the re-sequenced genome sequences and tested to predict the probability of polymorphism between the NIL-qPL7-25 line and the GLA4 cultivar. In the end, seven InDel markers were successfully developed (Table S1). Genotyping of all recombinant genes was performed using seven polymorphic markers, and the qPL7-25 was located between the two InDel markers, InDel-24591 and InDel-24710 on chromosome 7, with an interval of 119 kb, using a population of 1800 recessive plants derived from the crossing of NIL-qPL7-25 and GLA4 (Figure 4A). The target region contains 14 predicted genes based on the Rice Genome Annotation Project website (http://rice.plantbiology.msu.edu/), accessed on 10 August 2020 (Figure 4B, Table S2).
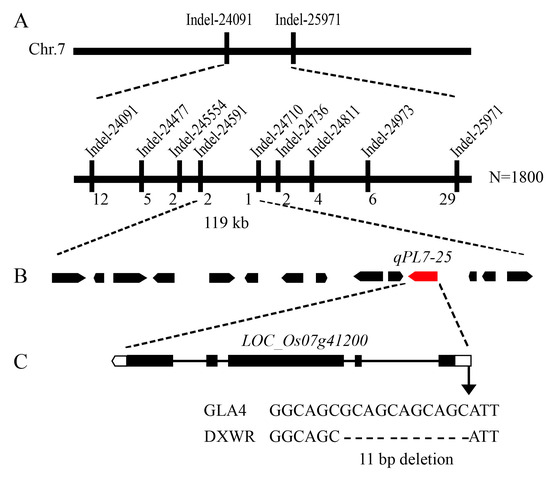
Figure 4.
Fine mapping of qPL7-25. (A) qPL7-25 was narrowed down to a 119 kb interval on chromosome 7, defined by the InDel markers InDel-24591 and InDel-24710. The numbers below the bars in the panel indicate the number of recombinant lines between each adjacent marker. (B) The positions of the 14 predicted genes in the target region. (C) The structure and allelic variation of the candidate gene LOC_Os07g41200/GL7. Introns are shown as lines, exons as black boxes, and the white boxes indicate non-coding regions. Black lines indicate deleted nucleotides. DXWR, Dongxiang wild rice; GLA4, Guangluai 4.
3.4. Candidate Gene Analysis of qPL7-25
Among these 14 predicted genes (Table S2), LOC_Os07g41100 encodes a conserved hypothetical protein, LOC_Os07g41110 encodes a retrotransposon protein, and LOC_Os07g41170 encodes an expressed protein. We used Genevestigator (https://genevestigator.com accessed on 10 August 2020) software to analyze the temporal and spatial expression of the remaining 11 genes. LOC_Os07g41220 has no expression data in this software, LOC_Os07g41150 showed pollen-specific expression, LOC_Os07g41180, and LOC_Os07g41190 were highly expressed in root tips, and LOC_Os07g41230 was a cloned gene related to disease resistance in rice. It is unlikely that the above genes were candidate genes. LOC_Os07g41090, LOC_Os07g41120, LOC_Os07g41140, and LOC_Os07g41200 were highly expressed in the panicle, LOC_Os07g41200 (GL7/GW7) was a previously cloned major QTL, controlling the grain length and width [45,46], and its expression showed a positive correlation with the critical period of spike development, both in terms of timing and tissue specificity (Figure S3A). In addition, NIL-Huazhan-GL7P13 and NIL-qPL7-25 also showed increased grain length compared to Huazhan and GLA4, respectively (Figure S3B). Therefore, we propose that LOC_Os07g41200/GL7 could be a candidate gene for qPL7-25.
To further confirm that LOC_Os07g41200/GL7 could be a candidate gene for qPL7-25, we sequenced and analyzed the coding sequence and promoter region of GL7 in the low GL7-expressing variety, Nipponbare (NPB) and the high GL7 expressing variety, landrace Ping13 (P13), DXWR, and GLA4. There were no SNP differences resulting in amino acid changes in the coding region of DXWR and GLA4, but there were SNP and InDel differences in the 5′ untranslated region (UTR) and promoter, especially in the region from -111 to -101 (transcription start site, +1), with an 11 bp deletion in the GL7 promoter of DXWR compared to GLA4, and P13 had an 8 bp deletion. In contrast, the sequence of GLA4 was similar to that of NPB (Figure 4C, Figure S4). This 11 bp deletion of GL7 was expressed 2.4-fold higher than GLA4 in young panicles of DXWR, as determined by qRT-PCR assays (Figure 5). These results suggest that this deletion could enhance the function of GL7 and thus affect the PL in rice. The GL7 gene from P13 was introduced into the indica rice variety “Huazhan” to construct NIL-Huazhan-GL7P13. The PH, PL, PBN, grain length, and grain length/width ratio of NIL-Huazhan-GL7P13 were significantly increased compared to Huazhan (Table S3), which was consistent with the comparative trend between NIL-qPL7-25 and GLA4 phenotypes (Table S4).
