Genetic Dissection of Tiller Number qTN4 in Rice
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Phenotype Measurement
2.3. Molecular Markers and Genotyping
2.4. Data Analysis
3. Results
3.1. Identification of QTLs, Especially qTN4 in the CSSLs Population
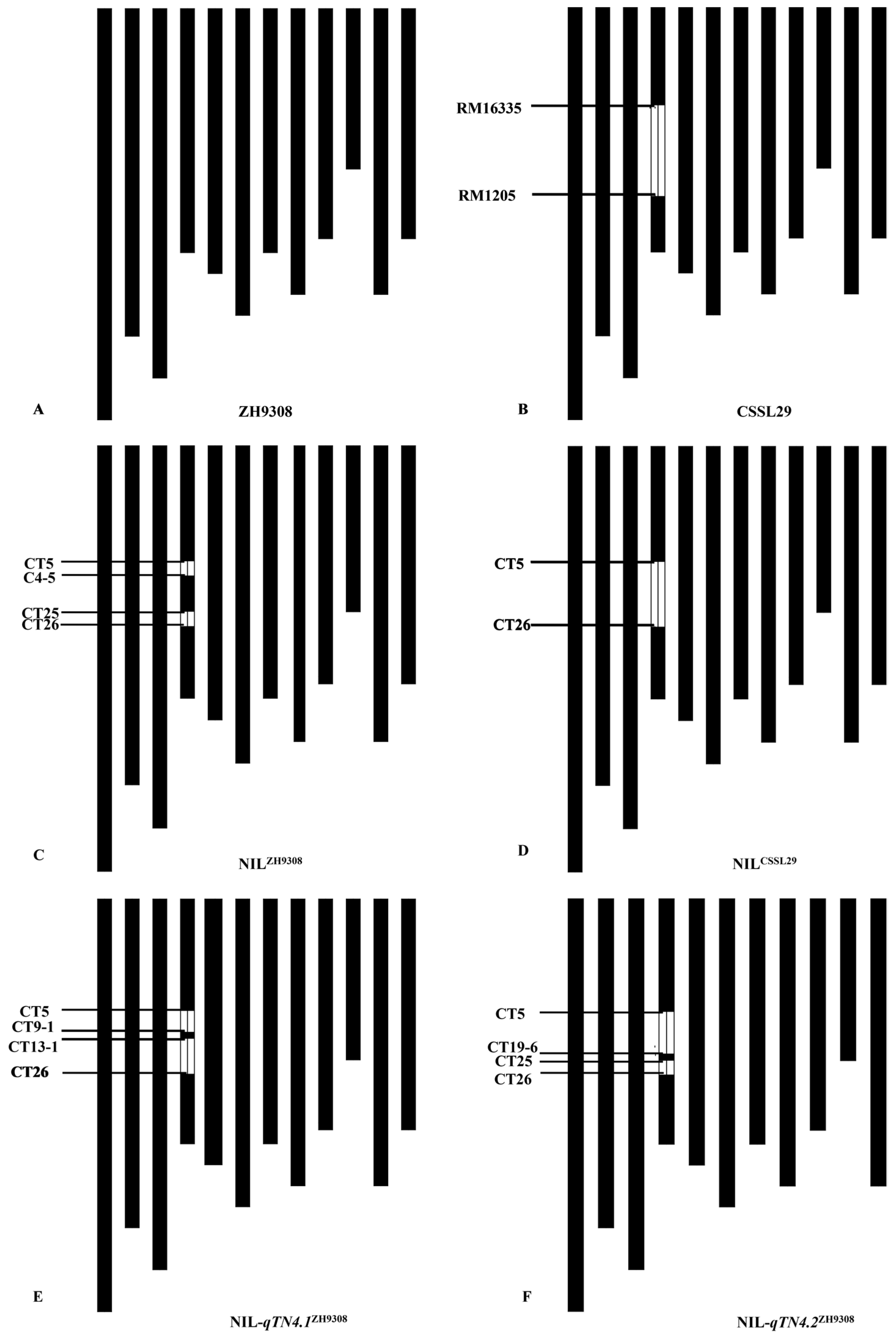
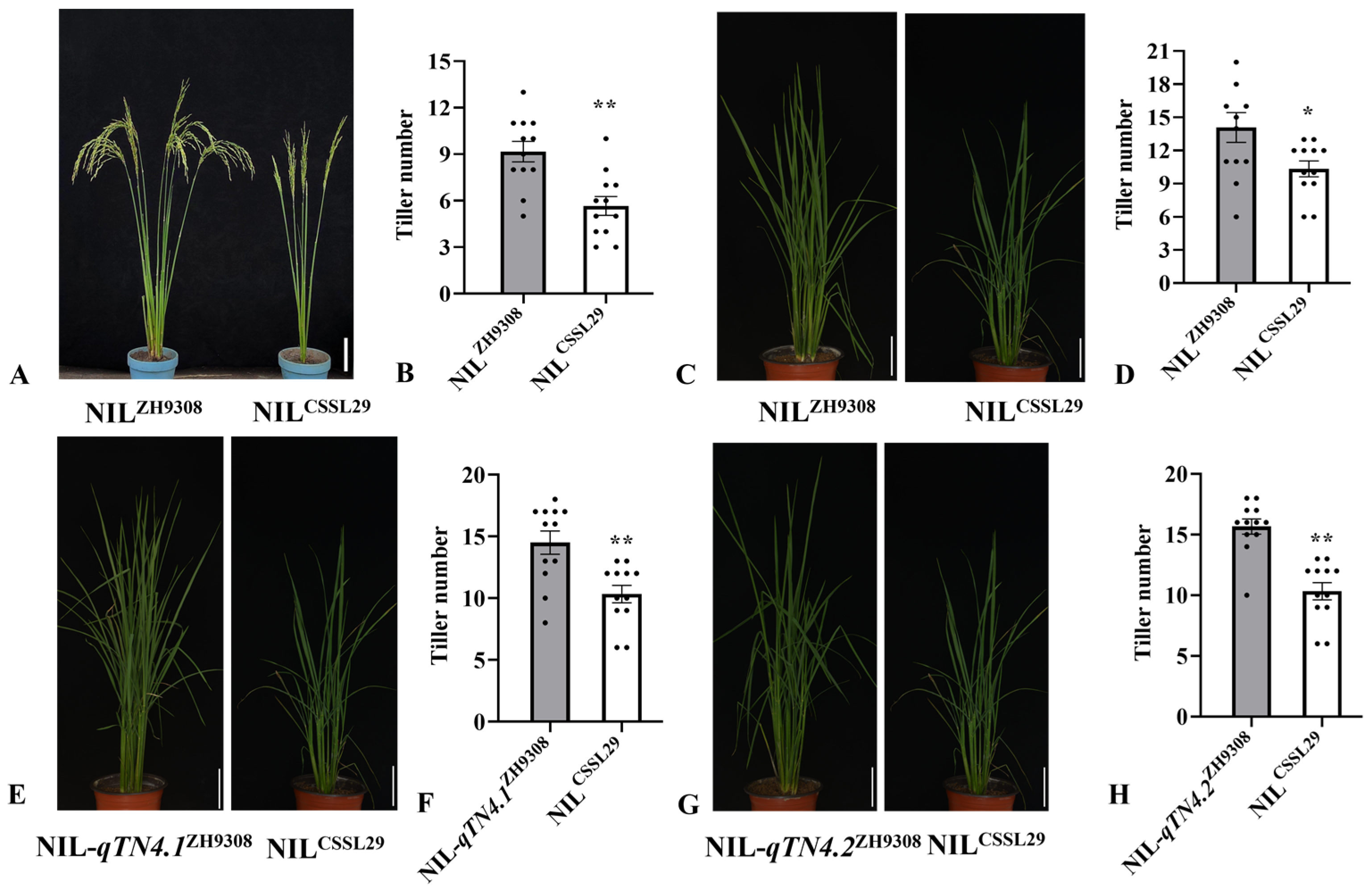
3.2. Validation and Mapping of the qTN4
3.3. Dissection of qTN4 into Two Quantitative Trait Loci
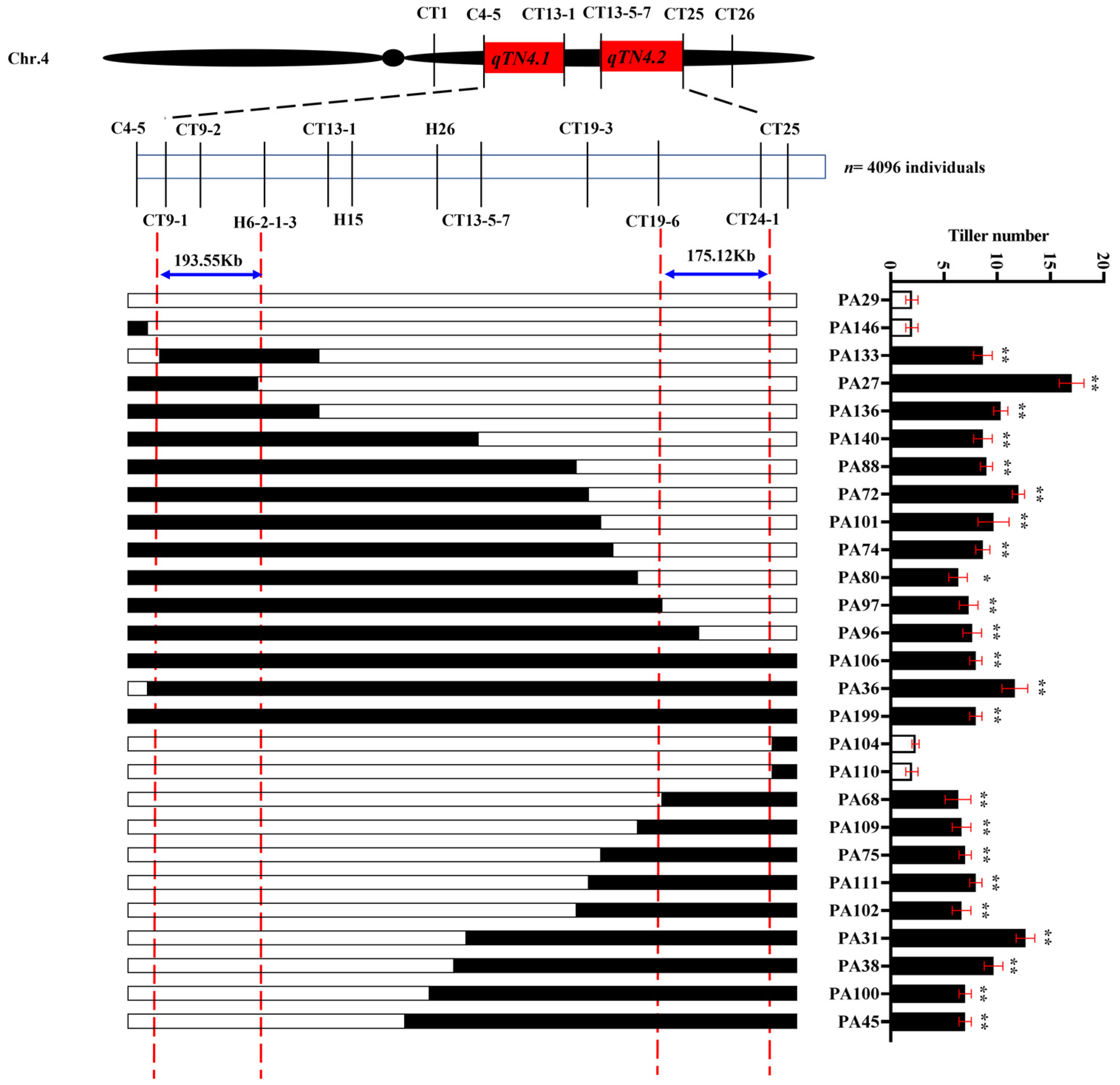
3.4. Fine Mapping of the qTN4.1 and qTN4.2
3.5. Candidate Genes of qTN4.1 and qTN4.2
4. Discussion
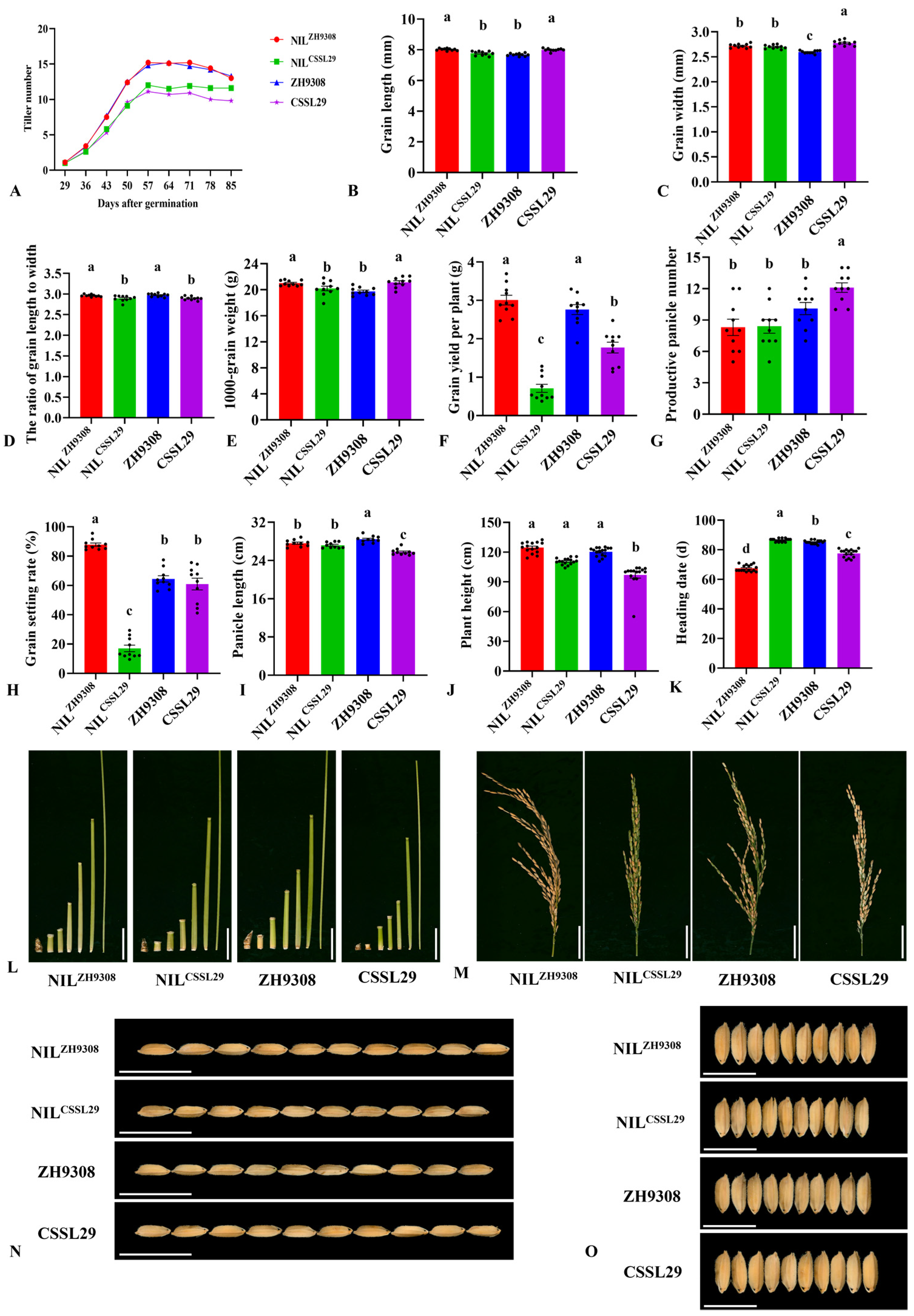
4.1. Phenotypic Variation among Important Agronomical Traits
4.2. Fine-Mapping for qTN4.1 and qTN4.2 and Analysis of Candidate Genes
4.3. The Prospect and Utilization of qTN4.1 and qTN4.2 in Rice Breeding
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Gayatri, G.; Kumar, G.M.; Ravindra, D.; Trilochan, M.; Ramakrishna, V.; Lambodar, B. Marker-assisted selection for grain number and yield-related traits of rice (Oryza Sativa L.). Physiol. Mol. Biol. Plants 2020, 26, 885–898. [Google Scholar]
- Godfray, H.C.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food security: The challenge of feeding 9 billion people. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Stamm, P.; Ramamoorthy, R.; Kumar, P.P. Feeding the extra billions: Strategies to improve crops and enhance future food security. Plant Biotechnol. Rep. 2011, 5, 107–120. [Google Scholar] [CrossRef]
