Negative Contrast: A Simple and Efficient Image Augmentation Method in Crop Disease Classification
Abstract
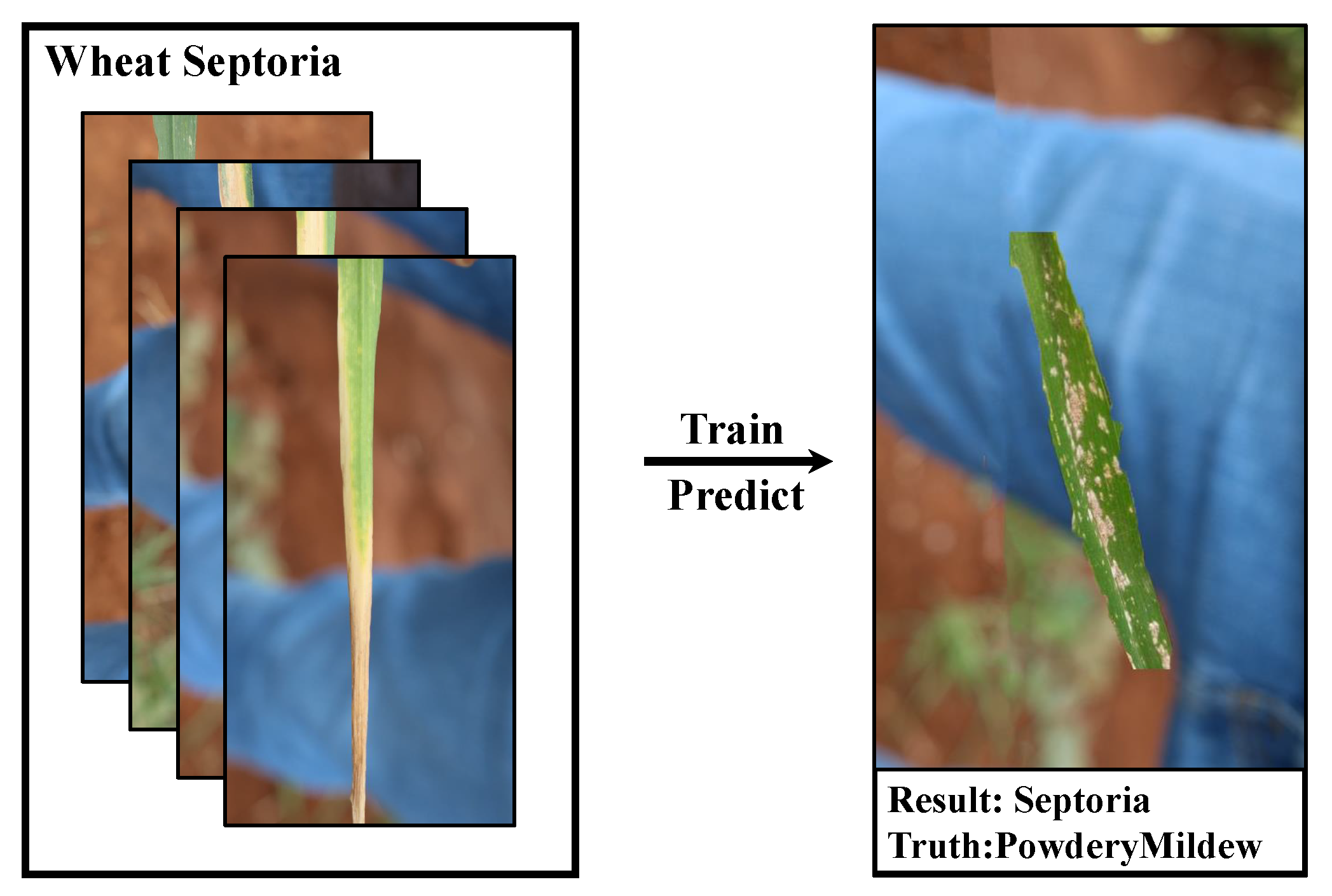
1. Introduction
- We present a compact and realistic dataset, Plant Real-World, for four crops, complete with training and testing sets.
- We introduce a strategy, health augmentation, that leverages healthy crop samples to enhance the performance of crop disease classification. This approach uses healthy crop samples as the negative sample input while making minor modifications to the softmax layer of the network, thereby considerably enhancing the recognition accuracy.
- Building on health augmentation, we further augment the model’s generalization performance by using disease samples from which diseased regions have been artificially removed as pseudo healthy samples. With a relatively small training set (5–20% of the original sample count), we obtained an average accuracy improvement of 30.8% across models.