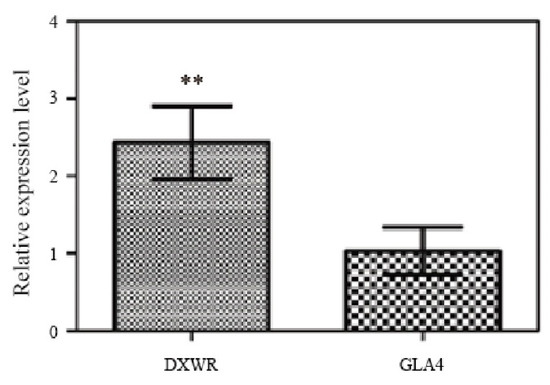
Figure 5.
Expression analysis of GL7 in young panicles of Dongxiang wild rice (DXWR) and Guangluai 4 (GLA4) by qRT-PCR; the rice’s Ubiquitin gene was used as an internal control. Data indicate means ± SD. (n = 3 biological replicates). ** indicates a significant difference between DXWR and GLA4 at the 0.01 levels, according to the t-test. DXWR, Dongxiang wild rice; GLA4, Guangluai 4.
We performed a haplotype network analysis of the GL7 gene promoter region and the 5′ UTR region according to the RiceVarMap V2.0 website (http://ricevarmap.ncpgr.cn/v2/), accessed on 15 October 2020 [55,56]. The results showed that NPB is type I, P13 is type II, DXWR is closest to type III, and GLA4 is closer to type IV (Figure 6). There are 1047 rice varieties in type I, and these include mainly temperate Japonica rice (547 haplotypes), some IndI (164 haplotypes), and AUS rice (106 haplotypes), as well as several other rice subspecies. Type II rice had 1010 haplotypes, including P13, tropical Japonica rice (471 haplotypes), and mixed Japonica (214 haplotypes). Type III had 1964 haplotypes and contained 882 IndIII Indica haplotypes, 353 IndII Indica haplotypes, and 443 mixed Indica haplotypes. Type IV rice contained 673 haplotypes and was dominated by IndI rice with 393 haplotypes and mixed indica with 216 haplotypes. As seen above, the promoter region and the 5′ UTR region of the GL7 gene are evolutionarily distinct, and different SNP haplotypes were derived from each of the Japonica and Indica rice species and are closely related to each rice subtype. The NPB and P13 belong to different haplotypes, and the GL7 genes of both have significantly different regulatory roles in grain length. DXWR and GLA4 also belong to different haplotypes, and their GL7 genes may also have similar differences leading to changes in the PL, and this type of difference may be widespread in indica rice with different typing. In conclusion, the LOC_Os07g41200 gene in DXWR can be tentatively identified as the candidate gene for qPL7-25 and is a new GL7 allele.
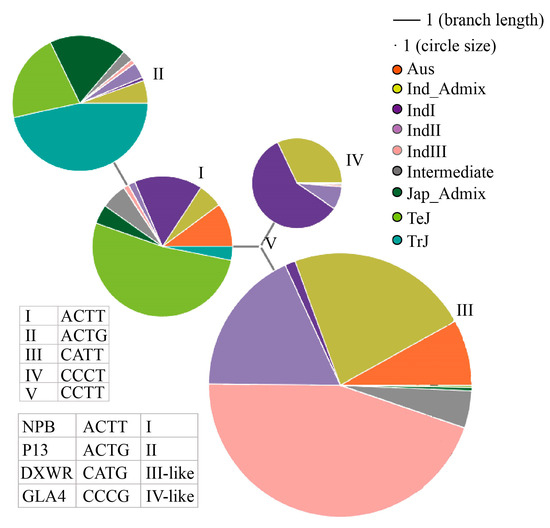
Figure 6.
Haplotype network analysis results using the single-nucleotide polymorphisms from the promoter and 5′ untranslated region of the GL7 gene.