- Xu, L.; Yuan, S.; Man, J.G. Changes in rice yield and yield stability in China during the past six decades. J. Sci. Food Agric. 2020, 100, 3560–3569. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.Z.; Zhang, Q.F. Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 2010, 61, 421–442. [Google Scholar] [CrossRef]
- Chen, R.Z.; Deng, Y.W.; Ding, Y.L.; Guo, J.X.; Qiu, J.; Wang, B.; Wang, C.S.; Xie, Y.Y.; Zhang, Z.H.; Chen, J.X.; et al. Rice functional genomics: Decades’ efforts and roads ahead. Sci. China Life Sci. 2022, 65, 33–92. [Google Scholar]
- Li, X.Y.; Qian, Q.; Fu, Z.M.; Wang, Y.H.; Xiong, G.S.; Zeng, D.L.; Wang, X.Q.; Liu, X.F.; Teng, S.; Hiroshi, F.; et al. Control of tillering in rice. Nature 2003, 422, 618–621. [Google Scholar] [CrossRef]
- Shao, G.N.; Lu, Z.F.; Xiong, J.S.; Wang, B.; Jing, Y.H.; Meng, X.B.; Liu, G.F.; Ma, H.Y.; Liang, Y.; Chen, F.; et al. Tiller bud formation regulators MOC1 and MOC3 cooperatively promote tiller bud outgrowth by activating FON1 expression in rice. Mol. Plant 2019, 12, 1090–1102. [Google Scholar] [CrossRef]
- Lei, L.; Zheng, H.L.; Wang, J.G.; Liu, H.L.; Sun, J.; Zhao, H.W.; Yang, L.M.; Zou, D.T. Genetic dissection of rice (Oryza Sativa L.) tiller, plant height, and grain yield based on QTL mapping and metaanalysis. Euphytica 2018, 214, 109. [Google Scholar] [CrossRef]
- Yan, J.Q.; Zhu, J.; He, C.X.; Benmoussa, M.; Wu, P. Quantitative trait loci analysis for the developmental behavior of tiller number in rice (Oryza Sativa L.). Theor. Appl. Genet. 1998, 97, 267–274. [Google Scholar] [CrossRef]
