Molecular Characterization and Haplotype Analysis of Low Phytic Acid-1 (lpa1) Gene Governing Accumulation of Kernel Phytic Acid in Subtropically-Adapted Maize
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genetic Materials
2.2. Genomic DNA Isolation, PCR Amplification and Sequencing of Lpa1
2.3. Alignment of Sequences and Functional Analysis of Lpa1 Gene
2.4. Gene-Based Diversity Analysis of Lpa1 among the Diverse Maize Genotypes
2.5. Retrieval of Gene and Protein Sequences of Lpa1 Orthologues
2.6. Gene Prediction and Phylogenetic Tree
2.7. Structural Analysis and Physicochemical Properties of LPA1 Protein
2.8. Homology Modeling of LPA1 Protein
3. Results
3.1. Sequence Characterization of Lpa1 Gene among Selected Maize Inbreds
3.2. Characterization of LPA1 Protein among Selected Maize Inbreds
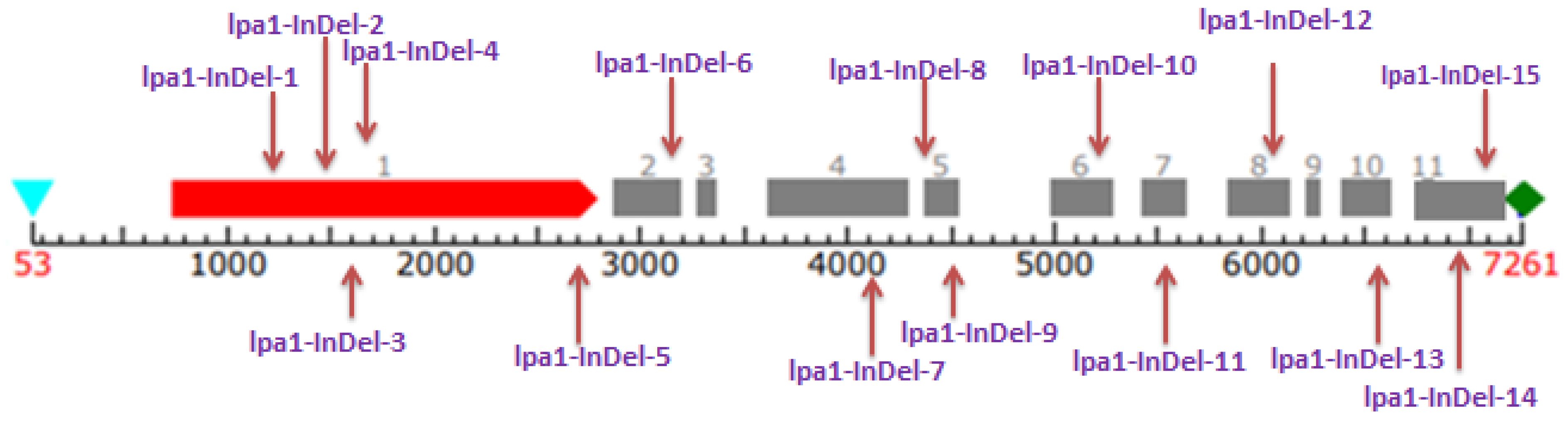
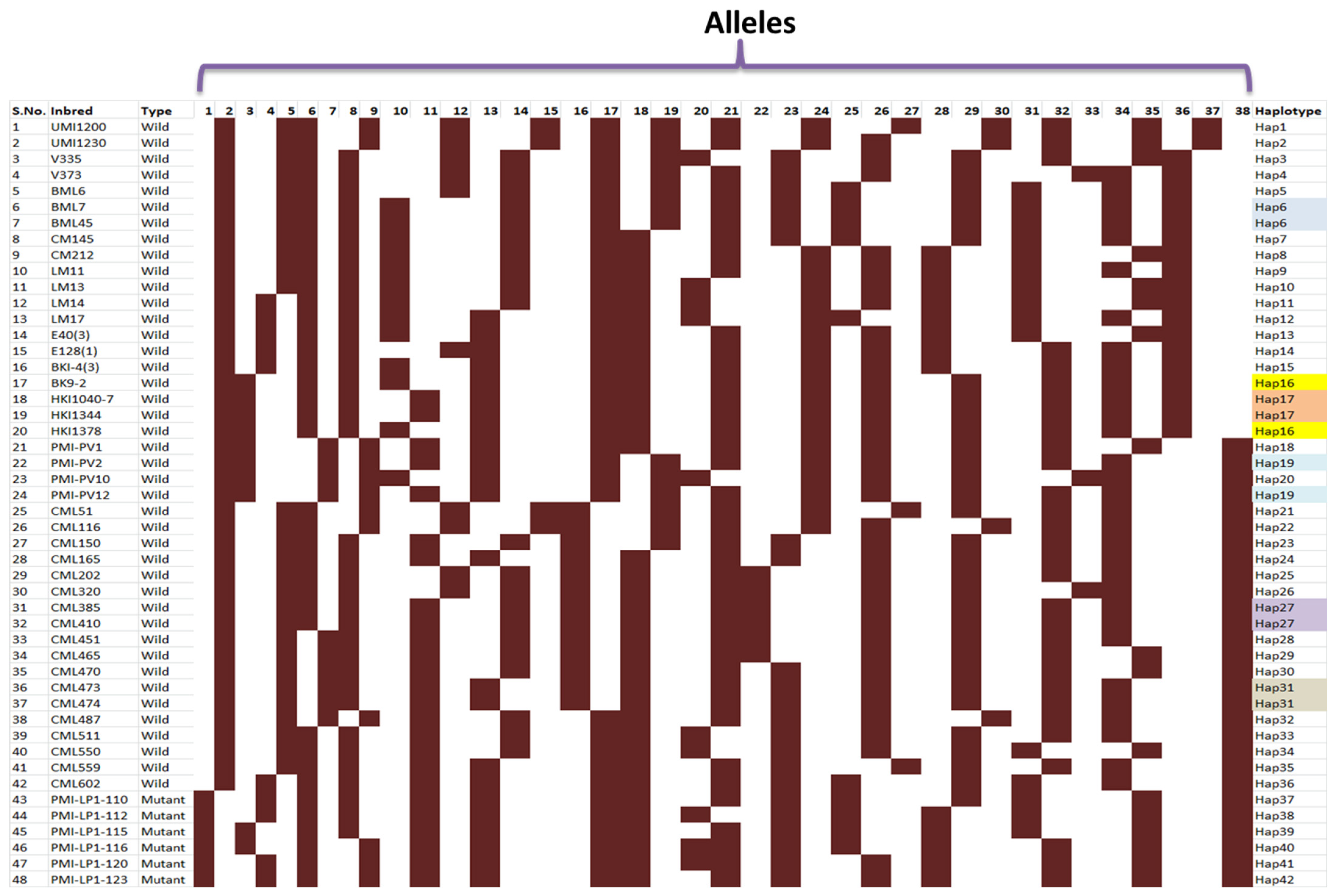
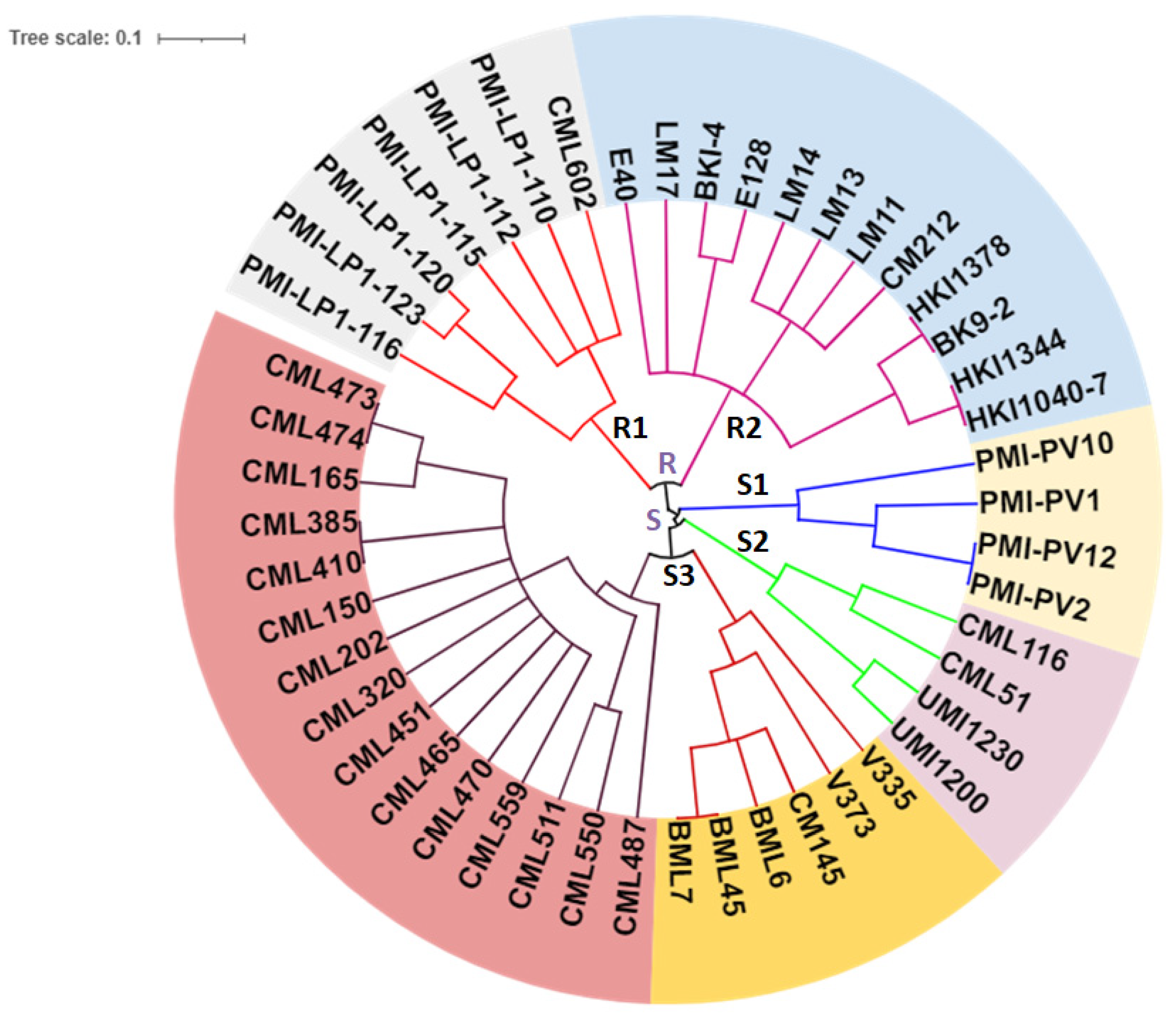
3.3. Gene-Based Diversity Analysis among Diverse Maize Inbreds Using InDel Markers
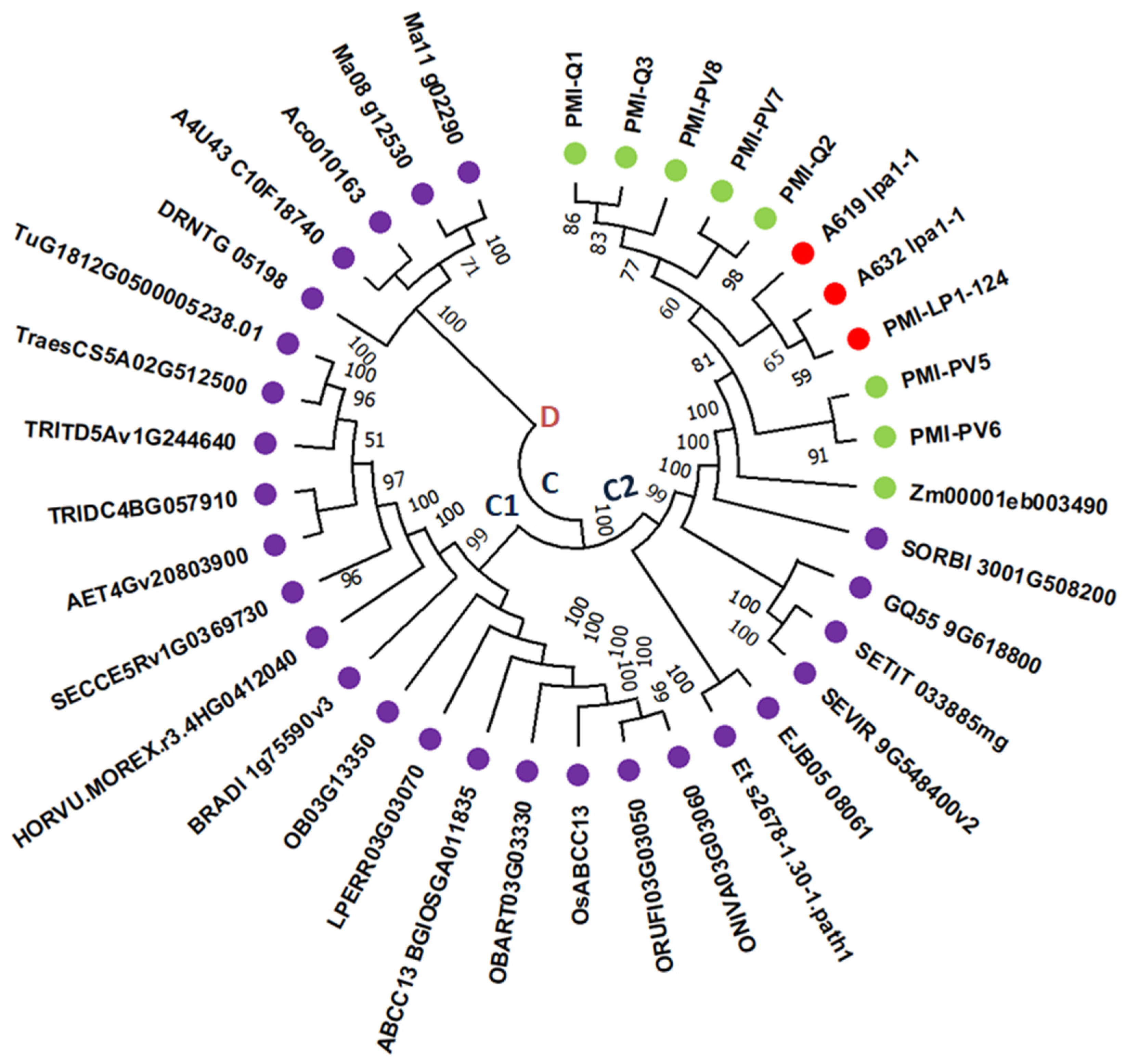
3.4. Structure of Lpa1 Gene in Maize and Its Orthologues