4. Discussion
PL is an important trait related to rice yield, and breeding long and large panicle rice varieties is an effective method to achieve higher yields. Therefore, the study of the PL QTL has important and far-reaching implications for improving rice yield and quality. The QTL of PL are controlled by multiple genes and influenced by environmental factors, which are difficult to map in individual plants [57]. Although a number of QTL affecting the rice PL have been localized by previous studies, few of them have been cloned, especially those located on chromosome 7. qPL6, LP1, qPL7, and qpl9 are some finely mapped QTL directly related to PL [30,58,59]. In this study, qPL7-25 was localized on chromosome 7 between InDel-24591 and InDel-24710 in a 119 kb interval (Figure 4A), which did not overlap with the previously reported QTL/gene position of PL. Therefore, qPL7-25 should be considered as a new PL QTL and as an enhanced allele from the DXWR parents. Previous studies have shown that the change in PL can be accompanied by a change in other panicle traits. Loss of function of SP1, SP3, and DEP3 not only shortened the PL but also reduced the number of branches and SPP [26,35,38]. DEP1, OsAPC6, and PT2 mutations result in reduced PL and grain size [21,25,60]. In our study, qPBN7-25, a QTL for PBN, was mapped to the same interval as qPL7-25 on chromosome 7, and both were potential alleles of the QTL from DXWR (Figure 3). Phenotypic analysis showed that both NIL-qPL7-25 and NIL-Huazhan-GL7P13 had more PBN than their parents (Tables S3 and S4). Our results were consistent with previous reports, and we propose that these two QTL were single-causal multiple effects of the same QTL/gene. Compared with other common wild and cultivated rice, DXWR has more abundant rare allelic variants (including SNPs and InDels), but many QTLs coexist with unfavorable genes and are masked. Gene editing technology can be used to quickly and efficiently break the tight linkage between favorable and unfavorable genes [61], thus enabling better and faster use of the favorable genes from DXWR.
There were 14 candidate genes in the fine mapping interval of qPL7-25 (Figure 4B, Table S2); LOC_Os07g41200 is a well-known cloned gene of GL7, which controls the grain length. Previous studies have shown that overexpression of GL7 in NPB and ZF802 (Zhefu802) backgrounds, and SLG7 in the NPB background, can increase the grain length and length/width ratio, and upregulation of GL7/GW7/SLG7 expression increases the cell length and decreases the cell width for epidermal cells of the outer and inner glumes [45,46,47]. We hypothesized that this cell division pattern is consistent with panicle elongation in the direction that leads to increased PL, and thus increased GL7 expression promotes increased PL. NIL-SLG7 showed a higher level of expression in the panicle and produced longer panicles (+14.2%) compared to NIL-slg7 [47], confirming our hypothesis. In addition, there were 14 bp and 36 bp deletions in the promoter of slg7 mutants constructed using CRISPR/Cas9, which resulted in an increased expression level of SLG7 and produced more slender grains compared with WYJ30 [62]. Sequence alignment analysis showed that there was also an 11 bp deletion in the GL7 promoter of DXWR compared with GLA4, and the expression level of GL7 was 2.4-fold higher in DXWR than in GLA4 performed by qRT-PCR analysis (Figure S4 and Figure 5). Unfortunately, we subsequently lost the seeds of NIL-qPL7-25 due to improper storage, so we could not provide data on the expression level of the GL7 gene in the young panicles of NIL-qPL7-25. However, another experiment provided strong support for our analysis: when we transferred GL7 from P13 (a long grain, long panicle, GL7 high expressing variety) to the relatively short panicle, short-grained Huazhan variety by backcrossing using MAS, the NIL-Huazhan-GL7P13 showed a significant increase in the PL, grain length, and grain length/width ratio, as well as a significant increase in the PBN compared to the Huazhan parent (Table S3). This showed a consistent trend with the comparison between the phenotypes of NIL-qPL7-25 and GLA4 (Table S4). Our results were consistent with the previous studies. In addition, DXWR and GLA4 belong to different haplotypes in the promoter and 5′ UTR regions of the GL7 gene (Figure 6). In view of the above, we propose that qPL7-25 is a new allele of GL7 with a greater breeding utilization value in improving rice quality rather than increasing rice yield. We are currently introducing GL7 into the promoted varieties to simultaneously improve the yield and quality of existing varieties through gene polymerization breeding.
In addition, the parents and RIL populations in this study were re-sequenced when the QTL was first located. Although the cost was high, a large amount of SNP and InDel information was obtained. Combined with the biological information analysis, it provides convenience for our subsequent future research and provides a good experimental basis for the further fine mapping or cloning of QTL genes on other chromosomes.