- Liang, W.H.; Shang, F.; Lin, Q.T.; Lou, C.; Zhang, J. Tillering and panicle branching genes in rice. Gene 2014, 537, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.Q.; Li, F.M.; Zhang, Q.; Wang, X.Q.; Guo, H.F.; Xie, J.Y.; Zhu, X.Y.; Ullah Khan, N.; Zhang, Z.Y.; Li, J.J.; et al. Genetic architecture to cause dynamic change in tiller and panicle numbers revealed by genome-wide association study and transcriptome profile in rice. Plant J. 2020, 104, 1603–1616. [Google Scholar] [CrossRef]
- Zhao, D.D.; Park, J.R.; Jang, Y.H.; Kim, E.G.; Du, X.X.; Farooq, M.; Yun, B.J.; Kim, K.M. Identification of one major QTL and a novel gene OsIAA17q5 associated with tiller number in rice using QTL analysis. Plants 2022, 11, 538. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.X.; Shang, L.G.; Yu, H.; Zeng, L.J.; Hu, J.; Ni, S.; Rao, Y.C.; Li, S.F.; Chu, J.F.; Meng, X.B.; et al. A strigolactone biosynthesis gene contributed to the Green Revolution in rice. Mol. Plant 2020, 13, 923–932. [Google Scholar] [CrossRef]
- Hua, L.; Wang, D.R.; Tan, L.B.; Fu, Y.C.; Liu, F.X.; Xiao, L.T.; Zhu, Z.F.; Fu, Q.; Sun, X.Y.; Gu, P.; et al. LABA1, a domestication gene associated with long, barbed awns in wild rice. Plant Cell 2015, 27, 1875–1888. [Google Scholar] [CrossRef]
- Gu, B.G.; Zhou, T.Y.; Luo, J.H.; Liu, H.; Wang, Y.C.; Shangguan, Y.Y.; Zhu, J.J.; Li, Y.; Sang, T.; Wang, Z.X.; et al. An-2 encodes a cytokinin synthesis enzyme that regulates awn length and grain production in rice. Mol. Plant 2015, 8, 1635–1650. [Google Scholar] [CrossRef]
- Monna, L.; Lin, X.; Kojima, S.; Sasaki, T.; Yano, M. Genetic dissection of a genomic region for a quantitative trait locus, Hd3, into two loci, Hd3a and Hd3b, controlling heading date in rice. Theor. Appl. Genet. 2022, 104, 772–778. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, H.; Tachibana, C.; Tamaki, S.; Taoka, K.; Kyozuka, J.; Shimamoto, K. Hd3a promotes lateral branching in rice. Plant J. 2015, 82, 256–266. [Google Scholar] [CrossRef]