3.5. Structure of LPA1 Protein in Maize and Its Orthologues
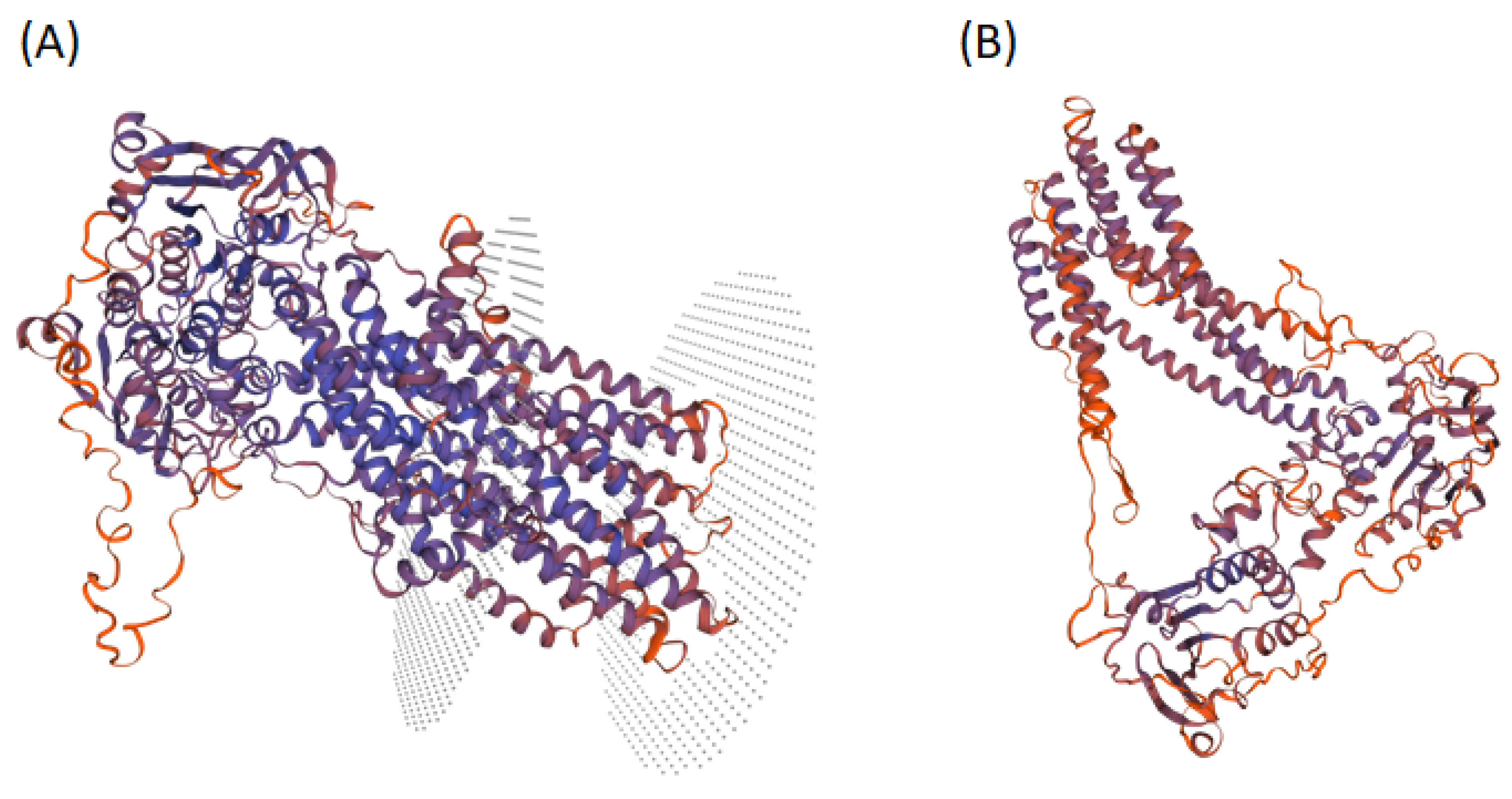
3.6. Homology Modeling of LPA1 Protein
3.7. Domains, Motifs and Features of LPA1 Protein
3.8. Physicochemical Properties of LPA1 Protein in Maize and Selected Orthologues
4. Discussion
4.1. Allelic Variations in Lpa1 Gene in Maize Genotypes
4.2. Variation in LPA1 Protein among Maize Genotypes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Huertas, R.; Karpinska, B.; Ngala, S.; Mkandawire, B.; Maling’a, J.; Wajenkeche, E.; Kimani, P.M.; Boesch, C.; Stewart, D.; Hancock, R.D.; et al. Biofortification of common bean (Phaseolus vulgaris L.) with iron and zinc: Achievements and challenges. Food Energy Secur. 2023, 12, e406. [Google Scholar] [CrossRef]
- Gupta, H.S.; Hossain, F.; Nepolean, T.; Vignesh, M.; Mallikarjuna, M.G. Understanding genetic and molecular bases of Fe and Zn accumulation towards development of micronutrient-enriched maize. In Nutrient Use Efficiency: From Basics to Advances; Springer: New Delhi, India, 2015; pp. 255–282. [Google Scholar] [CrossRef]