5. Conclusions
In this study, four PL QTL (qPL1-37, qPL4-26, qPL7-25, and qPL8-4) were detected using an RIL population consisting of 143 individuals derived from a cross between DXWR and GLA4. Among these, qPL7-25 showed great potential to increase the PL, which was further localized to a 119 kb region on chromosome 7 by a map-based cloning strategy using 1800 individuals derived from a cross between NIL-qPL7-25 and GLA4. Sequence alignment and qRT-PCR analysis suggested that qPL7-25 was probably a new allele of GL7, a major QTL regulating grain length. The introduction of qPL7-25 into the population varieties will help to improve the yield and quality of the existing varieties.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agriculture13081623/s1, Figure S1: The morphology and grain size comparisons of mature grains from GLA4 and DXWR; Figure S2: Construction of NIL and linkage map of qPL7-25 on chromosome 7 in the NIL; Figure S3: Expression patterns of candidate genes; Figure S4: Sequence alignment of promoter region and 5’ UTR region of GL7 gene; Table S1: InDel primers used in fine mapping of qPL7-25; Table S2: Predicted ORF on RGAP; Table S3: Data collection of main agronomic traits of Huazhan and NIL-Huazhan-GL7P13 in Hangzhou.; Table S4: Data collection of main agronomic traits of GLA4 and NIL-qPL7-25 in Hangzhou.
Author Contributions
Conceptualization, Y.M. and Y.W.; methodology, S.L., Z.W., P.H. and R.Y.; software, P.D. and Z.W.; validation, S.L. and Z.W.; formal analysis, C.L. (Chenxi Luo) and S.L.; investigation, M.T. and C.L. (Caolin Lu); resources, Y.M.; data curation, Y.M., S.L. and Y.R.; writing—original draft preparation, S.L., Y.R. and P.D.; writing—review and editing, S.L., Y.R., P.D. and Y.W.; supervision, Y.M. and Y.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Natural Science Foundation of Zhejiang Province (Grant No. LQ21C130004), the Zhejiang Provincial Natural Science Outstanding Youth Fund (Grant No. LR20C130001), Hainan Yazhou Bay Seed Lab (Grant No. B21HJ0219), and the Central Public-interest Scientific Institution Basal Research Fund (Grant No. CPSIBRF-CNRRI-202109).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The datasets presented in this study can be found in the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cho, Y.G.; Kang, H.J.; Lee, J.S.; Lee, Y.T.; Mccouch, S.R. Identification of quantitative trait loci in rice for yield, yield components, and agronomic traits across years and locations. Crop Sci. 2007, 47, 2403–2417. [Google Scholar] [CrossRef]
- Marathi, B.; Guleria, S.; Mohapatra, T.; Parsad, R.; Mariappan, N.; Kurungara, V.; Atwal, S.; Prabhu, K.; Singh, N.; Singh, A. QTL analysis of novel genomic regions associated with yield and yield related traits in new plant type based recombinant inbred lines of rice (Oryza sativa L.). BMC Plant Biol. 2012, 12, 137. [Google Scholar] [CrossRef] [PubMed]
- Moncada, P.; Martinez, C.P.; Borrero, J.; Chatel, M.; Gauch, H.; Guimaraes, E.; Tohme, J.; McCouch, S.R. Quantitative trait loci for yield and yield components in an Oryza sativa × Oryza rufipogon BC2F2 population evaluated in an upland environment. Theor. Appl. Genet. 2001, 102, 41–52. [Google Scholar] [CrossRef]
- Septiningsih, E.M.; Prasetiyono, J.; Lubis, E.; Tai, T.H.; Tjubaryat, T.; Moeljopawiro, S.; McCouch, S.R. Identification of quantitative trait loci for yield and yield components in an advanced backcross population derived from the Oryza sativa variety IR64 and the wild relative O. rufipogon. Theor. Appl. Genet. 2003, 107, 1419–1432. [Google Scholar] [CrossRef]