- Lu, Z.F.; Shao, G.N.; Xiong, J.S.; Jiao, Y.Q.; Wang, J.; Liu, G.F.; Meng, X.B.; Liang, Y.; Xiong, G.S.; Wang, Y.H.; et al. MONOCULM 3, an ortholog of WUSCHEL in rice, is required for tiller bud formation. J. Genet. Genom. 2015, 42, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.Y.; Chen, H.Q.; Dong, S.J.; Ma, Z.Y.; Ren, H.B.; Zhu, X.D.; Fang, X.H.; Chen, F. OSWUS promotes tiller bud growth by establishing weak apical dominance in rice. Plant J. 2020, 104, 1635–1647. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.; Jung, K.H.; Lee, D.E.; Lee, D.Y.; Lee, J.W.; An, K.; Kang, H.G.; An, G. The rice FON1 gene controls vegetative and reproductive development by regulating shoot apical meristem size. Mol. Cells 2006, 21, 147–152. [Google Scholar] [PubMed]
- Wen, X.X.; Sun, L.P.; Chen, Y.Y.; Xue, P.; Yang, Q.Q.; Wang, B.F.; Yu, N.; Cao, Y.R.; Zhang, Y.; Gong, K.; et al. Rice Dwarf and Low Tillering 10 (OsDLT10) regulates tiller number by monitoring auxin homeostasis. Plant Sci. 2020, 297, 110502. [Google Scholar] [CrossRef] [PubMed]
- Garoma, B. Effect of sowing density on tiller number, fresh biomass and soil cover of two green manure crops. Int. J. Agric. Biol. 2018, 8, 49–53. [Google Scholar]
- Liu, G.F.; Zhu, H.T.; Zhang, G.Q.; Li, L.H.; Ye, G.Y. Dynamic analysis of QTLs on tiller number in rice (Oryza sativa L.) with single segment substitution lines. Theor. Appl. Genet. 