Moderate Nitrogen Reduction Increases Nitrogen Use Efficiency and Positively Affects Microbial Communities in Agricultural Soils
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Experimental Treatment and Soil Sampling
2.3. Soil Properties and Nutrient Determination
2.4. 16 S rRNA and ITS Gene Sequencing
2.5. Data Analysis
3. Result
3.1. Soil Properties and Correlation Analysis
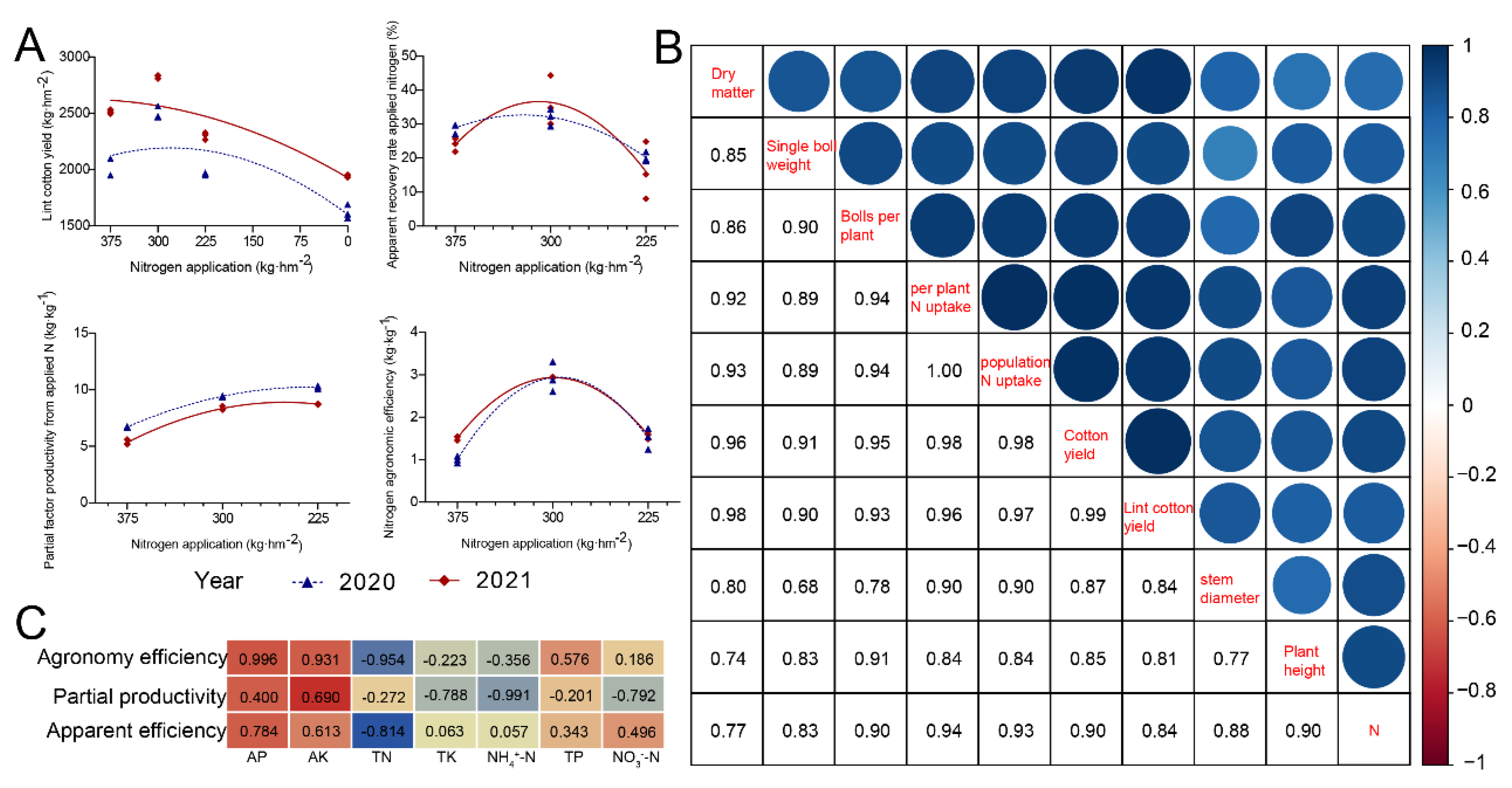
3.2. Plant Yield Components and Correlation Analysis
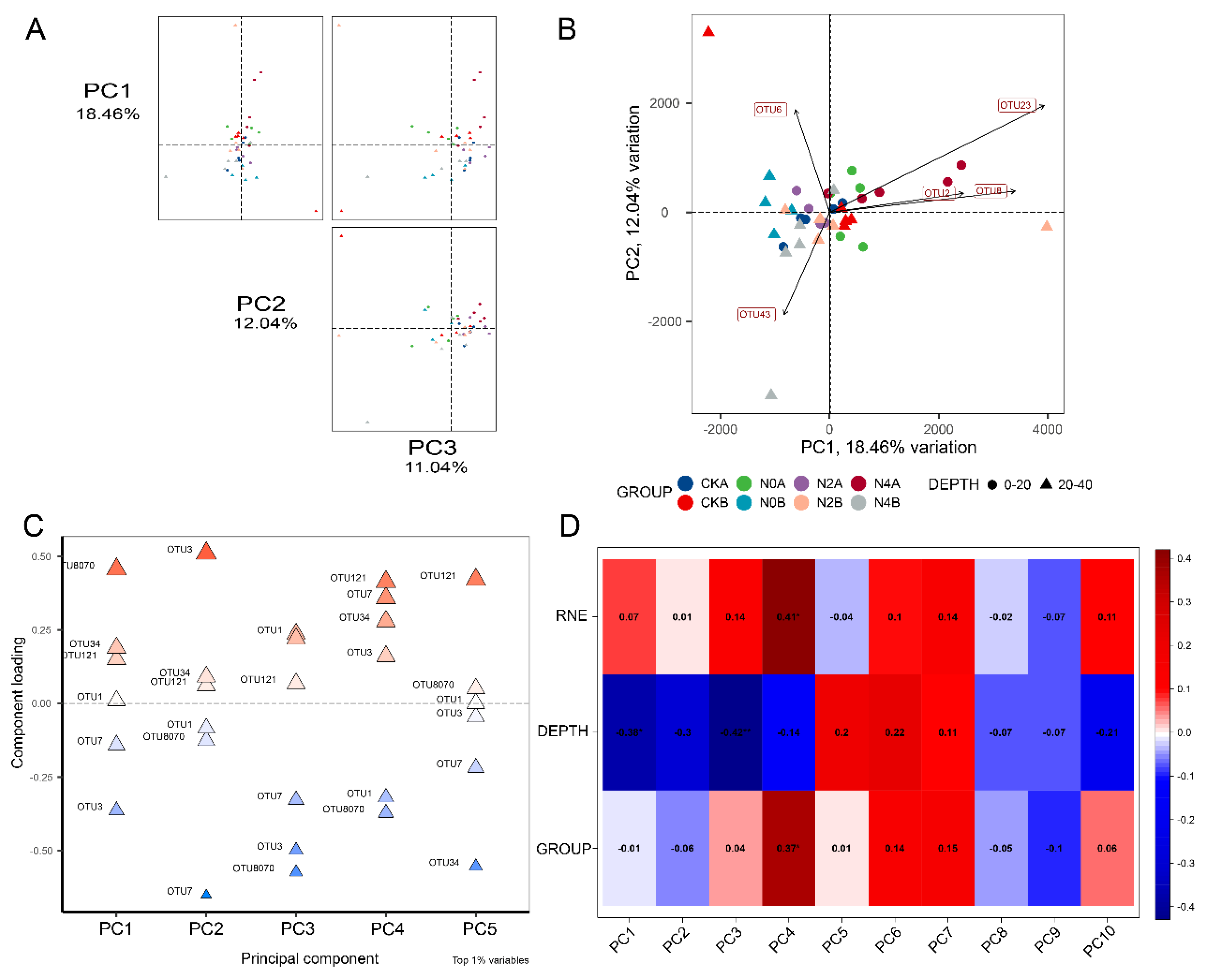
3.3. Grey Relational Analysis and Nitrogen Use Efficiency
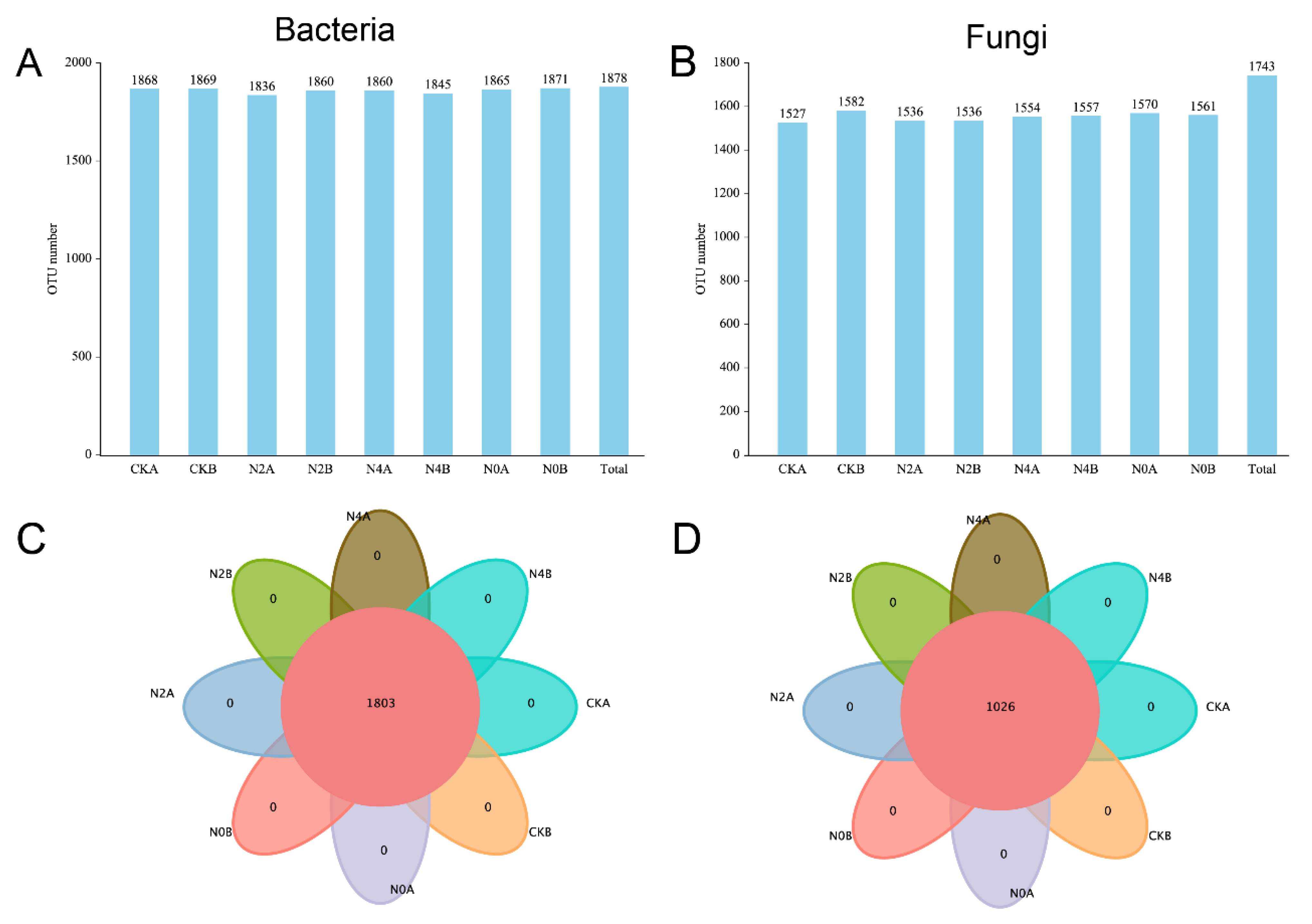
3.4. Identification of Soil Bacteria and Fungi in Different Soil Layers under RNE
3.5. Analysis of Microbial Community Diversity and Abundance
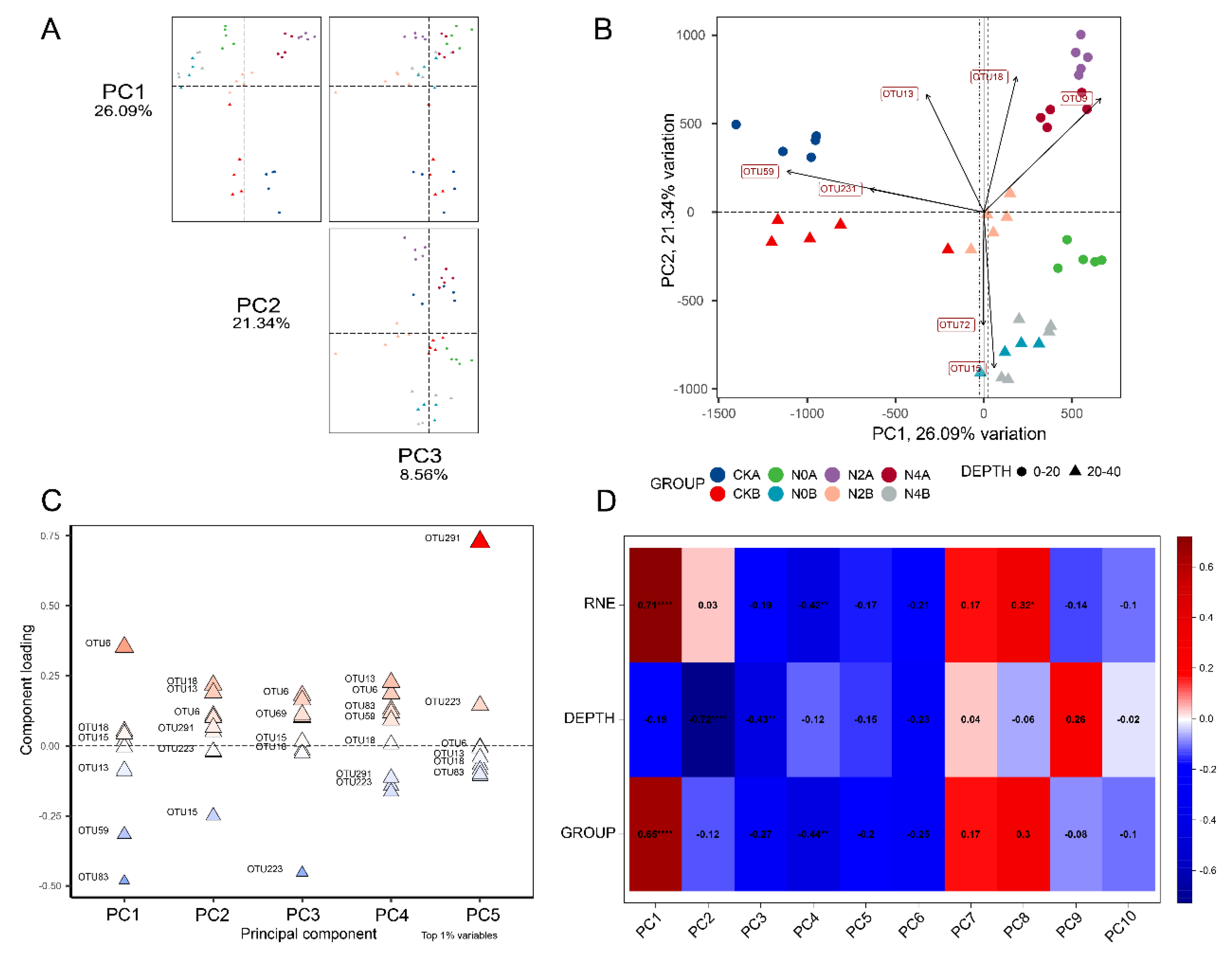