- Jaiswal, S.K.; Muthusamy, V.; Hossain, F.; Zunjare, R.U.; Goswami, R.; Chhabra, R.; Chand, G.; Dosad, S.; Bhowmick, R.; Guleria, S.K.; et al. Characterization of maize genotypes using microsatellite markers associated with QTLs for kernel iron and zinc. Indian J. Biotechnol. 2019, 18, 224–234. [Google Scholar]
- Pandey, N.; Hossain, F.; Kumar, K.; Vishwakarma, A.K.; Muthusamy, V.; Manjaiah, K.M.; Agrawal, P.K.; Guleria, S.K.; Reddy, S.S.; Thirunavukkarasu, N.; et al. Microsatellite marker-based genetic diversity among quality protein maize (QPM) inbreds differing for kernel iron and zinc. Mol. Plant Breed. 2015, 6, 1–10. [Google Scholar] [CrossRef]
- Gronli, O.; Kvamme, J.M.; Friborg, O.; Wynn, R. Zinc deficiency is common in several psychiatric disorders. PLoS ONE 2013, 8, e82793. [Google Scholar] [CrossRef] [Green Version]
- Pixley, K.; Palacios, N.R.; Glahn, R.P. The usefulness of iron bioavailability as a target trait for breeding maize (Zea mays L.) with enhanced nutritional value. Field Crop Res. 2011, 123, 153–160. [Google Scholar] [CrossRef]
- Global Nutrition Report 2022. Available online: https://globalnutritionreport.org (accessed on 30 March 2023).