- Septiningsih, E.M.; Trijatmiko, K.R.; Moeljopawiro, S.; McCouch, S.R. Identification of quantitative trait loci for grain quality in an advanced backcross population derived from the Oryza sativa variety IR64 and the wild relative O. rufipogon. Theor. Appl. Genet. 2003, 107, 1433–1441. [Google Scholar] [CrossRef]
- Thomson, M.J.; Tai, T.H.; McClung, A.M.; Lai, X.H.; Hinga, M.E.; Lobos, K.B.; Xu, Y.; Martinez, C.P.; McCouch, S.R. Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor. Appl. Genet. 2003, 107, 479–493. [Google Scholar] [CrossRef]
- Nguyen, B.D.; Brar, D.S.; Bui, B.C.; Nguyen, T.V.; Pham, L.N.; Nguyen, H.T. Identification and mapping of the QTL for aluminum tolerance introgressed from the new source, Oryza rufipogon Griff., into indica rice (Oryza sativa L.). Theor. Appl. Genet. 2003, 106, 583–593. [Google Scholar] [CrossRef]
- Marri, P.R.; Sarla, N.; Reddy, L.V.; Siddiq, E.A. Identification and mapping of yield and yield related QTHN from an Indian accession of Oryza rufipogon. BMC Genet. 2005, 6, 33–46. [Google Scholar] [CrossRef]
- Sweeney, M.; McCouch, S. The complex history of the domestication of rice. Ann. Bot. 2007, 100, 951–957. [Google Scholar] [CrossRef]
- Koseki, M.; Kitazawa, N.; Yonebayashi, S.; Maehara, Y.; Wang, Z.X.; Minobe, Y. Identification and fine mapping of a major quantitative trait locus originating from wild rice, controlling cold tolerance at the seedling stage. Mol. Genet. Genom. 2010, 284, 45–54. [Google Scholar] [CrossRef]
- Dong, X.X.; Wang, X.Y.; Zhang, L.S.; Yang, Z.T.; Xin, X.Y.; Wu, S.; Sun, C.Q.; Liu, J.X.; Yang, J.S.; Luo, X.J. Identification and characterization of OsEBS, a gene involved in enhanced plant biomass and spikelet number in rice. Plant Biotechnol. J. 2013, 11, 1044–1057. [Google Scholar] [CrossRef] [PubMed]
- Mao, D.; Yu, L.; Chen, D.; Li, L.; Zhu, Y.; Xiao, Y.; Zhang, D.; Chen, C. Multiple cold resistance loci confer the high cold tolerance adaptation of Dongxiang wild rice (Oryza rufipogon) to its high-latitude habitat. Theor. Appl. Genet. 2015, 128, 1359–1371. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, S.; Fu, Y.; Su, Z.; Wang, X.; Sun, C. Identification of a drought tolerant introgression line derived from Dongxiang common wild rice (O. rufipogon Griff.). Plant Mol. Biol. 2006, 62, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.W.; Peng, X.J.; Qian, M.J.; Cai, Y.C.; Ding, X.; Chen, Q.S.; Cai, Q.Y.; Zhu, Y.L.; Yan, L.G.; Cai, Y.H. The chimeric mitochondrial gene orf182 causes non-pollen-type abortion in Dongxiang cytoplasmic male-sterile rice. Plant J. 2018, 95, 715–726. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.K.; Pandey, P.; Kumar, B.; Suresh, B.G. Genetic architecture, inter-relationship and selection criteria for yield improvement in rice (Oryza sativa L.). Pak. J. Biol. Sci. 2011, 14, 540–545. [Google Scholar] [CrossRef]
- Miura, K.; Ashikari, M.; Matsuoka, M. The role of QTLs in the breeding of high-yielding rice. Trends Plant Sci. 2011, 16, 319–326. [Google Scholar] [CrossRef]
- Bai, X.F.; Wu, B.; Xing, Y.Z. Yield-related QTLs and their applications in rice genetic improvement. J. Integr. Plant Biol. 2012, 54, 300–311. [Google Scholar] [CrossRef]
- Khahani, B.; Tavakol, E.; Shariati, V.; Rossini, L. Meta-QTL and ortho-MQTL analyses identifed genomic regions controlling rice yield, yield-related traits and root architecture under water defcit conditions. Sci. Rep. 2021, 11, 6942. [Google Scholar] [CrossRef]