2. Materials and Methods
2.1. A New Dataset: Plant Real-World
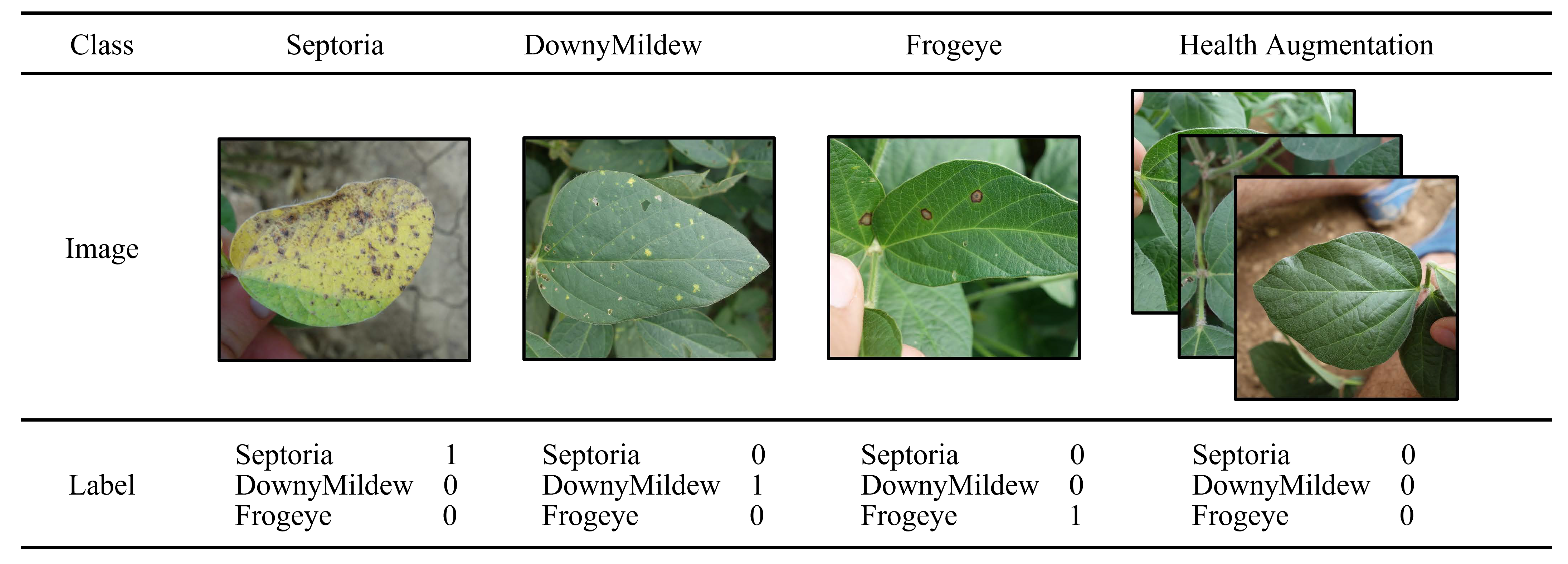
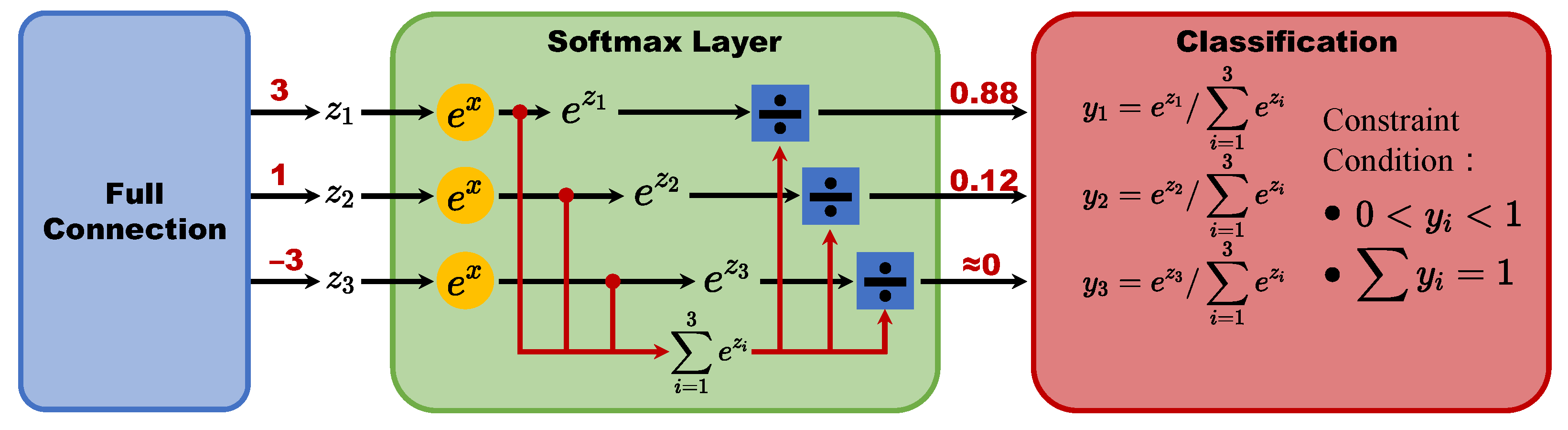
2.2. Health Augmentation
- (1)
- Remove the softmax layer.
- (2)
- Change the loss funciton from cross entropy to mean square error.