2012, 125, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.F.; Zeng, R.Z.; Zhu, H.T.; Zhang, Z.M.; Ding, X.H.; Zhao, F.M.; Li, W.T.; Zhang, G.Q. Dynamic expression of nine QTLs for tiller number detected with single segment substitution lines in rice. Theor. Appl. Genet. 2009, 118, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Leon, A.J.; Lee, M.; Andrade, F.H. Quantitative trait loci for growing degree days to flowering and photoperiod response in Sunflower (Helianthus annuus L.). Theor. Appl. Genet. 2001, 102, 497–503. [Google Scholar] [CrossRef]
- Yang, G.H.; Xing, Y.Z.; Li, S.Q.; Ding, J.Z.; Yue, B.; Deng, K.; Li, Y.S.; Zhu, Y.G. Molecular dissection of developmental behavior of tiller number and plant height and their relationship in rice (Oryza Sativa L.). Hereditas 2006, 143, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Hemamalini, G.S.; Shashidhar, H.E.; Hittalmani, S. Molecular marker assisted tagging of morphological and physiological traits under two contrasting moisture regimes at peak vegetative stage in rice (Oryza Sativa L.). Euphytica 2000, 112, 69–78. [Google Scholar] [CrossRef]
- Kwon, Y.H.; Kabange, N.R.; Lee, J.Y.; Lee, S.M.; Cha, J.K.; Shin, D.J.; Cho, J.H.; Kang, J.W.; Ko, J.M.; Lee, J.H. Novel QTL associated with shoot branching identified in doubled haploid rice (Oryza Sativa L.) under low nitrogen cultivation. Genes 2021, 12, 745. [Google Scholar] [CrossRef]
- Yu, S.B.; Li, J.X.; Xu, C.G.; Tan, Y.F.; Gao, Y.J.; Li, X.H.; Zhang, Q.; Saghai Maroof, M.A. Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. Proc. Natl. Acad. Sci. USA 1997, 94, 9226–9231. [Google Scholar] [CrossRef]