- Zhu, Y.Y.; Nomura, T.; Xu, Y.H.; Zhang, Y.Y.; Peng, Y.; Mao, B.Z.; Hanada, A.; Zhou, H.C.; Wang, R.X.; Li, P.J.; et al. ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 2006, 18, 442–456. [Google Scholar] [CrossRef]
- Kurakawa, T.; Ueda, N.; Maekawa, M.; Kobayashi, K.; Kojima, M.; Nagato, Y.; Sakakibara, H.; Kyozuka, J. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 2007, 445, 652–655. [Google Scholar] [CrossRef]
- Huang, X.Z.; Qian, Q.; Liu, Z.B.; Sun, H.Y.; He, S.Y.; Luo, D.; Xia, G.M.; Chu, C.C.; Li, J.Y.; Fu, X.D. Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 2009, 41, 494–497. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Tang, D.; Wang, K.J.; Wu, X.R.; Lu, L.L.; Yu, H.X.; Gu, M.H.; Yan, C.J.; Cheng, Z.K. Mutations in the F-box gene LARGER PANICLE improve the panicle architecture and enhance the grain yield in rice. Plant Biotechnol. J. 2011, 9, 1002–1013. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.W.; Coneva, V.; Casaretto, J.A.; Ying, S.; Mahmood, K.; Liu, F.; Nambara, E.; Bi, Y.M.; Rothstein, S.J. OsPIN5b modulates rice (Oryza sativa) plant architecture and yield by changing auxin homeostasis, transport and distribution. Plant J. 2015, 83, 913–925. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.P.; Fang, J.; Xu, F.; Wang, W.; Chu, C.C. Rice HOX12 regulates panicle exsertion by directly modulating the expression of ELONGATED UPPERMOST INTERNODE1. Plant Cell 2016, 28, 680–695. [Google Scholar] [CrossRef]
- Sun, P.Y.; Zhang, W.H.; Wang, Y.H.; He, Q.; Shu, F.; Liu, H.; Wang, J.; Wang, J.M.; Yuan, L.P.; Deng, H.F. OsGRF4 controls grain shape, panicle length and seed shattering in rice. J. Integr. Plant Biol. 2016, 58, 836–847. [Google Scholar] [CrossRef]
- Huang, Y.; Bai, Y.F.; Luo, M.F.; Ying, Y.Z. Short Panicle 3 controls panicle architecture by upregulating APO2/RFL and increasing cytokinin content in rice. J. Integr. Plant Biol. 2019, 61, 987–999. [Google Scholar] [CrossRef]
- Su, S.; Hong, J.; Chen, X.; Zhang, C.; Chen, M.; Luo, Z.; Chang, S.; Bai, S.; Liang, W.; Liu, Q.; et al. Gibberellins orchestrate panicle architecture mediated by DELLA-KNOX signalling in rice. Plant Biotechnol. J. 2021, 19, 2304–2318. [Google Scholar] [CrossRef]
- Xue, W.Y.; Xing, Y.Z.; Weng, X.Y.; Zhao, Y.; Tang, W.J.; Wang, L.; Zhou, H.J.; Yu, S.B.; Xu, C.G.; Li, X.H.; et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008, 40, 761–767. [Google Scholar] [CrossRef]
- Yan, W.H.; Wang, P.; Chen, H.X.; Zhou, H.J.; Li, Q.P.; Wang, C.R.; Ding, Z.H.; Zhang, Y.S.; Yu, S.B.; Xing, Y.Z.; et al. A Major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 2011, 4, 319–330. [Google Scholar] [CrossRef]
- Li, J.; Xu, R.; Wang, C.C.; Qi, L.; Zheng, X.M.; Wang, W.S.; Ding, Y.B.; Zhang, L.Z.; Wang, Y.Y.; Cheng, Y.L.; et al. A heading date QTL, qHD7.2, from wild rice (Oryza rufipogon) delays flowering and shortens panicle length under long-day conditions. Sci. Rep. 2018, 8, 2928. [Google Scholar]
- Ikeda, K.; Ito, M.; Nagasawa, N.; Kyozuka, J.; Nagato, Y. Rice ABERRANT PANICLE ORGANIZATION 1, encoding an F-box protein, regulates meristem fate. Plant J. 2007, 51, 1030–1040. [Google Scholar] [CrossRef] [PubMed]
- Ookawa, T.; Hobo, T.; Yano, M.; Murata, K.; Ando, T.; Miura, H.; Asano, K.; Ochiai, Y.; Ikeda, M.; Nishitani, R.; et al. New approach for rice improvement using a pleiotropic QTL gene for lodging resistance and yield. Nat. Commun. 2010, 1, 132. [Google Scholar] [CrossRef] [PubMed]
- Terao, T.; Nagata, K.; Morino, K.; Hirose, T. A gene controlling the number of primary rachis branches also controls the vascular bundle formation and hence is responsible to increase the harvest index and grain yield in rice. Theor. Appl. Genet. 2010, 120, 875–893. [Google Scholar] [CrossRef] [PubMed]