- Roy, C.; Kumar, S.; Ranjan, R.D.; Kumhar, S.R.; Govindan, V. Genomic approaches for improving grain zinc and iron content in wheat. Front. Genet. 2022, 13, 1045955. [Google Scholar] [CrossRef] [PubMed]
- Mehta, B.K.; Muthusamy, V.; Zunjare, R.U.; Baveja, A.; Chauhan, H.S.; Chhabra, R.; Singh, A.K.; Hossain, F. Biofortifcation of sweet corn hybrids for provitamin-a, lysine and tryptophan using molecular breeding. J. Cereal. Sci. 2020, 96, 103093. [Google Scholar] [CrossRef]
- Hossain, F.; Zunjare, R.U.; Muthusamy, V.; Bhat, J.S.; Mehta, B.K.; Sharma, D.; Talukder, Z.A.; Chhabra, R.; Katral, A.; Dutta, S.; et al. Biofortification of Maize for Nutritional Security. In Biofortification of Staple Crops; Springer: Singapore, 2022; pp. 147–174. [Google Scholar] [CrossRef]
- Raboy, V. myo-Inositol-1, 2, 3, 4, 5, 6-hexakisphosphate. Phytochemistry 2003, 64, 1033–1043. [Google Scholar] [CrossRef]
- Raboy, V. Low phytic acid crops: Observations based on four decades of research. Plants 2020, 9, 140. [Google Scholar] [CrossRef] [Green Version]
- Colombo, F.; Paolo, D.; Cominelli, E.; Sparvoli, F.; Nielsen, E.; Pilu, R. MRP transporters and low phytic acid mutants in major crops: Main pleiotropic effects and future perspectives. Front. Plant Sci. 2020, 11, 1301. [Google Scholar] [CrossRef]
- O’Dell, B.L.; De Boland, A.R.; Koirtyohann, S.R. Distribution of phytate and nutritionally important elements among the morphological components of cereal grains. J. Agric. Food Chem. 1972, 20, 718–723. [Google Scholar] [CrossRef]
- Cromwell, G.L.; Coffey, R.D. Phosphorus: A key essential nutrient, yet a possible major pollutant. Its central role in animal nutrition. In Biotechnology in the Feed Industry; Alltech Tech Publishers: Nicholasville, KY, USA, 1991; pp. 133–145. [Google Scholar]
- Puppala, K.R.; Buddhiwant, P.G.; Agawane, S.B.; Kadam, A.S.; Mote, C.S.; Lonkar, V.D.; Khire, J.M.; Dharne, M.S. Performance of Aspergillus niger (NCIM 563) phytase based feed supplement for broiler growth and phosphorus excretion. Biocatal. Agric. Biotechnol. 2021, 31, 101887. [Google Scholar] [CrossRef]
- Raboy, V. Approaches and challenges to engineering seed phytate and total phosphorus. Plant Sci. 2009, 177, 281–296. [Google Scholar] [CrossRef]
- Reddy, N.R.; Pierson, M.D.; Sathe, S.K.; Salunkhe, D.K. Nutritional consequences of phytates. In Phytates in Cereals and Legumes; Reddy, N.R., Pierson, M.D., Sathe, S.K., Salunkhe, D.K., Eds.; CRC Press, Inc.: Boca Raton, FL, USA, 1989; pp. 81–110. [Google Scholar]
- Bohlke, R.A.; Thaler, R.C.; Stein, H.H. Calcium, phosphorus, and amino acid digestibility in low-phytate corn, normal corn, and soybean meal by growing pigs. J. Anim. Sci. 2005, 83, 2396–2403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selle, P.H.; Ravindran, V.; Bryden, W.L.; Scott, T. Influence of dietary phytate and exogenous phytase on amino acid digestibility in poultry: A review. J. Poult. Sci. 2006, 43, 89–103. [Google Scholar] [CrossRef] [Green Version]
- Silva, E.O.; Bracarense, A.P.F. Phytic acid: From antinutritional to multiple protection factor of organic systems. J. Food Sci. 2016, 81, R1357–R1362. [Google Scholar] [CrossRef] [Green Version]
- Erenstein, O.; Jaleta, M.; Sonder, K.; Mottaleb, K.; Prasanna, B.M. Global maize production, consumption and trade: Trends and R&D implications. Food Secur. 2022, 14, 1295–1319. [Google Scholar] [CrossRef]
- Prasanna, B.M.; Palacios-Rojas, N.; Hossain, F.; Muthusamy, V.; Menkir, A.; Dhliwayo, T.; Ndhlela, T.; San Vicente, F.; Nair, S.K.; Vivek, B.S.; et al. Molecular breeding for nutritionally enriched maize: Status and prospects. Front. Genet. 2020, 10, 1392. [Google Scholar] [CrossRef] [Green Version]
- Hossain, F.; Rakshit, S.; Kumar, B.; Amalraj, J.J.; Muthusamy, V.; Prakash, B.; Zunjare, R.U.; Karjagi, C.; Khulbe, R.; Arora, A.; et al. 21 Molecular breeding for increasing nutrition quality in maize: Recent progress. In Molecular Breeding in Wheat, Maize and Sorghum: Strategies for Improving Abiotic Stress Tolerance and Yield; CABI: Wallingford, UK, 2021; p. 360. [Google Scholar]
- Schlemmer, U.; Frølich, W.; Prieto, R.M.; Grases, F. Phytate in foods and significance for humans: Food sources, intake, processing, bioavailability, protective role and analysis. Mol. Nutr. Food Res. 2009, 53, S330–S375. [Google Scholar] [CrossRef] [PubMed]
- Ragi, S.; Muthusamy, V.; Zunjare, R.U.; Bhatt, V.; Katral, A.; Abhijith, K.P.; Chand, G.; Mishra, S.J.; Sekhar, J.C.; Yadava, D.K.; et al. Genetic variation for grain phytate and molecular characterization of low phytic acid-2 (lpa2) gene-based maize (Zea mays L.) inbreds. Plant Breed. 2022, 141, 212–222. [Google Scholar] [CrossRef]