- Li, J.X.; Yu, S.B.; Xu, C.G.; Tan, Y.F.; Gao, Y.J.; Li, X.H.; Zhang, Q.F. Analyzing quantitative trait loci for yield using a vegetatively replicated F2 population from a cross between the parents of an elite rice hybrid. Theor. Appl. Genet. 2000, 101, 248–254. [Google Scholar] [CrossRef]
- Xing, Z.; Tan, F.; Hua, P.; Sun, L.; Xu, G.; Zhang, Q. Characterization of the main effects, epistatic effects and their environmental interactions of QTLs on the genetic basis of yield traits in rice. Theor. Appl. Genet. 2002, 105, 248–257. [Google Scholar] [CrossRef]
- Hua, J.P.; Xing, Y.Z.; Wu, W.R.; Xu, C.G.; Sun, X.L.; Yu, S.B.; Zhang, Q.F. Single-locus heterotic effects and dominance by dominance interactions can adequately explain the genetic basis of heterosis in an elite rice hybrid. Theor. Appl. Genet. 2002, 105, 248–257. [Google Scholar] [CrossRef]
- Hua, J.P.; Xing, Y.Z.; Xu, C.G.; Sun, X.L.; Yu, S.B.; Zhang, Q.F. Genetic dissection of an elite rice hybrid revealed that heterozygotes are not always advantageous for performance. Genetics 2002, 162, 1885–1895. [Google Scholar] [CrossRef]
- Aamir, R.; Wang, H.M.; Zhang, Z.H.; Anley, Z.W.; Li, Y.H.; Wang, H.; Xue, P.; Peng, Z.Q.; Shen, X.H.; Cheng, S.H.; et al. Development of chromosome segment substitution lines and genetic dissection of grain size related locus in rice. Rice Sci. 2021, 28, 322–324. [Google Scholar]
- Yang, H.L.; Yang, Q.Q.; Kang, Y.W.; Zhang, M.; Zhan, X.D.; Cao, L.Y.; Cheng, S.H.; Wu, W.W.; Zhang, Y.X. Finding stable QTL for plant height in super hybrid rice. Agriculture 2022, 12, 165. [Google Scholar] [CrossRef]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic. Acids. Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.Z.; Wen, D.Q.; Xie, J.H.; Liu, L.Q.; Wei, Y.Z.; Wang, Y.C.; Shi, S.Y. A rapid and effective method for silver staining of PCR products separated in polyacrylamide gels. Electrophoresis 2014, 35, 2520–2523. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.Y.; Xing, Y.Z.; Weng, X.Y.; Zhao, Y.; Tang, W.J.; Wang, L.; Zhou, H.J.; Yu, S.B.; Xu, C.G.; Li, X.H.; et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008, 40, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.S.; Luo, L.J.; Xu, C.G.; Zhang, Q.F.; Xing, Y.Z. Quantitative trait loci for panicle size, heading date and plant height co-segregating in trait-performance derived near-isogenic lines of rice (Oryza Sativa). Theor. Appl. Genet. 2006, 113, 361–368. [Google Scholar] [CrossRef]
- Hittalmani, S.; Parco, A.; Mew, T.V.; Zeigler, R.S.; Huang, N. Fine mapping and DNA marker-assisted pyramiding of the three major genes for blast resistance in rice. Theor. Appl. Genet. 2000, 100, 1121–1128. [Google Scholar] [CrossRef]
- Huang, C.L.; Hwang, S.Y.; Chiang, Y.C.; Lin, T.P. Molecular evolution of the Pi-Ta gene resistant to rice Blast in wild rice (Oryza rufipogon). Genetics 2008, 179, 1527–1538. [Google Scholar] [CrossRef]
- Wang, G.L.; Mackill, D.J.; Bonman, J.M.; McCouch, S.R.; Champoux, M.C.; Nelson, R.J. RFLP mapping of genes conferring complete and partial resistance to blast in a durably resistant rice cultivar. Genetics 1994, 136, 1421–1434. [Google Scholar] [CrossRef] [PubMed]
- Tu, D.B.; Wu, W.G.; Xi, M.; Zhou, Y.J.; Xu, Y.Z.; Chen, J.H.; Shao, C.H.; Zhang, Y.P.; Zhao, Q.Z. Effect of temperature and radiation on indica rice yield and quality in middle rice cropping system. Plants 2022, 11, 2697. [Google Scholar] [CrossRef]
