Research on the Method of Identifying the Severity of Wheat Stripe Rust Based on Machine Vision
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Source
2.2. Wheat Leaf Segmentation Method
2.2.1. Principle of SLIC Superpixel Segmentation
2.2.2. SLIC Segmentation Parameters
2.3. Wheat Stripe Rust Lesion Classification Method
2.3.1. Principles of Random Forest Classification Algorithm
2.3.2. Experimental Parameters Setting
2.4. Wheat Stripe Rust Severity Determination Method
2.4.1. Wheat Stripe Rust Severity Grading Standard
2.4.2. Wheat Stripe Rust Severity Identification Method
2.5. Comparison Methods
2.5.1. Methods for Wheat Stripe Rust Lesion Sub-Region Identification
2.5.2. Methods for Wheat Stripe Rust Lesion Extraction
2.6. Accuracy Evaluation Metrics
2.6.1. Accuracy Evaluation Metrics for the Identification Model
2.6.2. Accuracy Evaluation Metrics for Prediction Results
3. Results and Discussion
3.1. Results of the SLIC Superpixel Segmentation
3.2. Results of the Sub-Region Classification
3.2.1. Results of the Classification Model Training
3.2.2. Results of the Classification Model Predictions
3.3. Results of the Wheat Stripe Rust Lesion Extraction
3.4. Results of the Wheat Stripe Rust Severity Identification
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, W.; Wang, B.; Zhao, Z.; Li, Y.; Kang, Z. A historical review and suggestions for countermeasures against successive epidemics of wheat stripe rust in China. China Plant Prot. 2022, 42, 21–27. (In Chinese) [Google Scholar]
- Ma, Z. Researches and control of wheat stripe rust in China. J. Plant Prot. 2018, 45, 1–6. (In Chinese) [Google Scholar]
- Zhao, Y.; Jing, X.; Huang, W.; Dong, Y.; Li, C. Comparison of Sun-Induced Chlorophyll Fluorescence and Reflectance Data on Estimating Severity of Wheat Stripe Rust. Spectrosc. Spectr. Anal. 2019, 39, 2739–2745. (In Chinese) [Google Scholar]
- Zhao, Y.; Li, X.; Yu, K.; Cheng, F.; He, Y. Hyperspectral Imaging for Determining Pigment Contents in Cucumber Leaves in Response to Angular Leaf Lesion Disease. Sci. Rep. 2016, 6, 27790. [Google Scholar] [CrossRef]
- Bohnenkamp, D.; Behmann, J.; Mahlein, A.-K. In-Field Detection of Yellow Rust in Wheat on the Ground Canopy and UAV Scale. Remote Sens. 2019, 11, 2495. [Google Scholar] [CrossRef]
- Li, X.; Qin, F.; Zhao, L.; Li, J.; Ma, Z.; Wang, H. Grading identification of wheat stripe rust severity using near-infrared spectroscopy. Spectrosc. Spectr. Anal. 2015, 35, 367–371. (In Chinese) [Google Scholar]
- Wang, H.; Ma, Z.; Wang, T.; Cai, C.; An, H.; Zhang, L. Application of hyperspectral imaging in grading identification of wheat stripe rust severity. Spectrosc. Spectr. Anal. 2007, 9, 1811–1814. (In Chinese) [Google Scholar]
- Jiang, X. Study on the Estimation Method of the Severity of Wheat Stripe Rust by Near-Ground Remote Sensing. Master’s Thesis, An Hui University of Science and Technology, Huainan, China, 2023. (In Chinese). [Google Scholar]
- Zhang, N.; Yang, G.; Zhao, C.; Zhang, J.; Yang, X.; Pan, Y.; Huang, W.; Xu, B.; Li, M.; Zhu, X.; et al. Progress and prospect of hyperspectral remote sensing technology for crop diseases and pests. Natl. Remote Sens. Bull. 2021, 25, 403–422. (In Chinese) [Google Scholar] [CrossRef]
- Sladojevic, S.; Arsenovic, M.; Anderla, A.; Culibrk, D.; Stefanovic, D. Deep Neural Networks Based Recognition of Plant Diseases by Leaf Image Classification. Comput. Intell. Neurosci. 2016, 2016, 3289801. [Google Scholar] [CrossRef]
- Yuan, H.; Zhu, J.; Wang, Q.; Cheng, M.; Cai, Z. An Improved DeepLab v3+ Deep Learning Network Applied to the Segmentation of Grape Leaf Black Rot Spots. Front. Plant Sci. 2022, 13, 795410. [Google Scholar] [CrossRef]
- Wang, C.; Du, P.; Wu, H.-R.; Li, J.; Zhao, C.; Zhu, H. A cucumber leaf disease severity classification method based on the fusion of DeepLabV3+ and U-Net. Comput. Electron. Agric. 2021, 189, 106373. [Google Scholar] [CrossRef]