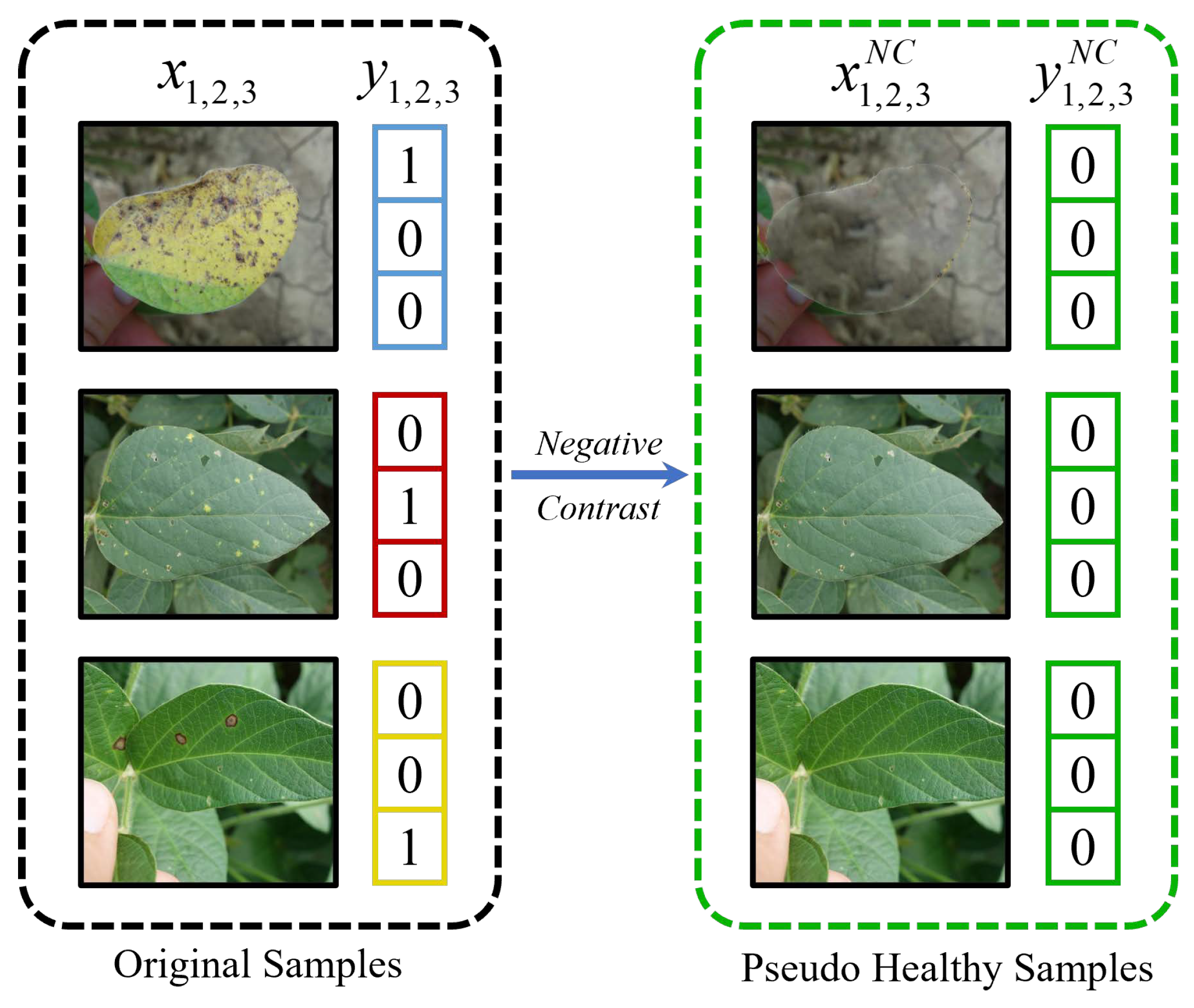
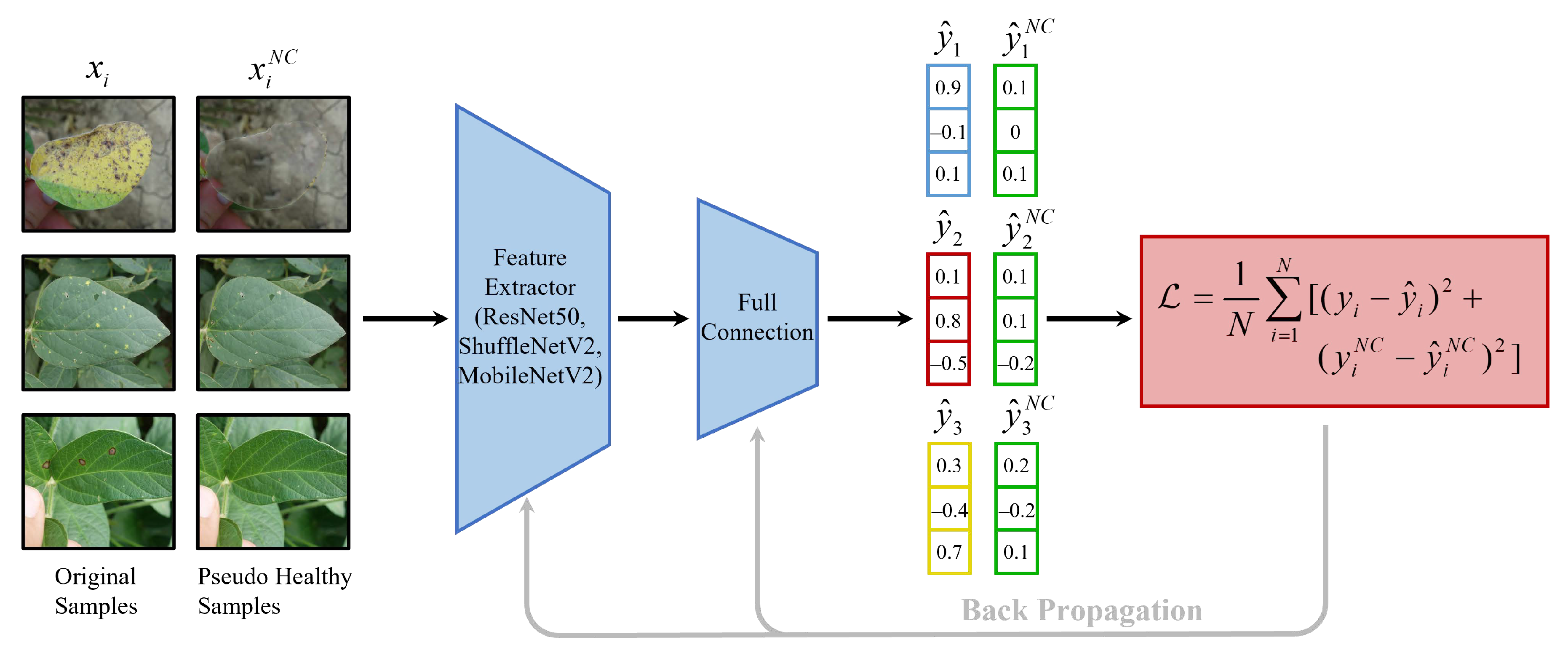
2.3. Negative Contrast
3. Results
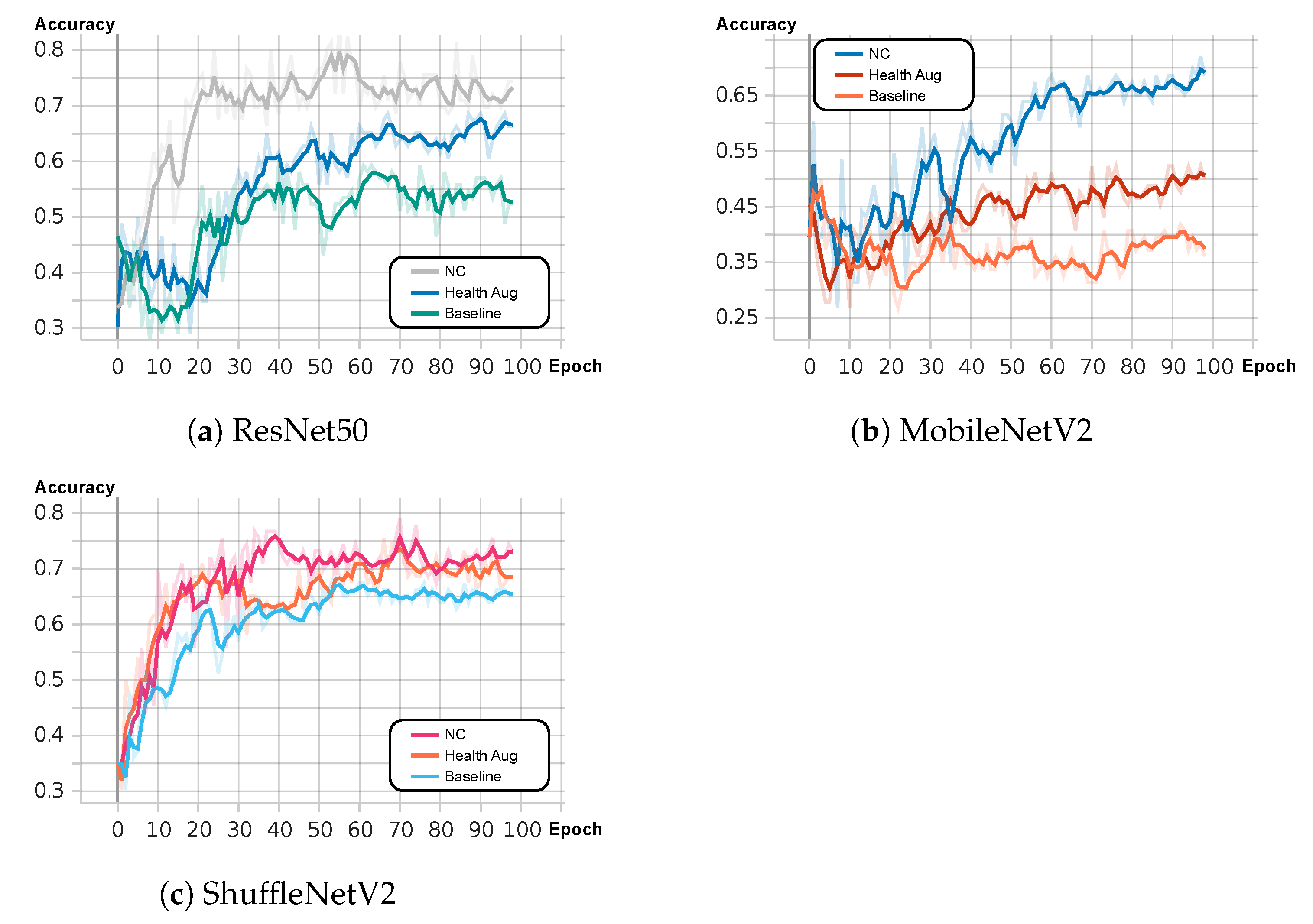
3.1. Health Augmentation
3.2. Negative Contrast
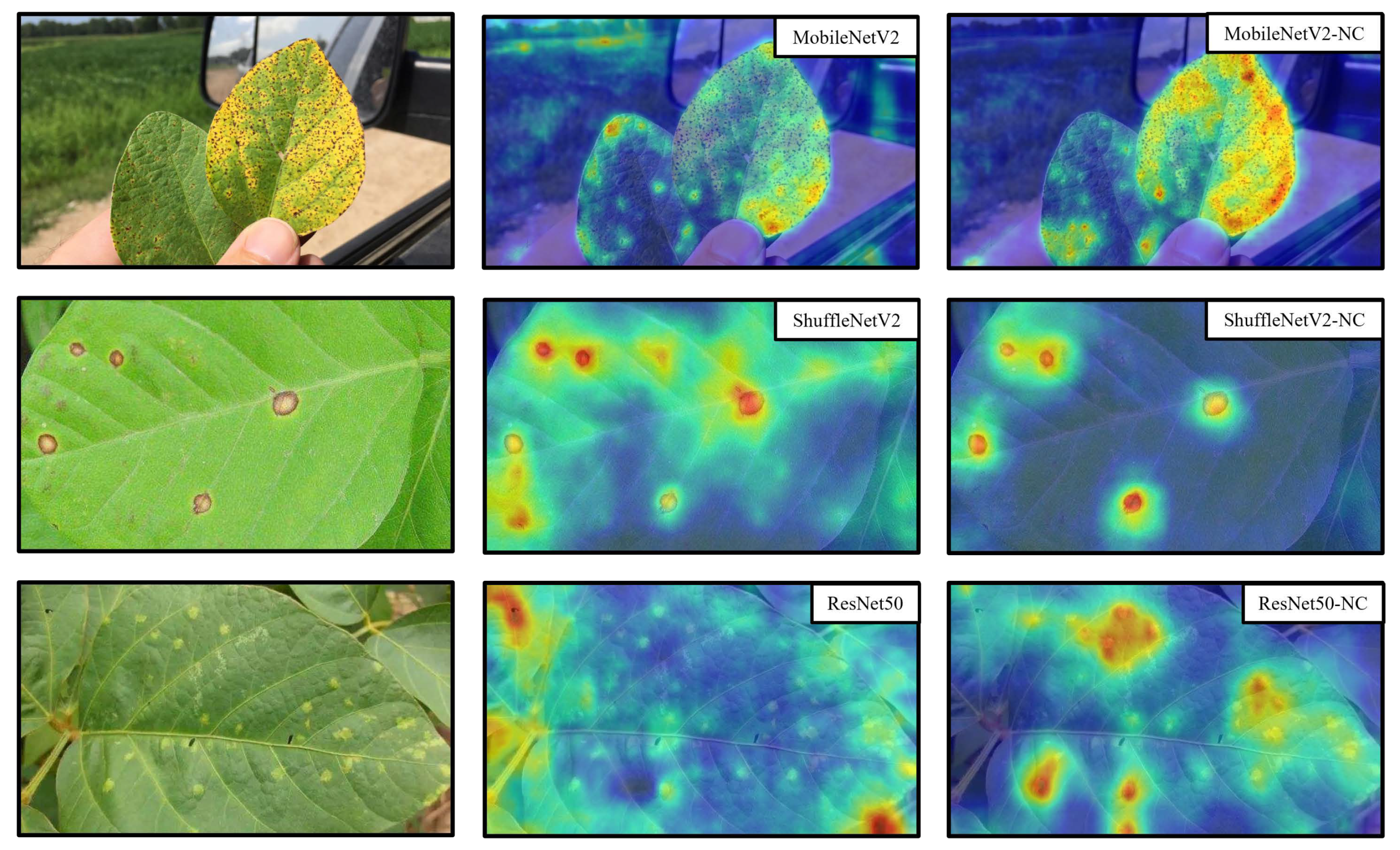
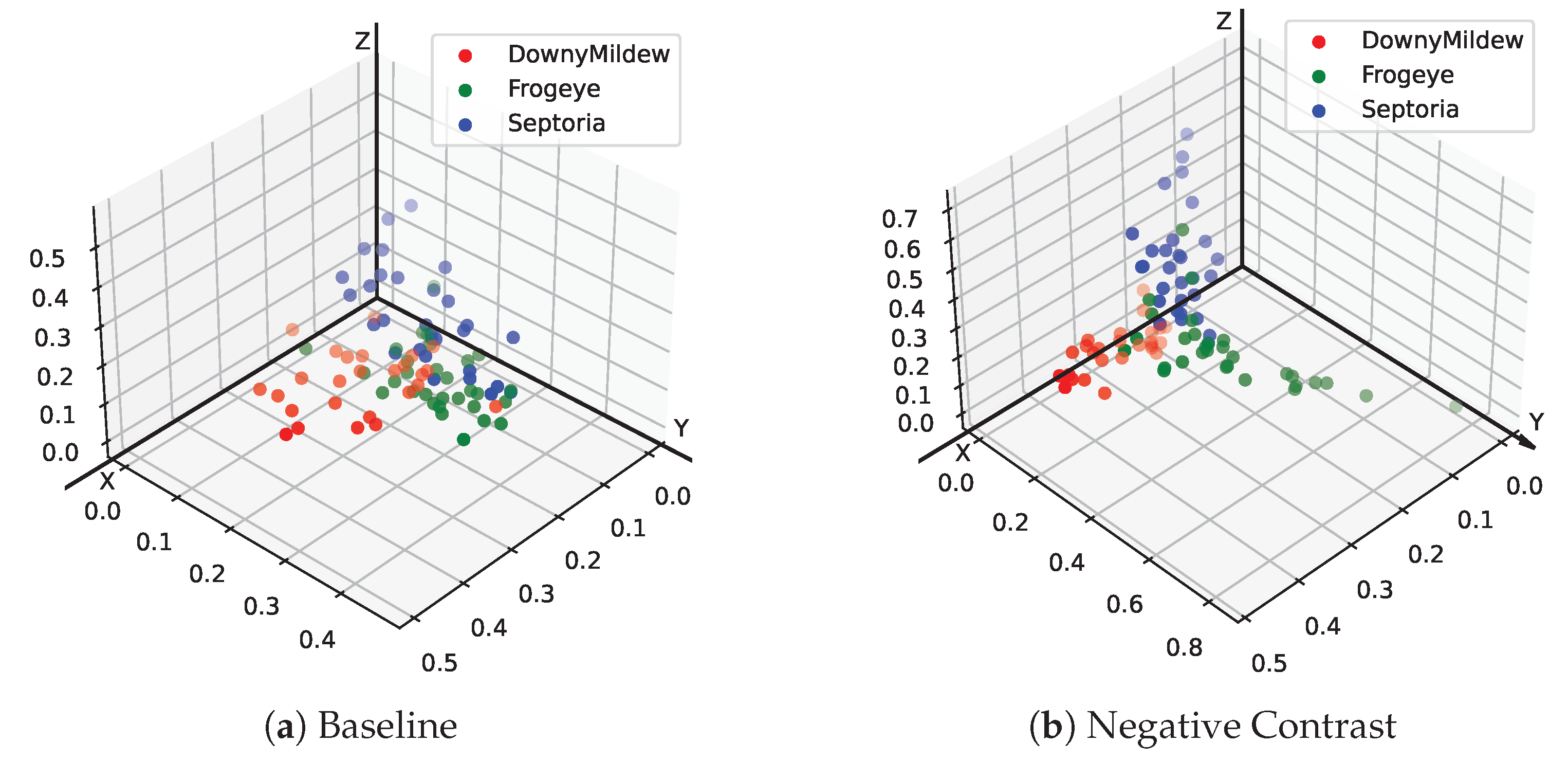
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2012, 60, 84–90. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.; Li, K.; Li, F. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Lowe, D.G. Object recognition from local scale-invariant features. In Proceedings of the Seventh IEEE International Conference on Computer Vision, Kerkyra, Greece, 20–27 September 1999; Volume 2, pp. 1150–1157. [Google Scholar]
- Dalal, N.; Triggs, B. Histograms of oriented gradients for human detection. In Proceedings of the 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05), San Diego, CA, USA, 20–25 June 2005; Volume 1, pp. 886–893. [Google Scholar]
- Lu, Y.; Yi, S.; Zeng, N.; Liu, Y.; Zhang, Y. Identification of rice diseases using deep convolutional neural networks. Neurocomputing 2017, 267, 378–384. [Google Scholar] [CrossRef]
- Ma, J.; Du, K.; Zheng, F.; Zhang, L.; Gong, Z.; Sun, Z. A recognition method for cucumber diseases using leaf symptom images based on deep convolutional neural network. Comput. Electron. Agric. 2018, 154, 18–24. [Google Scholar] [CrossRef]