- Ragi, S.; Muthusamy, V.; Zunjare, R.U.; Bhatt, V.; Katral, A.; Abhijith, K.P.; Kasana, R.; Gain, N.; Sekhar, J.C.; Yadava, D.K.; et al. Genetic and molecular characterization of sub-tropically adapted low phytate genotypes for utilization in mineral biofortification of maize (Zea mays L.). Crop Pasture Sci. 2021, 73, 104–115. [Google Scholar] [CrossRef]
- Pilu, R.; Landoni, M.; Cassani, E.; Doria, E.; Nielsen, E. The maize lpa241 mutation causes a remarkable variability of expression and some pleiotropic effects. Crop Sci. 2005, 45, 2096–2105. [Google Scholar] [CrossRef]
- Badone, F.C.; Amelotti, M.; Cassani, E.; Pilu, R. Study of low phytic acid1-7 (lpa1-7), a new ZmMRP4 mutation in maize. J. Hered. 2012, 103, 598–605. [Google Scholar] [CrossRef]
- Abhijith, K.P.; Muthusamy, V.; Chhabra, R.; Dosad, S.; Bhatt, V.; Chand, G.; Jaiswal, S.K.; Zunjare, R.U.; Vasudev, S.; Yadava, D.K.; et al. Development and validation of breeder-friendly gene-based marker for lpa1-1 and lpa2-1 genes conferring low phytic acid in maize kernel. 3 Biotech 2020, 10, 121. [Google Scholar] [CrossRef]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. Maize DNA miniprep. In Molecular Biology of Plants; Malberg, R., Messing, J., Sussex, I., Eds.; Cold Spring Harbor: New York, NY, USA, 1985; pp. 36–37. [Google Scholar]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Perrier, X.; Flori, A.; Bonnot, F. Data analysis methods. In Genetic Diversity of Cultivated Tropical Plants; Hamon, P., Seguin, M., Perrier, X., Glaszmann, J.C., Eds.; CRC Press: New York, NY, USA, 2003; pp. 43–76. [Google Scholar]
- Liu, K. PowerMarker: Integrated analysis environment for genetic marker data. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [Green Version]
- Kersey, P.J.; Allen, J.E.; Armean, I.; Boddu, S.; Bolt, B.J.; Carvalho-Silva, D.; Christensen, M.; Davis, P.; Falin, L.J.; Grabmueller, C.; et al. Ensembl genomes 2016: More genomes, more complexity. Nucleic Acids Res. 2016, 44, D574–D580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solovyev, V.; Kosarev, P.; Seledsov, I.; Vorobyev, D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 2006, 7, S10. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; DePauw, R.M.; Kumar, S.; Kumar, J.; Kumar, S.; Pandey, M.P. Breeding and adoption of biofortified crops and their nutritional impact on human health. Ann. N. Y. Acad. Sci. 2023, 1520, 5–19. [Google Scholar] [CrossRef] [PubMed]
- Raboy, V.; Young, K.A.; Dorsch, J.A.; Cook, A. Genetics and breeding of seed phosphorus and phytic acid. J. Plant Physiol. 2001, 158, 489–497. [Google Scholar] [CrossRef]
- Bhatt, V.; Muthusamy, V.; Panda, K.K.; Katral, A.; Chhabra, R.; Mishra, S.J.; Gopinath, I.; Zunjare, R.U.; Neeraja, C.N.; Rakshit, S.; et al. Expression dynamics of lpa1 gene and accumulation pattern of phytate in maize genotypes possessing opaque2 and crtrb1 genes at different stages of kernel development. Plants 2023, 12, 1745. [Google Scholar] [CrossRef]
- Raboy, V.; Gerbasi, P.F.; Young, K.A.; Stoneberg, S.D.; Pickett, S.G.; Bauman, A.T.; Murthy PP, N.; Sheridan, W.F.; Ertl, D.S. Origin and seed phenotype of maize low phytic acid 1–1 and low phytic acid 2–1. Plant Physiol. 2000, 124, 355–368. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Wang, H.; Schellin, K.; Li, B.; Faller, M.; Stoop, J.M.; Meeley, R.B.; Ertl, D.S.; Ranch, J.P.; Glassman, K. Embryo-specific silencing of a transporter reduces phytic acid content of maize and soybean seeds. Nat. Biotechnol. 2007, 25, 930–937. [Google Scholar] [CrossRef] [PubMed]
- Sood, S.; Flint-Garcia, S.; Willcox, M.C.; Holland, J.B. Mining natural variation for maize improvement: Selection on Phenotypes and Genes. In Genomics of Plant Genetic Resources; Tuberosa, R., Graner, A., Frison, E., Eds.; Springer: Amsterdam, The Netherlands, 2014; Volume 1, pp. 615–649. [Google Scholar] [CrossRef]
- Mayer, M.; Hölker, A.C.; González-Segovia, E.; Bauer, E.; Presterl, T.; Ouzunova, M.; Melchinger, A.E.; Schön, C.C. Discovery of benefcial haplotypes for complex traits in maize landraces. Nat. Commun. 2020, 11, 4954. [Google Scholar] [CrossRef]
- Bhatt, V.; Muthusamy, V.; Panda, K.K.; Zunjare, R.; Ragi, S.; Baveja, A.; Chhabra, R.; Kasana, R.; Mehta, B.; Guleria, S.K.; et al. Enrichment of multinutrients in maize using genomic assisted stacking of lpa-1, opaque2, and crtRB1 genes. In Proceedings of the Abstracts of 43rd Annual Meeting of Tissue Culture Association-India (PTCA-I), International Symposium on “Advances in Plant Biotechnology and Nutritional Security” (APBNS-2022), New Delhi, India, 28–30 April 2022; pp. 225–226. Available online: https://apbns2022.com (accessed on 21 March 2023).