- Guo, W.; Dang, M.; Jia, X.; He, Q.; Gao, C.; Dong, P. Wheat stripe rust disease severity identification based on deep learning. J. South China Agri. Univ. 2023, 44, 604–612. (In Chinese) [Google Scholar]
- Mi, Z. Study on Judging Method of Wheat Stripe Rust Infection Level Based on Machine Vision. Master’s Thesis, Northwest A&F University, Xianyang, China, 2022. (In Chinese). [Google Scholar]
- Bao, W.; Lin, Z.; Hu, G.; Liang, D.; Huang, L.; Yang, X. Severity Estimation of Wheat Leaf Diseases Based on RSTCNN. Trans. Chin. Soc. Agric. Mach. 2021, 52, 242–252. (In Chinese) [Google Scholar]
- Yu, X.; Xu, C.; Wang, D.; Zhang, W.; Qu, W.; Song, H. Identification of Wheat Leaf Diseases Based on SVM Method. J. Agric. Mech. Res. 2014, 36, 151–155. (In Chinese) [Google Scholar]
- Pulido Rojas, C.; Solaque Guzmán, L.; Velasco Toledo, N. Weed recognition by SVM texture feature classification in outdoor vegetable crops images. Ing. Investig. 2017, 37, 68–74. [Google Scholar] [CrossRef]
- Padol, P.B.; Yadav, A.A. SVM classifier based grape leaf disease detection. In Proceedings of the 2016 Conference on Advances in Signal Processing (CASP), Pune, India, 9–11 June 2016; pp. 175–179. [Google Scholar]
- Zhu, J.; Wu, A.; Wang, X.; Zhang, H. Identification of grape diseases using image analysis and BP neural networks. Multimed. Tools Appl. 2019, 79, 14539–14551. [Google Scholar] [CrossRef]
- Singh, R.N.; Krishnan, P.; Singh, V.K.; Banerjee, K. Application of thermal and visible imaging to estimate stripe rust disease severity in wheat using supervised image classification methods. Ecol. Inform. 2022, 71, 101774. [Google Scholar] [CrossRef]
- Jiang, X.; Feng, H.; Chang, H.; Yang, G.; Yang, X. Classification method of wheat stripe rust disease degree based on digital image. Jiangsu Agric. Sci. 2021, 49, 109–115. (In Chinese) [Google Scholar]
- Getachew, H.; Hawi, T. Wheat Leaf Dataset. Mendeley Data, V1.2021. Available online: https://data.mendeley.com/datasets/wgd66f8n6h/1 (accessed on 17 June 2023).
- Wang, C.; Chen, J.; Li, W. Review on superpixel segmentation algorithms. Appl. Res. Comput. 2014, 31, 6–12. (In Chinese) [Google Scholar]
- Song, X.; Zhou, L.; Li, Z.; Chen, J.; Zeng, L.; Yan, B. Review on superpixel methods in image segmentation. J. Image Graph. 2015, 20, 599–608. (In Chinese) [Google Scholar]
- Achanta, R.; Shaji, A.; Smith, K.; Lucchi, A.; Fua, P.; Süsstrunk, S. SLIC Superpixels Compared to State-of-the-Art Superpixel Methods. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 2274–2281. [Google Scholar] [CrossRef]
- Achanta, R.; Shaji, A.; Smith, K.; Lucchi, A.; Fua, P.; Süsstrunk, S. SLIC Superpixels; Technical Report; EPFL: Lausanne, Switzerland, 2010. [Google Scholar]
- Breiman, L. Random Forests. Mach Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Man, W.; Ji, Y.; Zhang, Z. Image classification based on improved random forest algorithm. In Proceedings of the 2018 IEEE 3rd International Conference on Cloud Computing and Big Data Analysis (ICCCBDA), Chengdu, China, 20–22 April 2018; pp. 346–350. [Google Scholar]
- Zhang, Z.; Li, S. Polarimetric SAR image classification based on AdaBoost improved random forest and SVM. J. Univ. Chin. Acad. Sci. 2022, 39, 776–782. (In Chinese) [Google Scholar]
- Xu, B.; Ye, Y.; Nie, L. An improved random forest classifier for image classification. In Proceedings of the 2012 IEEE International Conference on Information and Automation, Shenyang, China, 6–8 June 2012; pp. 795–800. [Google Scholar]
- Chaudhary, A.; Kolhe, S.; Kamal, R. An improved random forest classifier for multi-class classification. Inf. Process. Agric. 2016, 3, 215–222. [Google Scholar] [CrossRef]
- GB/T 15795-2011; Rules for monitoring and forecast of the wheat stripe rust. National Agro-Tech Extension and Service Center; Northwest A&F University: Xianyang, China, 2011.