- Too, E.C.; Yujian, L.; Njuki, S.; Yingchun, L. A comparative study of fine-tuning deep learning models for plant disease identification. Comput. Electron. Agric. 2019, 161, 272–279. [Google Scholar] [CrossRef]
- Hughes, D.P.; Salathé, M. An open access repository of images on plant health to enable the development of mobile disease diagnostics through machine learning and crowdsourcing. arXiv 2015, arXiv:1511.08060. [Google Scholar]
- Zhao, Y.; Chen, J.; Xu, X.; Lei, J.; Zhou, W. Sev-net: Residual network embedded with attention mechanism for plant disease severity detection. Concurr. Comput. Pract. Exp. 2021, 33, e6161. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Tiwari, V.; Joshi, R.C.; Dutta, M.K. Dense convolutional neural networks based multiclass plant disease detection and classification using leaf images. Ecol. Inform. 2021, 63, 101289. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, D.f.; Nanehkaran, Y.A.; Li, D. Detection of rice plant diseases based on deep transfer learning. J. Sci. Food Agric. 2020, 100, 3246–3256. [Google Scholar] [CrossRef]
- Karlekar, A.; Seal, A. Soynet: Soybean leaf diseases classification. Comput. Electron. Agric. 2020, 172, 105342. [Google Scholar] [CrossRef]