- Zunjare, R.U.; Chhabra, R.; Hossain, F.; Baveja, A.; Muthusamy, V.; Gupta, H.S. Molecular characterization of 5′ UTR of the lycopene epsilon cyclase (lcyE) gene among exotic and indigenous inbreds for its utilization in maize biofortification. 3 Biotech 2018, 8, 75. [Google Scholar] [CrossRef]
- Chhabra, R.; Muthusamy, V.; Gain, N.; Katral, A.; Prakash, N.R.; Zunjare, R.U.; Hossain, F. Allelic variation in sugary1 gene affecting kernel sweetness among diverse-mutant and-wild-type maize inbreds. Mol. Genet. Genom. 2021, 296, 1085–1102. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Kubicek, C.P.; Steindorff, A.S.; Chenthamara, K.; Manganiello, G.; Henrissat, B.; Zhang, J.; Cai, F.; Kopchinskiy, A.G.; Kubicek, E.M.; Kuo, A.; et al. Evolution and comparative genomics of the most common Trichoderma species. BMC Genom. 2019, 20, 485. [Google Scholar] [CrossRef] [Green Version]
- Katral, A.; Muthusamy, V.; Zunjare, R.U.; Chhabra, R.; Maman, S.; Yadava, D.K.; Hossain, F. Allelic variation in Zmfatb gene defines variability for fatty acids composition among diverse maize genotypes. Front. Nutr. 2022, 9, 845255. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Bielawski, J.P. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 2000, 15, 496–503. [Google Scholar] [CrossRef]
- Hossain, F.; Chhabra, R.; Devi, E.L.; Zunjare, R.U.; Jaiswal, S.K.; Muthusamy, V. Molecular analysis of mutant granule-bound starch synthaseI (waxy1) gene in diverse waxy maize inbreds. 3 Biotech 2019, 9, 3. [Google Scholar] [CrossRef]
- Chhabra, R.; Muthusamy, V.; Baveja, A.; Katral, A.; Mehta, B.; Zunjare, R.U.; Hossain, F. Allelic variation in shrunken2 gene affecting kernel sweetness in exotic-and indigenous-maize inbreds. PLoS ONE 2022, 17, e0274732. [Google Scholar] [CrossRef]
- Shin, J.H.; Kwon, S.J.; Lee, J.K.; Min, H.K.; Kim, N.S. Genetic diversity of maize kernel starch-synthesis genes with SNAPs. Genome 2006, 49, 1287–1296. [Google Scholar] [CrossRef]
- Delaneau, O.; Howie, B.; Cox, A.J.; Zagury, J.F.; Marchini, J. Haplotype estimation using sequencing reads. Am. J. Hum. Genet. 2013, 93, 687–696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; He, J.; Wang, D.; Shi, E.; Yang, W.; Geng, Q.; Wang, Z. Progress in research and application of InDel markers. Biodivers. Sci. 2016, 24, 237. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.Z.; Fang, J.H.; Yan, X.; Liu, J.; Bao, J.S.; Fransson, G.; Andersson, R.; Jansson, C.; Aman, P.; Sun, C. Molecular insights into how a defciency of amylose affects carbon allocation-carbohydrate and oil analyses and gene expression profling in the seeds of a rice waxy mutant. BMC Plant Biol. 2012, 12, 230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaedeke, N.; Klein, M.; Kolukisaoglu, U.; Forestier, C.; Müller, A.; Ansorge, M.; Becker, D.; Mamnun, Y.; Kuchler, K.; Schulz, B.; et al. The Arabidopsis thaliana ABC transporter at MRP5 controls root development and stomata movement. EMBO J. 2001, 20, 1875–1887. [Google Scholar] [CrossRef] [Green Version]
- Prakash, N.R.; Chhabra, R.; Zunjare, R.U.; Muthusamy, V.; Hossain, F. Molecular characterization of teosinte branched1 gene governing branching architecture in cultivated maize and wild relatives. 3 Biotech 2020, 10, 77. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Rai, A.K.; Kanwar, S.S.; Sharma, T.R. Comparative analysis of zinc finger proteins involved in plant disease resistance. PLoS ONE 2012, 7, 42578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aznar-Moreno, J.A.; Sánchez, R.; Gidda, S.K.; Martínez-Force, E.; Moreno-Pérez, A.J.; Calerón, M.V.; Garcés, R.; Mullen, R.T.; Salas, J.J. New insights into sunflower (Helianthus annuus L.) FatA and FatB thioesterases, their regulation, structure and distribution. Front. Plant Sci. 2018, 9, 1496. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| S. No. | Inbred | Code | Type | Source |
|---|---|---|---|---|
| 1 | PMI-PV5 | Lpa1-wild-1 | Wild-type | ICAR-IARI, New Delhi |
| 2 | PMI-PV6 | Lpa1-wild-2 | Wild-type | ICAR-IARI, New Delhi |
| 3 | PMI-PV7 | Lpa1-wild-3 | Wild-type | ICAR-IARI, New Delhi |
| 4 | PMI-PV8 | Lpa1-wild-4 | Wild-type | ICAR-IARI, New Delhi |
| 5 | PMI-Q1 | Lpa1-wild-5 | Wild-type | ICAR-IARI, New Delhi |
| 6 | PMI-Q2 | Lpa1-wild-6 | Wild-type | ICAR-IARI, New Delhi |
| 7 | PMI-Q3 | Lpa1-wild-7 | Wild-type | ICAR-IARI, New Delhi |
| 8 | PMI-LP1-124 | lpa1-1-mutant-1 | Mutant-type | ICAR-IARI, New Delhi |