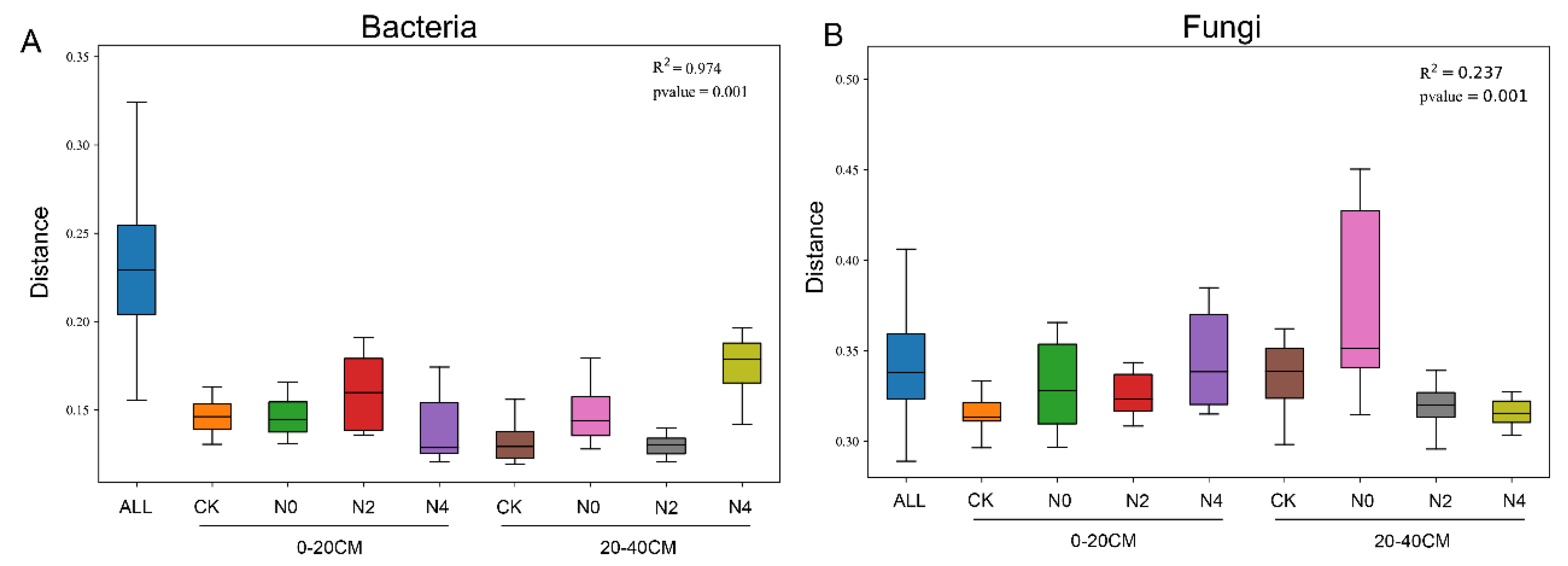
3.6. Analysis of Microbial Community Structure of Soils
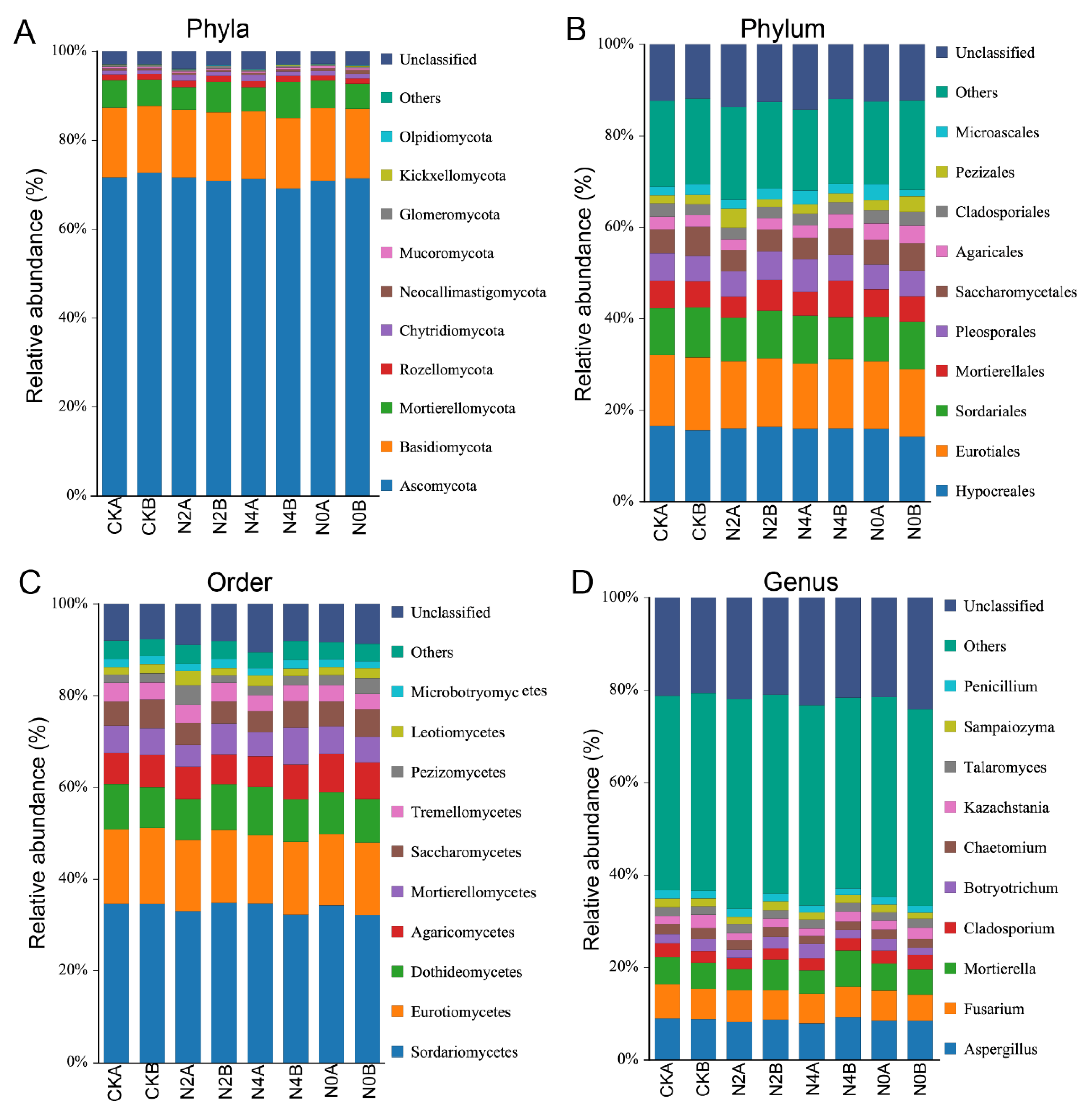
3.7. Bacterial Species Composition of Soils
3.8. Fungal Species Composition of Soils
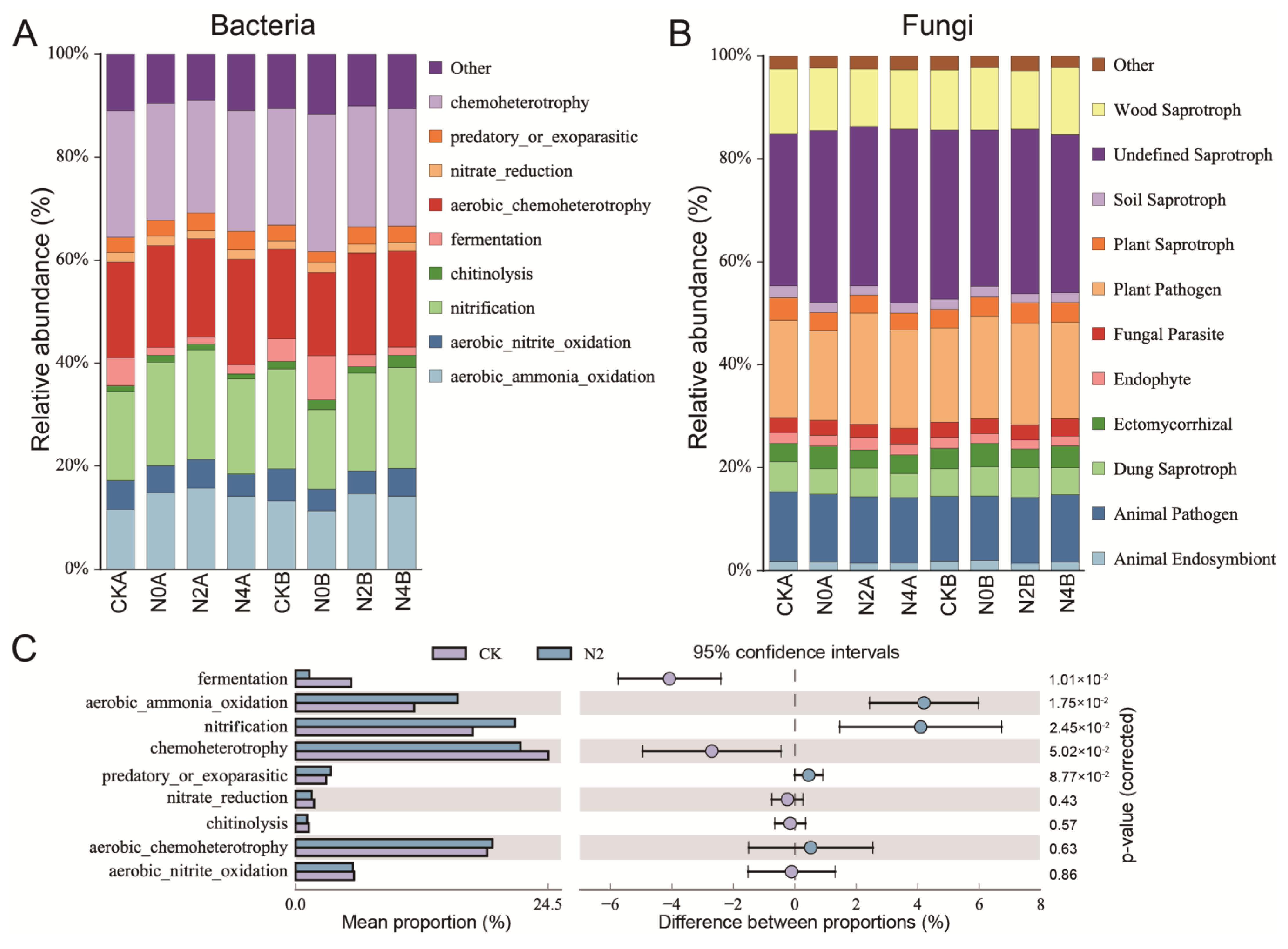
3.9. Functional Prediction of Bacteria and Fungi in Soils
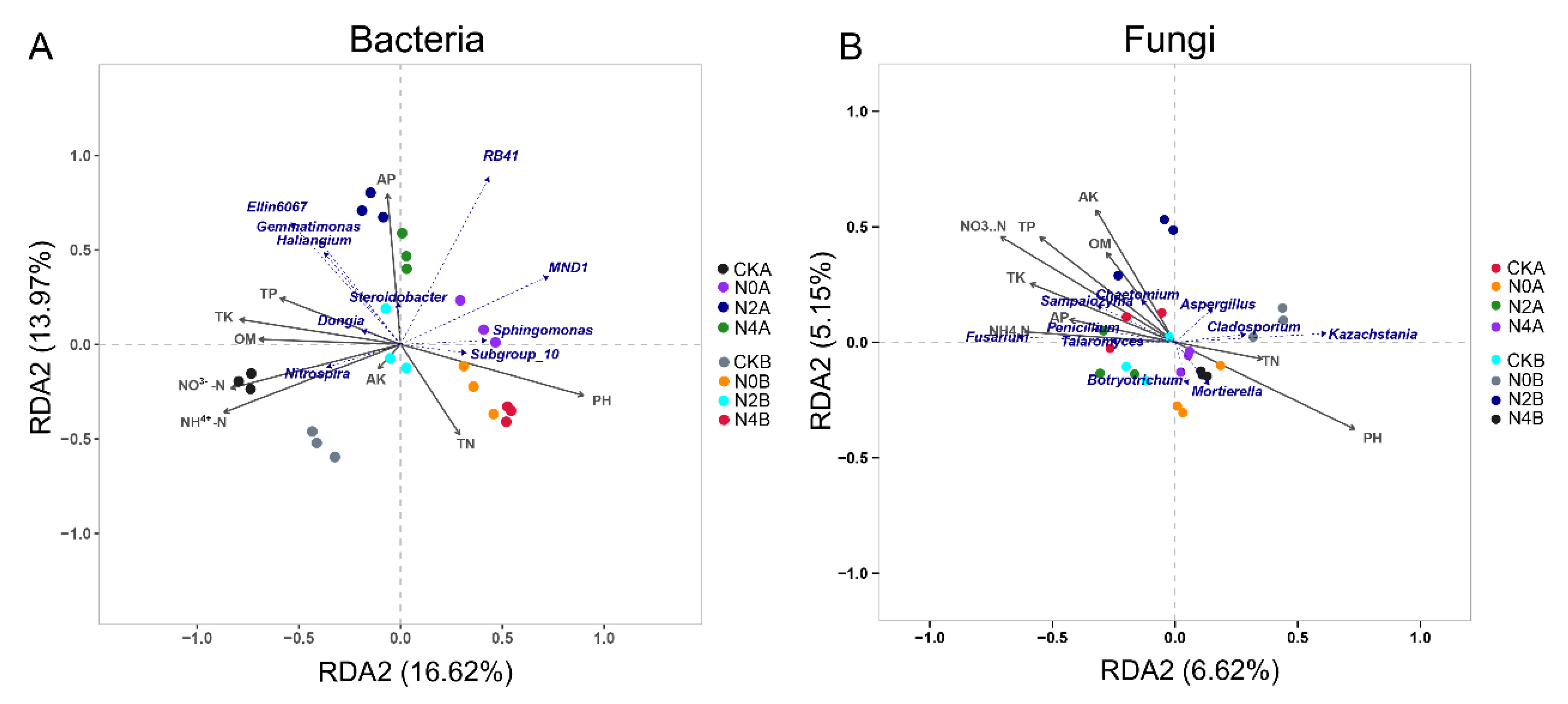
3.10. Correlation Analysis between Microorganisms and Soil Physicochemical Properties
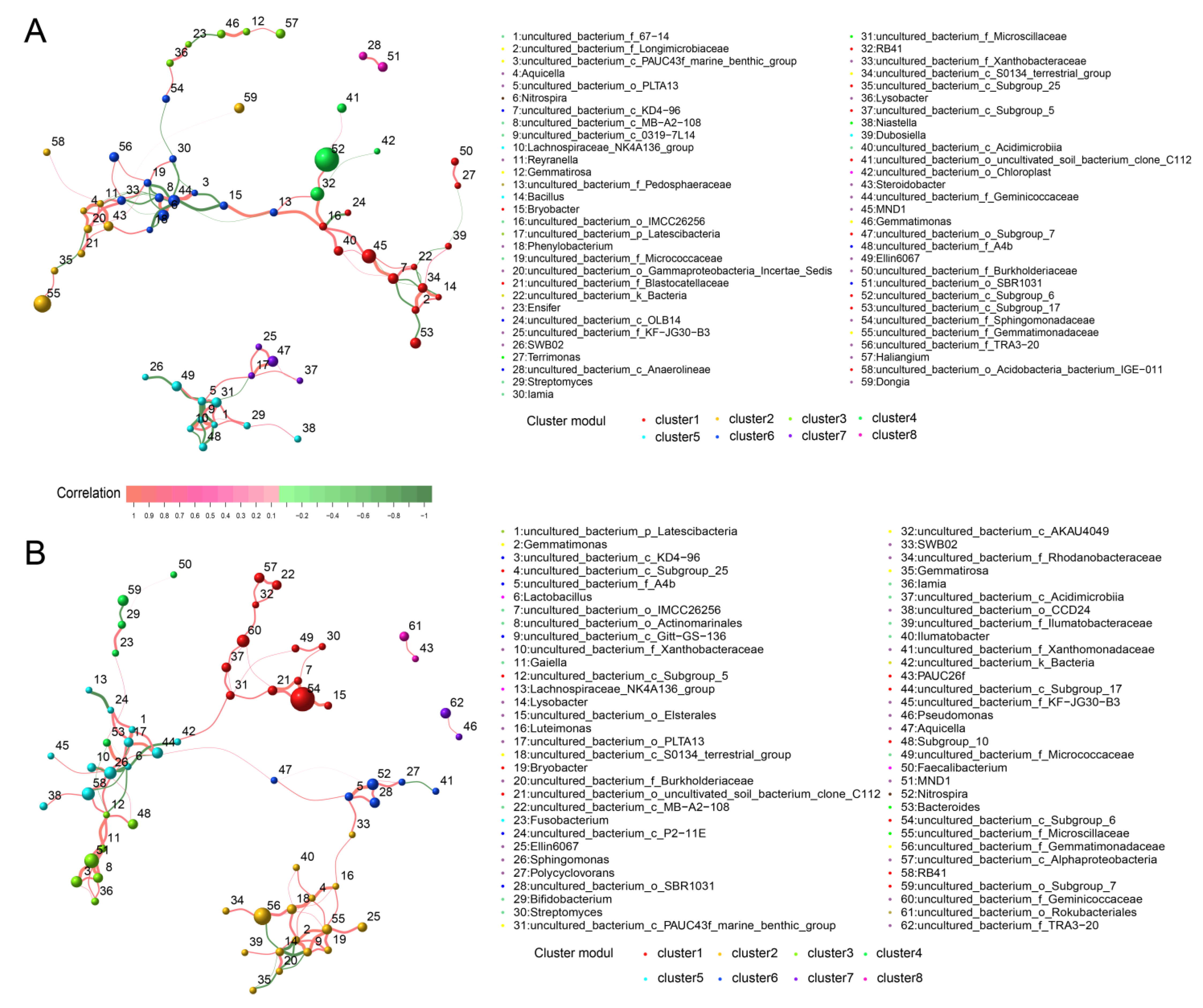
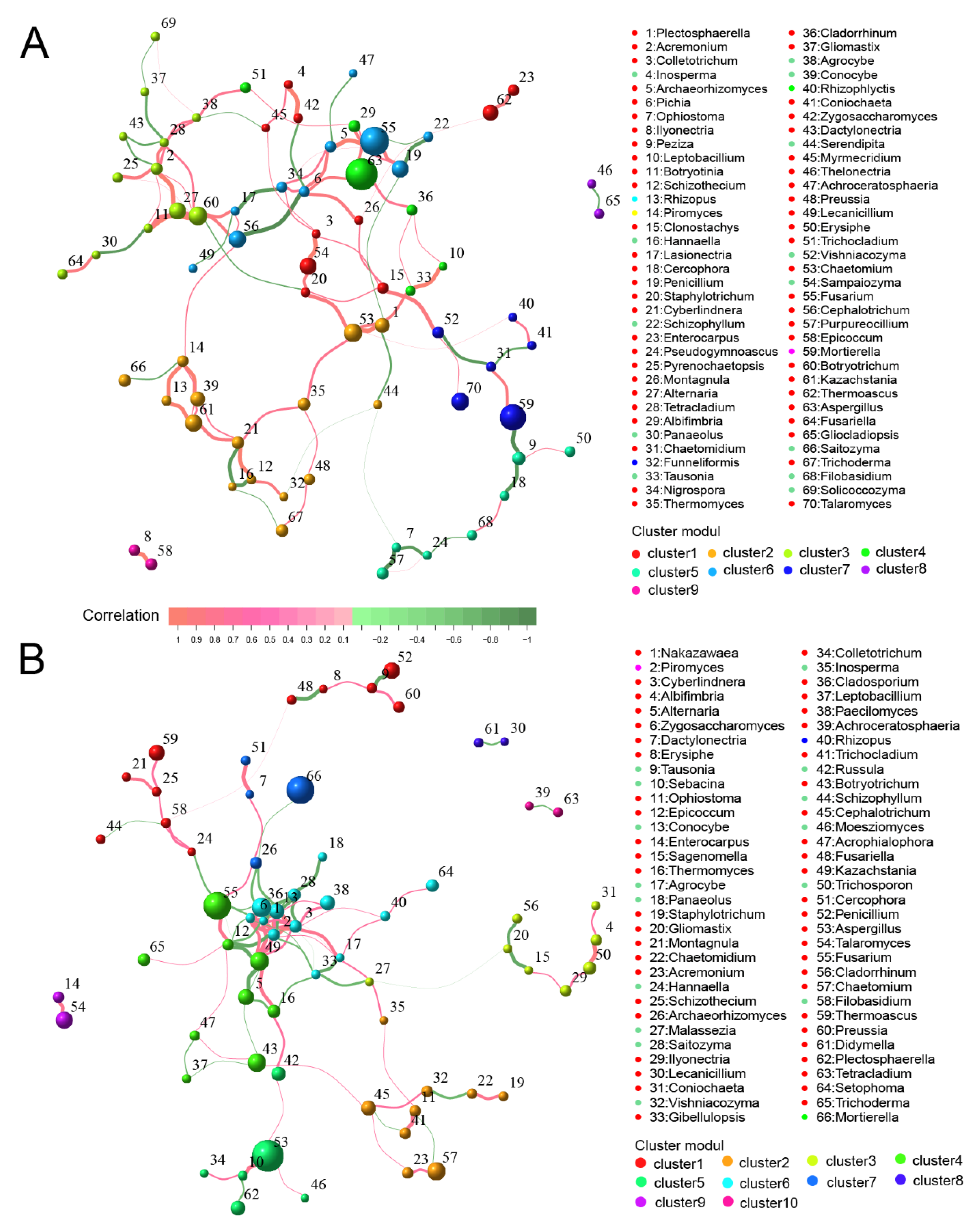
3.11. Co-Occurrence Network of Bacterial and Fungal Communities
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Lehmann, J.; Kleber, M. The contentious nature of soil organic matter. Nature 2015, 528, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Pan, G.; Ding, Y.; Chen, S.; Sun, J.; Feng, X.; Zhang, C.; Marios, D.; Zheng, J.; Zhang, X.; Cheng, K.; et al. Exploring the nature of soil organic matter from humic substances isolation to SOMics of molecular assemblage. Adv. Earth Sci. 2019, 34, 451–470. [Google Scholar] [CrossRef]
- Xu, Y.D. Conservation agriculture-mediated soil carbon sequestration: A review. Chin. J. Ecol. 2022, 30, 658–670. [Google Scholar] [CrossRef]
- Zhong, Y.; Yan, W.; Shangguan, Z. Impact of long-term N additions upon coupling between soil microbial community structure and activity, and nutrient-use efficiencies. Soil Biol. Biochem. 2015, 91, 151–159. [Google Scholar] [CrossRef]
- Ganugi, P.; Fiorini, A.; Ardenti, F.; Caffi, T.; Bonini, P.; Taskin, E.; Puglisi, E.; Tabaglio, V.; Trevisan, M.; Lucini, L. Nitrogen use efficiency, rhizosphere bacterial community, and root metabolome reprogramming due to maize seed treatment with microbial biostimulants. Physiol. Plant. 2022, 174, e13679. [Google Scholar] [CrossRef]
- Martínez-Dalmau, J.; Berbel, J.; Ordóñez-Fernández, R. Nitrogen fertilization. A review of the risks associated with the inefficiency of its use and policy responses. Sustainability 2021, 13, 5625. [Google Scholar] [CrossRef]
- Yang, X.-D.; Ni, K.; Shi, Y.-Z.; Yi, X.-Y.; Zhang, Q.-F.; Fang, L.; Ma, L.-F.; Ruan, J. Effects of long-term nitrogen application on soil acidification and solution chemistry of a tea plantation in China. Agric. Ecosyst. Environ. 2018, 252, 74–82. [Google Scholar] [CrossRef]