- Atila, Ü.; Uçar, M.; Akyol, K.; Uçar, E. Plant leaf disease classification using efficientnet deep learning model. Ecol. Inform. 2021, 61, 101182. [Google Scholar] [CrossRef]
- Singh, D.P.; Jain, N.; Jain, P.; Kayal, P.; Kumawat, S.; Batra, N. Plantdoc: A dataset for visual plant disease detection. In Proceedings of the 7th ACM IKDD CoDS and 25th COMAD, Hyderabad, India, 5–7 January 2019. [Google Scholar]
- Xu, M.; Yoon, S.; Fuentes, A.; Park, D.S. A comprehensive survey of image augmentation techniques for deep learning. arXiv 2022, arXiv:2205.01491. [Google Scholar] [CrossRef]
- Devries, T.; Taylor, G.W. Improved regularization of convolutional neural networks with cutout. arXiv 2017, arXiv:1708.04552. [Google Scholar]
- Zhong, Z.; Zheng, L.; Kang, G.; Li, S.; Yang, Y. Random erasing data augmentation. In Proceedings of the AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017. [Google Scholar]
- Chen, P.; Liu, S.; Zhao, H.; Jia, J. Gridmask data augmentation. arXiv 2020, arXiv:2001.04086. [Google Scholar]
- Zhang, H.; Cissé, M.; Dauphin, Y.; Lopez-Paz, D. mixup: Beyond empirical risk minimization. arXiv 2017, arXiv:1710.09412. [Google Scholar]
- Yun, S.; Han, D.; Oh, S.J.; Chun, S.; Choe, J.; Yoo, Y.J. Cutmix: Regularization strategy to train strong classifiers with localizable features. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Republic of Korea, 27 October–2 November 2019; pp. 6022–6031. [Google Scholar]