| 9 | A619 lpa1-1 | lpa1-1-mutant-2 | Mutant-type | USDA-ARS, Aberdeen, ID, USA |
| 10 | A632 lpa1-1 | lpa1-1-mutant-3 | Mutant-type | USDA-ARS, Aberdeen, ID, USA |
| S. No. | Accessions | Gene ID | Protein ID | |
|---|---|---|---|---|
| 1 | Lpa1-wild-1 | Zea mays | Nucleotide sequence generated in the present study | Protein sequence translated from nucleotide sequence |
| 2 | Lpa1-wild-2 | |||
| 3 | Lpa1-wild-3 | |||
| 4 | Lpa1-wild-4 | |||
| 5 | Lpa1-wild-5 | |||
| 6 | Lpa1-wild-6 | |||
| 7 | Lpa1-wild-7 | |||
| 8 | lpa1-1-mutant-1 | |||
| 9 | lpa1-1-mutant-2 | |||
| 10 | lpa1-1-mutant-3 | |||
| 11 | ZmMRP4-B73-Ref | Zm00001eb003490 | A7KVC2 | |
| 12 | Aegilops tauschi | AET4Gv20803900 | M8CWG8 | |
| 13 | Ananas comosus | Aco010163 | A0A199URG9 | |
| 14 | Asparagus officinalis | A4U43_C10F18740 | A0A5P1E459 | |
| 15 | Brachypodium distachyon | BRADI_1g75590v3 | I1H9W0 | |
| 16 | Dioscorea rotundata | DRNTG_05198 | DRNTG_05198.1 | |
| 17 | Eragrostis curvula | EJB05_08061 | TVU48425 | |
| 18 | Hordeum vulgare | HORVU.MOREX.r3.4HG0412040 | A0A287PXI6 | |
| 19 | Leersia perrieri | LPERR03G03070 | A0A0D9VPF0 | |
| 20 | Musa acuminata | Ma08_g12530 | Ma08_t12530.1 | |
| 21 | Musa acuminata | Ma11_g02290 | Ma11_t02290.2 | |
| 22 | Oryza barthii | OBART03G03330 | A0A0D3FDN4 | |
| 23 | Oryza brachyantha | OB03G13350 | J3LJV9 | |
| 24 | Panicum hallii HAL2 | GQ55_9G618800 | A0A2T7CHT9 | |
| 25 | Secale cereale | SECCE5Rv1G0369730 | SECCE5Rv1G0369730.1 | |
| 26 | Setaria viridis | SEVIR_9G548400v2 | A0A4U6TCN8 | |
| 27 | Sorghum bicolor | SORBI_3001G508200 | A0A1Z5SBX3 | |
| 28 | Triticum aestivum | TraesCS5A02G512500 | A0A1D5YDM9 | |
| 29 | Triticum dicoccoides | TRIDC4BG057910 | TRIDC4BG057910.7 | |
| 30 | Triticum turgidum | TRITD5Av1G244640 | TRITD5Av1G244640.2 | |
| 31 | Triticum urartu | TuG1812G0500005238.01 | M8AP62 | |
| 32 | Oryza sativa Japonica Group | OsABCC13 | Q10RX7 | |
| 33 | Oryza sativa Indica Group | ABCC13 BGIOSGA011835 | A2XCD4 | |
| 34 | Oryza rufipogon | ORUFI03G03050 | A0A0E0NPI1 | |
| 35 | Oryza nivara | ONIVA03G03060 | A0A0E0GGM3 | |
| 36 | Eragrostis tef | Et_s2678-1.30-1.path1 | Et_s2678-1.30-1.mrna1 | |
| 37 | Setaria italica | SETIT_033885mg | K4A4T1 | |
| S. No. | Marker | Major Allele Frequency | No. of Alleles | Gene Diversity | Heterozygosity | PIC |
|---|---|---|---|---|---|---|
| 1 | lpa1-InDel-1 | 0.8750 | 2.00 | 0.2188 | 0.0000 | 0.1948 |
| 2 | lpa1-InDel-2 | 0.5833 | 3.00 | 0.5729 | 0.0000 | 0.5101 |
| 3 | lpa1-InDel-3 | 0.7917 | 2.00 | 0.3299 | 0.0000 | 0.2755 |
| 4 | lpa1-InDel-4 | 0.7500 | 2.00 | 0.3750 | 0.0000 | 0.3047 |
| 5 | lpa1-InDel-5 | 0.5208 | 3.00 | 0.6120 | 0.0000 | 0.5423 |
| 6 | lpa1-InDel-6 | 0.4792 | 3.00 | 0.5720 | 0.0000 | 0.4783 |
| 7 | lpa1-InDel-7 | 0.7292 | 2.00 | 0.3950 | 0.0000 | 0.3170 |
| 8 | lpa1-InDel-8 | 0.7292 | 2.00 | 0.3950 | 0.0000 | 0.3170 |
| 9 | lpa1-InDel-9 | 0.8125 | 2.00 | 0.3047 | 0.0417 | 0.2583 |
| 10 | lpa1-InDel-10 | 0.4583 | 3.00 | 0.6007 | 0.0000 | 0.5158 |
| 11 | lpa1-InDel-11 | 0.7292 | 3.00 | 0.4210 | 0.0000 | 0.3704 |
| 12 | lpa1-InDel-12 | 0.6458 | 3.00 | 0.5026 | 0.0000 | 0.4346 |
| 13 | lpa1-InDel-13 | 0.6250 | 3.00 | 0.5078 | 0.0000 | 0.4277 |
| 14 | lpa1-InDel-14 | 0.6458 | 2.00 | 0.4575 | 0.0000 | 0.3528 |
| 15 | lpa1-InDel-15 | 0.5833 | 3.00 | 0.5174 | 0.0000 | 0.4200 |
| Mean | 0.6639 | 2.53 | 0.4521 | 0.0028 | 0.3813 | |
| S. No. | Sequence | Crop Species | Amino Acid | Molecular Weight | Isoelectric Point (pI) | Negatively Charged aa (Asp + Glu) | Positively Charged aa (Arg + Lys) | Instability Index | Aliphatic Index | GRAVY |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | A7KVC2 | Zea mays | 1510 | 166,790.19 | 8.44 | 145 | 155 | 46.28 | 107.19 | 0.19 |
| 2 | PMI-PV5 | 1196 | 131,732.1 | 7.16 | 129 | 128 | 47.47 | 101.41 | −0.006 | |
| 5 | PMI-PV6 | 1127 | 122,784.47 | 8.53 | 111 | 121 | 42.87 | 105.98 | 0.094 | |
| 4 | PMI-PV7 | 1076 | 118,397.43 | 9.3 | 105 | 135 | 48.05 | 102.49 | −0.02 | |
| 3 | PMI-PV8 | 1275 | 140,824.85 | 9.36 | 105 | 145 | 49.87 | 105.86 | 0.174 | |
| 7 | PMI-Q1 | 1085 | 119,470.42 | 7.87 | 101 | 104 | 47.4 | 108.47 | 0.229 | |
| 6 | PMI-Q2 | 1186 | 130,285.93 | 6.7 | 115 | 111 | 47.28 | 106.69 | 0.173 | |
| 8 | PMI-Q3 | 1176 | 129,741.83 | 8.76 | 117 | 134 | 50.07 | 102.82 | 0.055 | |
| 9 | A632 lpa 1-1 | 1134 | 124,915.64 | 7.66 | 115 | 117 | 48.58 | 109 | 0.186 | |
| 10 | PMI-LP1-124 | 1004 | 111,797.38 | 8.89 | 98 | 114 | 48.23 | 96.74 | −0.024 | |