- Liu, X.-P.; Bi, Q.-F.; Qiu, L.-L.; Li, K.-J.; Yang, X.-R.; Lin, X.-Y. Increased risk of phosphorus and metal leaching from paddy soils after excessive manure application: Insights from a mesocosm study. Sci. Total Environ. 2019, 666, 778–785. [Google Scholar] [CrossRef]
- Schweitzer, J.A.; Bailey, J.K.; Fischer, D.G.; LeRoy, C.J.; Lonsdorf, E.V.; Whitham, T.G.; Hart, S.C. Plant–soil–microorganism interactions: Heritable relationship between plant genotype and associated soil microorganisms. Ecology 2008, 89, 773–781. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Y.; Ouyang, D.; Lei, J.; Tan, Q.; Xie, L.; Li, Z.; Liu, T.; Xiao, Y.; Farooq, T.H.; et al. Systematical review of interactions between microplastics and microorganisms in the soil environment. J. Hazard. Mater. 2021, 418, 126288. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Lan, Y.; Wang, Q.; Cole, J.R.; Rosen, G.L. Using the rdp classifier to predict taxonomic novelty and reduce the search space for finding novel organisms. PLoS ONE 2012, 7, e32491. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microb. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Liang, S.; Deng, J.; Jiang, Y.; Wu, S.; Zhou, Y.; Zhu, W.-X. Functional distribution of bacterial community under different land use patterns based on FaProTax function prediction. Pol. J. Environ. Stud. 2019, 29, 1245–1261. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An open source software for exploring and manipulating networks. In Proceedings of the International AAAI Conference on Web and Social Media, San Jose, CA, USA, 17–20 May 2009; Available online: https://ojs.aaai.org/index.php/ICWSM/article/view/13937 (accessed on 2 March 2022).