- Luan, W.J.; Liu, Y.Q.; Zhang, F.X.; Song, Y.L.; Wang, Z.Y.; Peng, Y.K.; Sun, Z.X. OsCD1 encodes a putative member of the cellulose synthase-like D sub-family and is essential for rice plant architecture and growth. Plant Biotech. J. 2011, 9, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.L.; Piao, R.H.; Shi, J.X.; Lee, S.I.; Jiang, W.Z.; Kim, B.K.; Lee, J.; Han, L.Z.; Ma, W.B.; Koh, H.J. Fine mapping and candidate gene analysis of dense and erect panicle 3, DEP3, which confers high grain yield in rice (Oryza sativa L.). Theor. Appl. Genet. 2011, 122, 1439–1449. [Google Scholar] [CrossRef]
- Ma, X.F.; Cheng, Z.J.; Qin, R.Z.; Qiu, Y.; Heng, Y.Q.; Yang, H.; Ren, Y.L.; Wang, X.L.; Bi, J.C.; Ma, X.D.; et al. OsARG encodes an arginase that plays critical roles in panicle development and grain production in rice. Plant J. 2013, 73, 190–200. [Google Scholar] [CrossRef]
- Huang, L.J.; Hua, K.; Xu, R.; Zeng, D.L.; Wang, R.C.; Dong, G.J.; Zhang, G.Z.; Lu, X.L.; Fang, N.; Wang, D.K.; et al. The LARGE2-APO1/APO2 regulatory module controls panicle size and grain number in rice. Plant Cell 2021, 33, 1212–1228. [Google Scholar] [CrossRef]
- Li, S.B.; Qian, Q.; Fu, Z.M.; Zeng, D.L.; Meng, X.B.; Kyozuka, J.; Maekawa, M.; Zhu, X.D.; Zhang, J.; Li, J.Y.; et al. Short panicle1 encodes a putative PTR family transporter and determines rice panicle size. Plant J. 2009, 58, 592–605. [Google Scholar] [CrossRef]
- Miura, K.; Ikeda, M.A.; Matsubara, A.; Song, X.J.; Ito, M.; Asano, K.; Matsuoka, M.; Kitano, H.; Ashikari, M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 2010, 42, 545–549. [Google Scholar] [CrossRef]
- Li, F.; Liu, W.B.; Tang, J.Y.; Chen, J.F.; Tong, H.N.; Hu, B.; Li, C.L.; Fang, J.; Chen, M.S.; Chu, C.C. Rice DENSE AND ERECT PANICLE 2 is essential for determining panicle outgrowth and elongation. Cell Res. 2010, 20, 838–849. [Google Scholar] [CrossRef]
- Nakagawa, H.; Tanaka, A.; Tanabata, T.; Ohtake, M.; Fujioka, S.; Nakamura, H.; Ichikawa, H.; Mori, M. SHORT GRAIN1 decreases organ elongation and brassinosteroid response in rice. Plant Physiol. 2012, 158, 1208–1219. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, A.; Ohmori, Y.; Kitano, H.; Taguchi-Shiobara, F.; Hirano, H.Y. Aberrant spikelet and panicle1, encoding a TOPLESS-related transcriptional co-repressor, is involved in the regulation of meristem fate in rice. Plant J. 2012, 70, 327–339. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, A.; Sasao, M.; Yasuno, N.; Takagi, K.; Daimon, Y.; Chen, R.H.; Yamazaki, R.; Tokunaga, H.; Kitaguchi, Y.; Sato, Y.; et al. TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proc. Natl. Acad. Sci. USA 2013, 110, 767–772. [Google Scholar] [CrossRef] [PubMed]
- Léran, S.; Varala, K.; Boyer, J.C.; Chiurazzi, M.; Crawford, N.; Daniel-Vedele, F.; David, L.; Dickstein, R.; Fernandez, E.; Forde, B.; et al. A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci. 2014, 19, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.X.; Xiong, G.S.; Hu, J.; Jiang, L.; Yu, H.; Xu, J.; Fang, Y.X.; Zeng, L.J.; Xu, E.B.; Xu, J.; et al. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 2015, 47, 944–948. [Google Scholar] [CrossRef]
- Wang, S.K.; Li, S.; Liu, Q.; Wu, K.; Zhang, J.Q.; Wang, S.S.; Wang, Y.; Chen, X.B.; Zhang, Y.; Gao, C.X.; et al. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 2015, 47, 949–954. [Google Scholar] [CrossRef]