- Kim, J.-H.; Choo, W.; Jeong, H.; Song, H.O. Co-mixup: Saliency guided joint mixup with supermodular diversity. arXiv 2021, arXiv:2102.03065. [Google Scholar]
- Singh, K.K.; Lee, Y.J. Hide-and-seek: Forcing a network to be meticulous for weakly-supervised object and action localization. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 3544–3553. [Google Scholar]
- Zheng, Z.; Yu, Z.; Wu, Y.; Zheng, H.; Zheng, B.; Lee, M. Generative adversarial network with multi-branch discriminator for imbalanced cross-species image-to-image translation. Neural Netw. Off. J. Int. Neural Netw. Soc. 2021, 141, 355–371. [Google Scholar] [CrossRef]
- Ali-Gombe, A.; Elyan, E. Mfc-gan: Class-imbalanced dataset classification using multiple fake class generative adversarial network. Neurocomputing 2019, 361, 212–221. [Google Scholar] [CrossRef]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.C.; Bengio, Y. Generative adversarial nets. In Proceedings of the Conference on Neural Information Processing Systems NIPS, Montreal, QC, Canada, 8–13 December 2014. [Google Scholar]
- Wah, C.; Branson, S.; Welinder, P.; Perona, P.; Belongie, S.J. The Caltech-Ucsd Birds-200-2011 Dataset. 2011. Available online: https://paperswithcode.com/dataset/cub-200-2011 (accessed on 24 June 2023).