- Zangani, E.; Afsahi, K.; Shekari, F.; Mac Sweeney, E.; Mastinu, A. Nitrogen and phosphorus addition to soil improves seed yield, foliar stomatal conductance, and the photosynthetic response of rapeseed (Brassica napus L.). Agriculture 2021, 11, 483. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, X.; Xu, W.; Liu, X.; Li, Y.; Wei, J. Ammonia volatilization as the major nitrogen loss pathway in dryland agro-ecosystems. Environ. Pollut. 2020, 265, 114862. [Google Scholar] [CrossRef]
- Ma, R.; Yu, K.; Xiao, S.; Liu, S.; Ciais, P.; Zou, J. Data-driven estimates of fertilizer-induced soil NH3, NO and N2O emissions from croplands in China and their climate change impacts. Glob. Chang. Biol. 2022, 28, 1008–1022. [Google Scholar] [CrossRef]
- Wang, Y.; Ying, H.; Yin, Y.; Zheng, H.; Cui, Z. Estimating soil nitrate leaching of nitrogen fertilizer from global meta-analysis. Sci. Total Environ. 2019, 657, 96–102. [Google Scholar] [CrossRef]
- Tautges, N.E.; Chiartas, J.L.; Gaudin, A.C.; O’Geen, A.T.; Herrera, I.; Scow, K.M. Deep soil inventories reveal that impacts of cover crops and compost on soil carbon sequestration differ in surface and subsurface soils. Glob. Chang. Biol. 2019, 25, 3753–3766. [Google Scholar] [CrossRef]
- Chen, J.; van Groenigen, K.J.; Hungate, B.A.; Terrer, C.; van Groenigen, J.W.; Maestre, F.T.; Ying, S.C.; Luo, Y.; Jørgensen, U.; Sinsabaugh, R.L.; et al. Long-term nitrogen loading alleviates phosphorus limitation in terrestrial ecosystems. Glob. Chang. Biol. 2020, 26, 5077–5086. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, R.H.; Stewart, J.W.B.; Dormaar, J.F.; Schaalje, G.B. Long-term crop rotation and fertilizer effects on phosphorus transformations: I. In a chernozemic soil. Can. J. Soil. Sci. 1992, 72, 569–579. [Google Scholar] [CrossRef]
- Tian, M.Y.; Yu, C.J.; Wang, J.K.; Ding, F.; Chen, Z.H.; Jiang, N.; Jiang, H.; Chen, L.J. Effect of nitrogen additions on soil pH, phosphorus contents and phosphatase activities in grassland. Chin. J. Appl. Ecol. 2020, 31, 2985–2992. [Google Scholar] [CrossRef]
- Jing, Y.; Zhu, H.; Ding, H.; Bi, R. Spatial variation in soil available potassium and temporal changes due to intrinsic and extrinsic factors: A 10-year study. J. Soil Sci. Plant Nutr. 2022, 22, 1305–1314. [Google Scholar] [CrossRef]
- Wang, J.; Li, R.; Zhang, H.; Wei, G.; Li, Z. Beneficial bacteria activate nutrients and promote wheat growth under conditions of reduced fertilizer application. BMC Biol. 2020, 20, 38. [Google Scholar] [CrossRef]
- Liao, L.; Wang, X.; Wang, J.; Liu, G.; Zhang, C. Nitrogen fertilization increases fungal diversity and abundance of saprotrophs while reducing nitrogen fixation potential in a semiarid grassland. Plant Soil 2021, 465, 515–532. [Google Scholar] [CrossRef]
- Beltran-Garcia, M.J.; Martínez-Rodríguez, A.; Olmos-Arriaga, I.; Valdes-Salas, B.; Di Mascio, P.; White, J.F. Nitrogen fertilization and stress factors drive shifts in microbial diversity in soils and plants. Symbiosis 2021, 84, 379–390. [Google Scholar] [CrossRef]
- Chen, L.; Li, K.K.; Shi, W.J.; Wang, X.L.; Wang, E.T.; Liu, J.F.; Sui, X.H.; Mi, G.H.; Tian, C.F.; Chen, W.X. Negative impacts of excessive nitrogen fertilization on the abundance and diversity of diazotrophs in black soil under maize monocropping. Geoderma 2021, 393, 114999. [Google Scholar] [CrossRef]
- Liu, S.; Qin, T.; Dong, B.; Shi, X.; Lv, Z.; Zhang, G. The influence of climate, soil properties and vegetation on soil nitrogen in sloping farmland. Sustainability 2021, 13, 1480. [Google Scholar] [CrossRef]
- Li, S.; Zhao, L.; Zhang, S.; Liu, Q.; Li, H. Effects of nitrogen level and soil moisture on sweet potato root distribution and soil chemical properties. J. Soil Sci. Plant Nutr. 2021, 21, 536–546. [Google Scholar] [CrossRef]
- Yu, M. Bacterial community structure and putative nitrogen-cycling functional traits along a charosphere gradient under waterlogged conditions. Soil Biol. Biochem. 2021, 162, 108420. [Google Scholar] [CrossRef]
- Banerjee, S.; Schlaeppi, K.; van der Heijden, M.G.A. Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol 2018, 16, 567–576. [Google Scholar] [CrossRef]
- Kalam, S.; Basu, A.; Ahmad, I.; Sayyed, R.Z.; El-Enshasy, H.A.; Dailin, D.J.; Suriani, N.L. Recent understanding of soil acidobacteria and their ecological significance: A critical review. Front. Microbiol. 2020, 11, 580024. [Google Scholar] [CrossRef]
- Asaf, S.; Numan, M.; Khan, A.L.; Al-Harrasi, A. Sphingomonas: From diversity and genomics to functional role in environmental remediation and plant growth. Crit. Rev. Biotechnol. 2020, 40, 138–152. [Google Scholar] [CrossRef]
- Aswini, C. A review on Chaetomium globosum is versatile weapons for various plant pathogens. J. Pharmacogn. Phytochem. 2019, 8, 946–949. [Google Scholar]
- Hao, X.; Zhu, Y.-G.; Nybroe, O.; Nicolaisen, M.H. The composition and phosphorus cycling potential of bacterial communities associated with hyphae of Penicillium in soil are strongly affected by soil origin. Front. Microbiol. 2020, 10, 2951. [Google Scholar] [CrossRef]