| 11 | A619 lpa1-1 | 632 | 70,300.03 | 6.59 | 76 | 74 | 47.23 | 100.55 | −0.112 | |
| 12 | M8CWG8 | Aegilops tauschi | 1504 | 166,057.78 | 8.76 | 150 | 166 | 46.28 | 102.51 | 0.096 |
| 13 | A0A199URG9 | Ananas comosus | 1522 | 168,424.55 | 8.27 | 152 | 159 | 42.97 | 103.21 | 0.168 |
| 14 | A0A5P1E459 | Asparagus officinalis | 583 | 65,439.23 | 8.28 | 58 | 61 | 44.06 | 103.22 | 0.108 |
| 15 | I1H9W0 | Brachypodium distachyon | 1505 | 165,978.66 | 8.1 | 151 | 156 | 43.57 | 104.51 | 0.162 |
| 16 | DRNTG_05198.1 | Dioscorea rotundata | 1522 | 170,326.47 | 8.36 | 148 | 156 | 40.46 | 106.74 | 0.185 |
| 17 | TVU48425 | Eragrostis curvula | 1502 | 165,632.69 | 7.62 | 151 | 153 | 43.09 | 105.88 | 0.185 |
| 18 | A0A287PXI6 | Hordeum vulgare | 1080 | 120,082.52 | 8.22 | 115 | 120 | 43.63 | 104.93 | 0.096 |
| 19 | A0A0D9VPF0 | Leersia perrieri | 1505 | 166,391.42 | 7.23 | 154 | 154 | 44.29 | 106.35 | 0.177 |
| 20 | Ma08_t12530.1 | Musa acuminata | 1511 | 168,530.23 | 8.18 | 152 | 158 | 42.51 | 107.4 | 0.184 |
| 21 | Ma11_t02290.2 | 1502 | 168,023.89 | 8.56 | 151 | 163 | 43.58 | 106.76 | 0.183 | |
| 22 | A0A0D3FDN4 | Oryza barthii | 1385 | 154,216.68 | 6.28 | 152 | 144 | 45.36 | 103.34 | 0.089 |
| 23 | J3LJV9 | Oryza brachyantha | 1128 | 125,411.6 | 5.88 | 135 | 121 | 44.12 | 102.15 | −0.015 |
| 24 | A0A2T7CHT9 | Panicum hallii HAL2 | 1504 | 165,872.84 | 8.19 | 146 | 152 | 44.38 | 106.8 | 0.178 |
| 25 | SECCE5Rv1G0369730.1 | Secale cereale | 1509 | 165,870.61 | 7.97 | 152 | 156 | 43.63 | 105.15 | 0.173 |
| 26 | A0A4U6TCN8 | Setaria viridis | 1507 | 166,595.77 | 8.48 | 145 | 155 | 45.28 | 106.77 | 0.177 |
| 27 | A0A1Z5SBX3 | Sorghum bicolor | 1512 | 166,559.92 | 8.31 | 146 | 154 | 44.67 | 107.38 | 0.203 |
| 28 | A0A1D5YDM9 | Triticum aestivum | 1441 | 159,222.76 | 7.06 | 150 | 149 | 42.19 | 104.74 | 0.155 |
| 29 | TRIDC4BG057910.7 | Triticum dicoccoides | 1483 | 163,430.9 | 8.09 | 150 | 155 | 42.55 | 105.14 | 0.161 |
| 30 | TRITD5Av1G244640.2 | Triticum turgidum | 1080 | 119,946.28 | 8.1 | 115 | 119 | 45.13 | 104.57 | 0.091 |
| 31 | M8AP62 | Triticum urartu | 1346 | 149,368.05 | 6.57 | 145 | 141 | 41.59 | 103.4 | 0.1 |
| 32 | Q10RX7 | Oryza sativa Japonica Group | 1446 | 159,766.09 | 7.78 | 153 | 155 | 43.48 | 102.95 | 0.085 |
| 33 | A2XCD4 | Oryza sativa Indica Group | 1357 | 149,630.27 | 6.39 | 148 | 141 | 44.49 | 104.52 | 0.11 |
| 34 | A0A0E0NPI1 | Oryza rufipogon | 1448 | 160,012.38 | 7.47 | 154 | 155 | 43.74 | 102.81 | 0.084 |
| 35 | A0A0E0GGM3 | Oryza nivara | 1411 | 155,399.84 | 7.28 | 149 | 149 | 44.42 | 102.54 | 0.075 |
| 36 | Et_s2678-1.30-1.mrna1 | Eragrostis tef | 711 | 79,605.95 | 5.09 | 94 | 76 | 47.16 | 105.88 | −0.005 |
| 37 | K4A4T1 | Setaria italica | 1435 | 158,725.42 | 8.53 | 142 | 153 | 45.2 | 105.74 | 0.136 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhatt, V.; Muthusamy, V.; Chhabra, R.; Katral, A.; Ragi, S.; Rojaria, V.; Chand, G.; Sarma, G.R.; Zunjare, R.U.; Panda, K.K.; et al. Molecular Characterization and Haplotype Analysis of Low Phytic Acid-1 (lpa1) Gene Governing Accumulation of Kernel Phytic Acid in Subtropically-Adapted Maize. Agriculture 2023, 13, 1286. https://doi.org/10.3390/agriculture13071286
Bhatt V, Muthusamy V, Chhabra R, Katral A, Ragi S, Rojaria V, Chand G, Sarma GR, Zunjare RU, Panda KK, et al. Molecular Characterization and Haplotype Analysis of Low Phytic Acid-1 (lpa1) Gene Governing Accumulation of Kernel Phytic Acid in Subtropically-Adapted Maize. Agriculture. 2023; 13(7):1286. https://doi.org/10.3390/agriculture13071286
Chicago/Turabian StyleBhatt, Vinay, Vignesh Muthusamy, Rashmi Chhabra, Ashvinkumar Katral, Shridhar Ragi, Vinay Rojaria, Gulab Chand, Govinda Rai Sarma, Rajkumar Uttamrao Zunjare, Kusuma Kumari Panda, and et al. 2023. "Molecular Characterization and Haplotype Analysis of Low Phytic Acid-1 (lpa1) Gene Governing Accumulation of Kernel Phytic Acid in Subtropically-Adapted Maize" Agriculture 13, no. 7: 1286. https://doi.org/10.3390/agriculture13071286
APA StyleBhatt, V., Muthusamy, V., Chhabra, R., Katral, A., Ragi, S., Rojaria, V., Chand, G., Sarma, G. R., Zunjare, R. U., Panda, K. K., Singh, A. K., & Hossain, F. (2023). Molecular Characterization and Haplotype Analysis of Low Phytic Acid-1 (lpa1) Gene Governing Accumulation of Kernel Phytic Acid in Subtropically-Adapted Maize. Agriculture, 13(7), 1286. https://doi.org/10.3390/agriculture13071286