- Khosla, A.; Jayadevaprakash, N.; Yao, B.; Li, F. Novel Dataset for Fine-Grained Image Categorization: Stanford Dogs. 2012. Available online: https://paperswithcode.com/dataset/stanford-dogs (accessed on 24 June 2023).
- Snell, J.; Swersky, K.; Zemel, R.S. Prototypical networks for few-shot learning. arXiv 2017, arXiv:1703.05175. [Google Scholar]
- Lin, T.-Y.; RoyChowdhury, A.; Maji, S. Bilinear cnn models for fine-grained visual recognition. In Proceedings of the 2015 IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 7–13 December 2015; pp. 1449–1457. [Google Scholar]
- Maji, S.; Rahtu, E.; Kannala, J.; Blaschko, M.B.; Vedaldi, A. Fine-grained visual classification of aircraft. arXiv 2013, arXiv:1306.5151. [Google Scholar]
- Ma, N.; Zhang, X.; Zheng, H.; Sun, J. Shufflenet v2: Practical guidelines for efficient cnn architecture design. arXiv 2018, arXiv:1807.11164. [Google Scholar]
- Sandler, M.; Howard, A.G.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- Lee, S.H.; Goëau, H.; Bonnet, P.; Joly, A. New perspectives on plant disease characterization based on deep learning. Comput. Electron. Agric. 2020, 170, 105220. [Google Scholar] [CrossRef]
- Zhu, X.; Anguelov, D.; Ramanan, D. Capturing long-tail distributions of object subcategories. In Proceedings of the 2014 IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 915–922. [Google Scholar]
- Dwibedi, D.; Misra, I.; Hebert, M. Cut, paste and learn: Surprisingly easy synthesis for instance detection. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 1310–1319. [Google Scholar]
- Selvaraju, R.R.; Das, A.; Vedantam, R.; Cogswell, M.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. Int. J. Comput. Vis. 2017, 128, 336–359. [Google Scholar] [CrossRef]
- van der Maaten, L.; Hinton, G.E. Visualizing data using t-sne. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Du, R.; Chang, D.; Bhunia, A.K.; Xie, J.; Song, Y.-Z.; Ma, Z.; Guo, J. Fine-grained visual classification via progressive multi-granularity training of jigsaw patches. In Proceedings of the European Conference on Computer Vision ECCV, Glasgow, UK, 23–28 August 2020. [Google Scholar]
- Zhuang, P.; Wang, Y.; Qiao, Y. Learning attentive pairwise interaction for fine-grained classification. In Proceedings of the Association for the Advancement of Artificial Intelligence AAAI, New York, NY, USA, 7–12 February 2020. [Google Scholar]
- Min, S.; Yao, H.; Xie, H.; Zha, Z.; Zhang, Y. Multi-objective matrix normalization for fine-grained visual recognition. IEEE Trans. Image Process. 2020, 29, 4996–5009. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, S.; Yang, S.; Li, H.; Li, J.; Li, Z. Weakly supervised fine-grained image classification via guassian mixture model oriented discriminative learning. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; pp. 9746–9755. [Google Scholar]
- Liu, C.; Xie, H.; Zha, Z.; Ma, L.; Yu, L.; Zhang, Y. Filtration and distillation: Enhancing region attention for fine-grained visual categorization. In Proceedings of the Association for the Advancement of Artificial Intelligence AAAI, New York, NY, USA, 7–12 February 2020. [Google Scholar]
- Ji, R.; Wen, L.; Zhang, L.; Du, D.; Wu, Y.; Zhao, C.; Liu, X.; Huang, F. Attention convolutional binary neural tree for fine-grained visual categorization. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; pp. 10465–10474. [Google Scholar]
- Huang, G.; Liu, Z.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar]
- Zhang, H.; Wu, C.; Zhang, Z.; Zhu, Y.; Zhang, Z.-L.; Lin, H.; Sun, Y.; He, T.; Mueller, J.; Manmatha, R.; et al. Resnest: Split-attention networks. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), New Orleans, LA, USA, 19–20 June 2022; pp. 2735–2745. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the 2021 IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 10–17 October 2021; pp. 9992–10002. [Google Scholar]