- Ray, J.D.; Yu, L.; McCouch, S.R.; Champoux, M.C.; Wang, G.; Nguyen, H.T. Mapping quantitative trait loci associated with root penetration ability in rice (Oryza Sativa L.). Theor. Appl. Genet. 1996, 92, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.W.; Yang, C.; Cao, J.D.; Chen, H.; Pang, J.H.; Zhao, Q.Q.; Wang, Z.Y.; Fu, Z.Q.; Liu, J. A BHLH transcription activator regulates defense signaling by nucleo-cytosolic trafficking in rice. J. Integr. Plant Biol. 2020, 62, 1552–1573. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.; Fan, Y.; Li, D.; Sun, Z.; Ruan, Z.; Yang, H.; Kang, Y.; Wu, W.; Liu, Q.; Zhan, X.; et al. Genetic Dissection of Tiller Number qTN4 in Rice. Agriculture 2023, 13, 411. https://doi.org/10.3390/agriculture13020411
Chen H, Fan Y, Li D, Sun Z, Ruan Z, Yang H, Kang Y, Wu W, Liu Q, Zhan X, et al. Genetic Dissection of Tiller Number qTN4 in Rice. Agriculture. 2023; 13(2):411. https://doi.org/10.3390/agriculture13020411
Chicago/Turabian StyleChen, Hongmei, Yongyi Fan, Dian Li, Zhihao Sun, Zheyan Ruan, Huali Yang, Yiwei Kang, Weixun Wu, Qunen Liu, Xiaodeng Zhan, and et al. 2023. "Genetic Dissection of Tiller Number qTN4 in Rice" Agriculture 13, no. 2: 411. https://doi.org/10.3390/agriculture13020411
APA StyleChen, H., Fan, Y., Li, D., Sun, Z., Ruan, Z., Yang, H., Kang, Y., Wu, W., Liu, Q., Zhan, X., Cao, L., Zhou, M., Cheng, S., & Zhang, Y. (2023). Genetic Dissection of Tiller Number qTN4 in Rice. Agriculture, 13(2), 411. https://doi.org/10.3390/agriculture13020411