- Zhou, Y.; Miao, J.; Gu, H.Y.; Peng, X.R.; Leburu, M.; Yuan, F.H.; Gu, H.W.; Gao, Y.; Tao, Y.J.; Zhu, J.Y.; et al. Natural variations in SLG7 regulate grain shape in rice. Genetics 2015, 201, 1591–1599. [Google Scholar] [CrossRef]
- Ikeda-Kawakatsu, K.; Maekawa, M.; Izawa, T.; Itoh, J.I.; Nagato, Y. ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant J. 2015, 69, 168–180. [Google Scholar] [CrossRef]
- Ikeda-Kawakatsu, K.; Yasuno, N.; Oikawa, T.; Iida, S.; Nagato, Y.; Maekawa, M.; Kyozuka, J. Expression level of ABERRANT PANICLE ORGANIZATION1 determines rice inflorescence form through control of cell proliferation in the meristem. Plant Physiol. 2009, 150, 736–747. [Google Scholar] [CrossRef]
- Sun, X.W.; Liu, D.Y.; Zhang, X.F.; Li, W.B.; Liu, H.; Hong, W.G.; Jiang, C.B.; Guan, N.; Ma, C.X.; Zeng, H.P.; et al. SLAF-seq: An efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS ONE 2013, 8, e58700. [Google Scholar] [CrossRef]
- Vision, T.J.; Brown, D.G.; Shmoys, D.B.; Durrett, R.T.; Tanksley, S.D. Selective mapping: A strategy for optimizing the construction of high-density linkage maps. Genetics 2000, 155, 407–420. [Google Scholar] [CrossRef]
- McCouch, S.R.; Cho, Y.G.; Yano, M.; Paul, E.; Blinstrub, M.; Morishima, H.; Kinoshita, T. Suggestions for QTL nomenclature for rice, Rice Genet. News 1997, 14, 11–13. [Google Scholar]
- Yang, J.; Zhu, J. Methods for predicting superior genotypes under multiple environments based on QTL effects. Theor. Appl. Genet. 2005, 110, 1268–1274. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Yao, W.; Ouyang, Y.D.; Yang, W.N.; Wang, G.W.; Lian, X.M.; Xing, Y.Z.; Chen, L.L.; Xie, W.B. RiceVarMap: A comprehensive database of rice genomic variations. Nucleic Acids Res. 2015, 43, 1018–1022. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Li, J.C.; Yang, L.; Qin, G.; Xia, C.J.; Xu, X.B.; Su, Y.M.; Liu, Y.M.; Ming, L.C.; Chen, L.L.; et al. An inferred functional impact map of genetic variants in rice. Mol. Plant. 2021, 14, 1584–1599. [Google Scholar] [CrossRef]
- Adriani, D.E.; Dingkuhn, M.; Dardou, A.; Adam, H.; Luquet, D.; Lafarge, T. Rice panicle plasticity in near isogenic lines carrying a QTL for larger panicle is genotype and environment dependent. Rice 2016, 9, 28. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, J.J.; Wang, J.M.; Wang, L.Y.; He, Z.H. Quantitative trait locus analysis and fine mapping of the qPL6 locus for panicle length in rice. Theor. Appl. Genet. 2015, 128, 1151–1161. [Google Scholar] [CrossRef]
- Liu, E.B.; Liu, Y.; Wu, G.C.; Zeng, S.Y.; Thi, T.; Liang, L.J.; Liang, Y.F.; Dong, Z.Y.; She, D.; Wang, H.; et al. Identification of a candidate gene for panicle length in rice (Oryza sativa L.) via association and linkage analysis. Front. Plant Sci. 2016, 7, 596. [Google Scholar] [CrossRef]
- Kumar, M.; Basha, P.O.; Puri, A.; Rajpurohit, D.; Randhawa, G.S.; Sharma, T.R.; Dhaliwal, H.S. A candidate gene OsAPC6 of anaphase-promoting complex of rice identified through T-DNA insertion. Funct. Integr. Genom. 2010, 10, 349–358. [Google Scholar] [CrossRef]
- Li, S.F.; Shen, L.; Hu, P.; Wu, X.M.; Yuan, Q.L.; Rao, Y.C.; Qian, Q.; Wang, K.J.; Zhu, X.D.; Shang, L.G.; et al. A method for effectively overcoming tight functional linkage between genes in rice by CRISPR/Cas9 system. Rice Sci. 2020, 27, 180–183. [Google Scholar]
- Tan, W.C.; Miao, J.; Xu, B.; Zhou, C.T.; Wang, Y.R.; Gu, X.Q.; Liang, S.N.; Wang, B.X.; Chen, C.; Zhu, J.Y.; et al. Rapid production of novel beneficial alleles for improving rice appearance quality by targeting a regulatory element of SLG7. Plant Biotechnol. J. 2023, 21, 1305–1307. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).