- Win, T.T.; Bo, B.; Malec, P.; Khan, S.; Fu, P. Newly isolated strain of Trichoderma asperellum from disease suppressive soil is a potential bio-control agent to suppress Fusarium soil borne fungal phytopathogens. J. Plant Pathol. 2021, 103, 549–561. [Google Scholar] [CrossRef]
- Cline, L.C.; Schilling, J.S.; Menke, J.; Groenhof, E.; Kennedy, P.G. Ecological and functional effects of fungal endophytes on wood decomposition. Funct. Ecol. 2018, 32, 181–191. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, G.; Yan, Z.; Wang, Z.; Fu, W. Effects of Furfural residues on community composition and diversity of rhizosphere microbe of seed production. Research Square 2020. [Google Scholar] [CrossRef]
- Xun, W.; Liu, Y.; Li, W.; Ren, Y.; Xiong, W.; Xu, Z.; Zhang, N.; Miao, Y.; Shen, Q. Specialized metabolic functions of keystone taxa sustain soil microbiome stability. Microbiome 2021, 9, 35. [Google Scholar] [CrossRef]
- Tang, R.; Li, X.; Mo, Y.; Ma, Y.; Ding, C.; Wang, J.; Zhang, T.; Wang, X. Toxic responses of metabolites, organelles and gut microorganisms of Eisenia fetida in a soil with chromium contamination. Environ. Pollut. 2019, 251, 910–920. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, R.; Lu, B.; Guerin-Laguette, A.; He, X.; Yu, F. Mycorrhization of Quercus mongolica seedlings by Tuber melanosporum alters root carbon exudation and rhizosphere bacterial communities. Plant Soil 2021, 467, 391–403. [Google Scholar] [CrossRef]
- Fähnrich, P.; Irrgang, K. Conversion of cellulose to sugars and cellobionic acid by the extracellular enzyme system of Chaetomium cellulolyticum. Biotechnol. Lett. 1982, 4, 775–780. [Google Scholar] [CrossRef]
- Rochefort, A.; Simonin, M.; Marais, C.; Guillerm-Erckelboudt, A.-Y.; Barret, M.; Sarniguet, A. Transmission of seed and soil microbiota to seedling. Msystems 2021, 6, e00446-21. [Google Scholar] [CrossRef]


| Category | Depth (cm) | Sample | ACE | Chao1 | Simpson | Shannon | Coverage |
|---|---|---|---|---|---|---|---|
| Bacteria | 0–20 | CK | 1789.56 ± 11.80 a | 1808.45 ± 12.36 a | 0.9965 ± 0.00007 a | 9.2153 ± 0.0191 a | 0.9986 |
| N0 | 1772.00 ± 5.34 ab | 1785.72 ± 4.89 ab | 0.9964 ± 0.00005 ab | 9.1571 ± 0.0194 ab | 0.9986 | ||
| N2 | 1725.51 ± 5.10 b | 1740.90 ± 8.02 b | 0.9959 ± 0.00011 b | 9.0625 ± 0.0316 b | 0.9986 | ||
| N4 | 1787.51 ± 6.00 a | 1801.99 ± 9.09 a | 0.9964 ± 0.00010 a | 9.231 ± 0.0303 a | 0.9987 | ||
| 20–40 | CK | 1796.32 ± 9.70 a | 1801.98 ± 7.62 a | 0.9968 ± 0.00007 a | 9.30208 ± 0.0213 a | 0.9988 | |
| N0 | 1713.39 ± 46.74 a | 1720.80 ± 53.05 a | 0.9921 ± 0.00435 a | 8.72612 ± 0.3914 a | 0.9971 | ||
| N2 | 1796.58 ± 5.88 a | 1809.07 ± 6.89 a | 0.9965 ± 0.00011 a | 9.2138 ± 0.0246 a | 0.9987 | ||
| N4 | 1771.34 ± 6.00 a | 1785.09 ± 9.78 a | 0.9966 ± 0.00005 a | 9.20336 ± 0.0133 a | 0.9986 | ||
| Fungi | 0–20 | CK | 1115.32 ± 45.34 a | 1182.79 ± 44.33 ab | 0.9916 ± 0.00017 a | 8.2311 ± 0.0355 a | 0.9987 |
| N0 | 1112.21 ± 60.46 a | 1230.61 ± 47.83 a | 0.9914 ± 0.00013 a | 8.2144 ± 0.0097 a | 0.9986 | ||
| N2 | 954.14 ± 14.19 a | 1024.35 ± 28.64 b | 0.9859 ± 0.00533 a | 8.0255 ± 0.1694 a | 0.999 | ||
| N4 | 1048.67 ± 64.67 a | 1116.14 ± 50.66 ab | 0.9905 ± 0.00065 a | 8.1488 ± 0.0610 a | 0.9988 | ||
| 20–40 | CK | 1046.40 ± 34.43 b | 1144.51 ± 43.98 a | 0.9904 ± 0.00097 a | 8.1777 ± 0.0520 a | 0.9988 | |
| N0 | 1067.76 ± 39.73 b | 1146.86 ± 32.23 a | 0.9903 ± 0.00088 a | 8.1708 ± 0.0549 a | 0.9988 | ||
| N2 | 1033.32 ± 27.14 b | 1117.48 ± 22.75 a | 0.9906 ± 0.00108 a | 8.1500 ± 0.0881 a | 0.9988 | ||
| N4 | 1260.62 ± 23.60 a | 1239.77 ± 30.63 a | 0.9904 ± 0.00048 a | 8.1757 ± 0.0359 a | 0.9982 |
| Category | Result | TN | NO3−-N | NH4+-N | AP | TP | TK | AK | OM | PH |
|---|---|---|---|---|---|---|---|---|---|---|
| Bacteria | R value | 0.129 | 0.496 | 0.689 | 0.198 | 0.088 | 0.396 | −0.082 | 0.304 | 0.535 |
| p-value | 0.058 | 0.001 | 0.001 | 0.008 | 0.160 | 0.001 | 0.807 | 0.003 | 0.001 | |
| Fungi | R value | −0.007 | −0.030 | −0.085 | 0.219 | −0.053 | 0.084 | −0.138 | 0.132 | −0.020 |
| p-value | 0.434 | 0.681 | 0.695 | 0.046 | 0.624 | 0.183 | 0.853 | 0.214 | 0.450 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, J.; Su, L.; Fang, Y.; Wang, C.; Meng, L.; Wang, J.; Zhang, J.; Xu, W. Moderate Nitrogen Reduction Increases Nitrogen Use Efficiency and Positively Affects Microbial Communities in Agricultural Soils. Agriculture 2023, 13, 796. https://doi.org/10.3390/agriculture13040796
Tang J, Su L, Fang Y, Wang C, Meng L, Wang J, Zhang J, Xu W. Moderate Nitrogen Reduction Increases Nitrogen Use Efficiency and Positively Affects Microbial Communities in Agricultural Soils. Agriculture. 2023; 13(4):796. https://doi.org/10.3390/agriculture13040796
Chicago/Turabian StyleTang, Jianghua, Lili Su, Yanfei Fang, Chen Wang, Linyi Meng, Jiayong Wang, Junyao Zhang, and Wenxiu Xu. 2023. "Moderate Nitrogen Reduction Increases Nitrogen Use Efficiency and Positively Affects Microbial Communities in Agricultural Soils" Agriculture 13, no. 4: 796. https://doi.org/10.3390/agriculture13040796
APA StyleTang, J., Su, L., Fang, Y., Wang, C., Meng, L., Wang, J., Zhang, J., & Xu, W. (2023). Moderate Nitrogen Reduction Increases Nitrogen Use Efficiency and Positively Affects Microbial Communities in Agricultural Soils. Agriculture, 13(4), 796. https://doi.org/10.3390/agriculture13040796