- Zoph, B.; Vasudevan, V.; Shlens, J.; Le, Q.V. Learning transferable architectures for scalable image recognition. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 8697–8710. [Google Scholar]
- Touvron, H.; Cord, M.; Douze, M.; Massa, F.; Sablayrolles, A.; J’egou, H. Training data-efficient image transformers & distillation through attention. In Proceedings of the International Conference on Machine Learning, Virtual, 13–18 July 2020. [Google Scholar]
- Tran, T.; Pham, T.T.; Carneiro, G.; Palmer, L.J.; Reid, I.D. A bayesian data augmentation approach for learning deep models. arXiv 2017, arXiv:1710.10564. [Google Scholar]
- Cubuk, E.D.; Zoph, B.; Mané, D.; Vasudevan, V.; Le, Q.V. Autoaugment: Learning augmentation strategies from data. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 113–123. [Google Scholar]
- Hataya, R.; Zdenek, J.; Yoshizoe, K.; Nakayama, H. Faster autoaugment: Learning augmentation strategies using backpropagation. In Proceedings of the European Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019. [Google Scholar]
- Takase, T.; Karakida, R.; Asoh, H. Self-paced data augmentation for training neural networks. Neurocomputing 2020, 442, 296–306. [Google Scholar] [CrossRef]
- Bochkovskiy, A.; Wang, C.-Y.; Liao, H.-Y.M. Yolov4: Optimal speed and accuracy of object detection. arXiv 2020, arXiv:2004.10934. [Google Scholar]


| Crops | Diseases | Training | Testing |
|---|---|---|---|
| Wheat | Flag Smut | 14 | 6 |
| Mildew | 7 | 1 | |
| Powdery Mildew | 21 | 35 | |
| Septoria | 97 | 29 | |
| Stipe Rust | 207 | 99 | |
| Corn | Blight | 75 | 32 |
| Common Rust | 70 | 17 | |
| Gray Leaf Spot | 58 | 57 | |
| Soybean | Downy Mildew | 124 | 26 |
| Frogeye | 140 | 30 | |
| Septoria | 104 | 30 | |
| Rice | Blast | 27 | 25 |
| Brown Spot | 75 | 21 | |
| Leaf Scald | 53 | 13 | |
| Sheath Blight | 64 | 9 | |
| Tungro | 46 | 8 |
| Crops | ResNet50 | MobileNetV2 | ShuffleNetV2 | |||
|---|---|---|---|---|---|---|
| Baseline | Health Aug | Baseline | Health Aug | Baseline | Health Aug | |
| Wheat | 65.9 | 71.2 | 65.3 | 70.0 | 71.2 | 72.9 |
| Corn | 50.9 | 83.0 | 45.3 | 78.3 | 50.9 | 85.9 |
| Soybean | 84.9 | 87.2 | 81.4 | 80.2 | 86.0 | 87.2 |
| Rice | 32.9 | 48.7 | 34.2 | 46.1 | 39.5 | 43.4 |
| Crops | ResNet50 | MobileNetV2 | ShuffleNetV2 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Baseline | Health Aug | NC | Baseline | Health Aug | NC | Baseline | Health Aug | NC | |
| Wheat (10%) | 63.4 | 62.5 | 67.4 | 59.1 | 60.0 | 73.4 | 63.4 | 66.9 | 71.6 |
| Corn (20%) | 40.7 | 43.4 | 52.8 | 38.8 | 38.8 | 46.2 | 45.9 | 45.9 | 53.8 |
| Soybean (5%) | 59.7 | 69.2 | 84.3 | 41.3 | 53.6 | 72.1 | 68.2 | 74.9 | 78.8 |
| Rice (10%) | 19.8 | 44.6 | 47.2 | 33.5 | 33.7 | 47.2 | 32.4 | 34.1 | 42.5 |
| Crops | Baseline | Zoom | Rotate | Color | Brightness | Contrast | Erasing | NC |
|---|---|---|---|---|---|---|---|---|
| Wheat (10%) | 62.2 | 62.4 | 62.2 | 61.0 | 63.3 | 60.8 | 64.7 | 70.8 |
| Corn (20%) | 48.1 | 46.9 | 48.1 | 42.8 | 49.7 | 49.1 | 49.1 | 50.9 |
| Soybean (5%) | 61.2 | 63.6 | 61.2 | 55.8 | 61.2 | 64.7 | 61.6 | 78.4 |
| Rice (10%) | 36.8 | 36.4 | 34.2 | 40.8 | 35.1 | 33.8 | 32.9 | 45.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Yin, Z.; Li, D.; Zhao, Y. Negative Contrast: A Simple and Efficient Image Augmentation Method in Crop Disease Classification. Agriculture 2023, 13, 1461. https://doi.org/10.3390/agriculture13071461
Li J, Yin Z, Li D, Zhao Y. Negative Contrast: A Simple and Efficient Image Augmentation Method in Crop Disease Classification. Agriculture. 2023; 13(7):1461. https://doi.org/10.3390/agriculture13071461
Chicago/Turabian StyleLi, Jiqing, Zhendong Yin, Dasen Li, and Yanlong Zhao. 2023. "Negative Contrast: A Simple and Efficient Image Augmentation Method in Crop Disease Classification" Agriculture 13, no. 7: 1461. https://doi.org/10.3390/agriculture13071461
APA StyleLi, J., Yin, Z., Li, D., & Zhao, Y. (2023). Negative Contrast: A Simple and Efficient Image Augmentation Method in Crop Disease Classification. Agriculture, 13(7), 1461. https://doi.org/10.3390/agriculture13071461