- Li, C.; Li, Y.; Tan, H.; Wang, X.; Zhai, C. Grading Detection Method of Grape Downy Mildew Based on K-means Clustering and Random Forest Algorithm. Trans. Chin. Soc. Agric. Mach. 2022, 53, 225–236. (In Chinese) [Google Scholar]
- Chen, T.; Guestrin, C. XG Boost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016. [Google Scholar]
- Freund, Y.; Schapire, R.E. A Decision-Theoretic Generalization of On-Line Learning and an Application to Boosting. J. Comput. Syst. Sci. 1997, 55, 119–139. [Google Scholar] [CrossRef]
- Meyer, F. Color image segmentation. In Proceedings of the 1992 International Conference on Image Processing and its Applications, Maastricht, The Netherlands, 7–9 April 1992; pp. 303–306. [Google Scholar]
- Chinrungrueng, C.; Séquin, C.H. Optimal adaptive k-means algorithm with dynamic adjustment of learning rate. In IJCNN-91-Seattle International Joint Conference on Neural Networks; IEEE: New York, NY, USA, 1991; Volume 851, pp. 855–862. [Google Scholar]








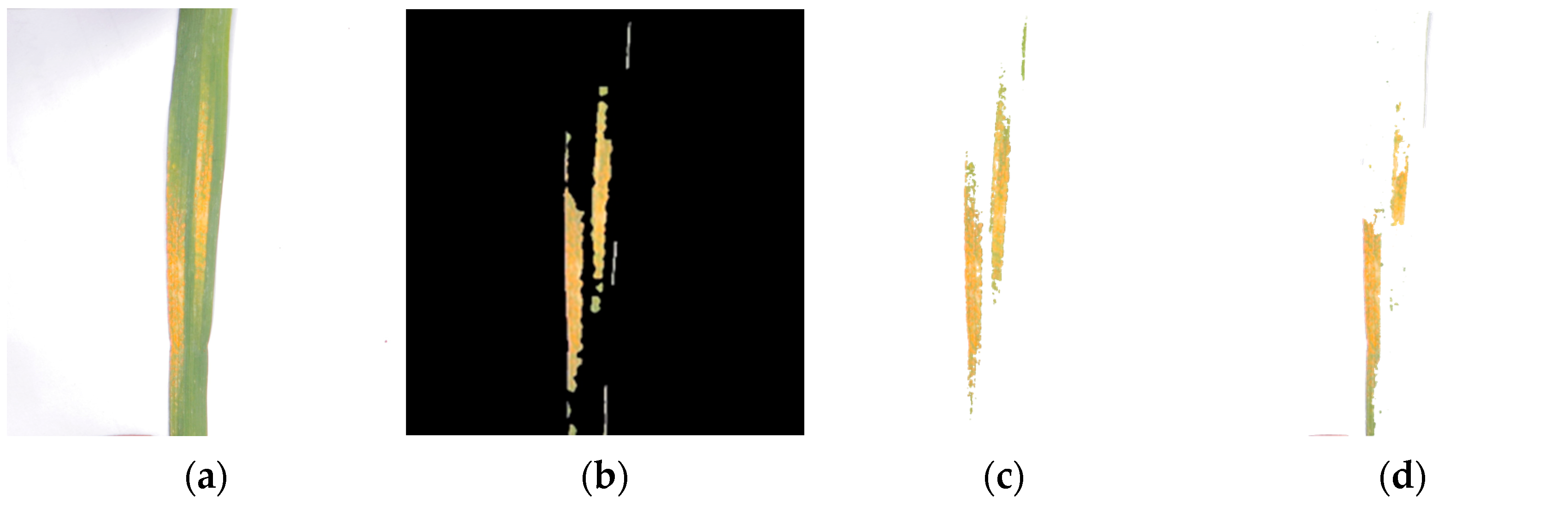
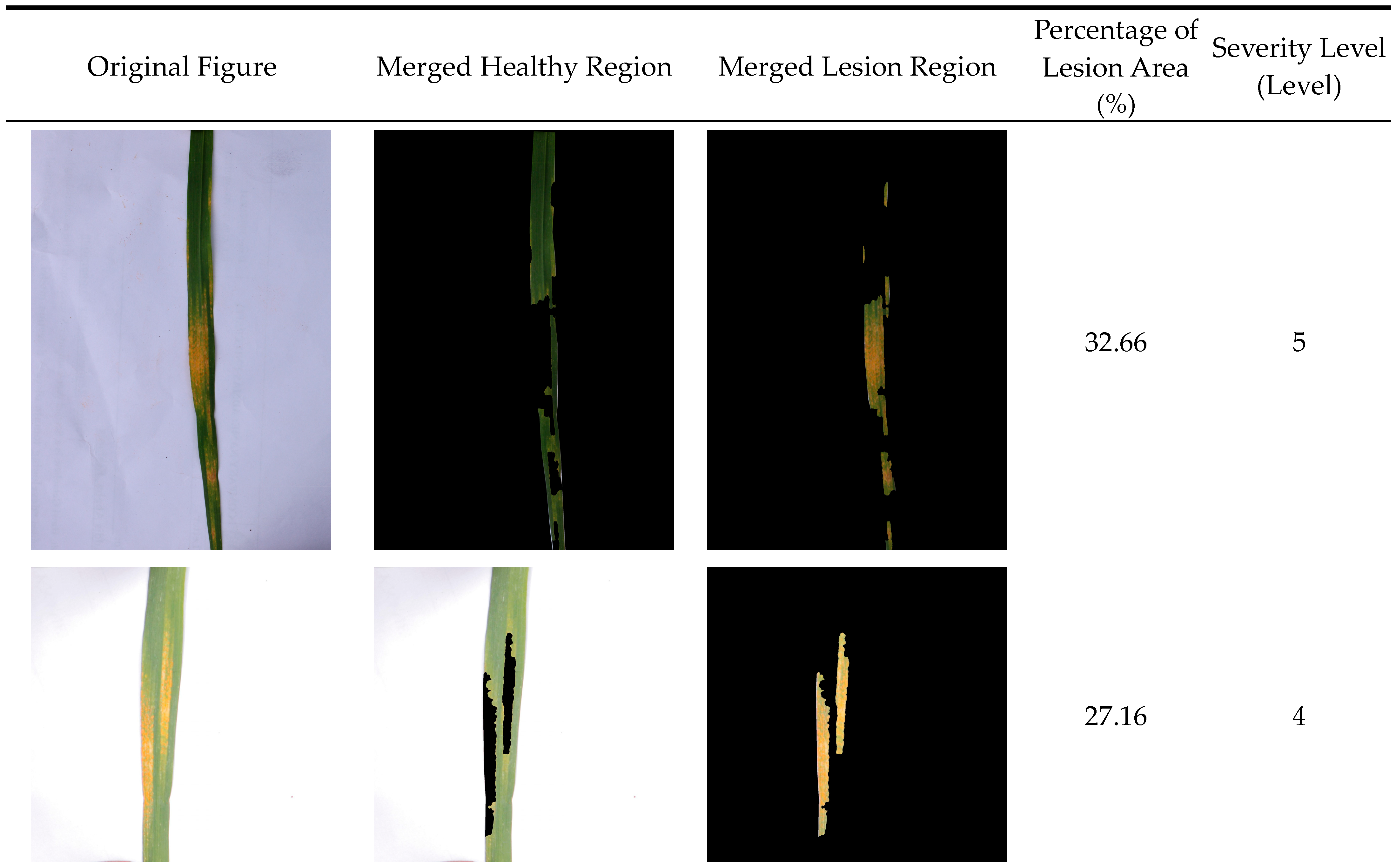

| Disease Severity | 0 Level | 1 Level | 2 Level | 3 Level | 4 Level | 5 Level | 6 Level | 7 Level | 8 Level |
|---|---|---|---|---|---|---|---|---|---|
| Percentage of diseased area relative to total leaf area (%) | 0 | 1 | 5 | 10 | 20 | 40 | 60 | 80 | 100 |
| Model | Label | Precision (%) | Recall (%) | F1 Score (%) | Accuracy (%) | Cross-Validation Score (%) |
|---|---|---|---|---|---|---|
| Random forest | Disease | 94.57 | 91.67 | 93.10 | 93.22 | 90.47 |
| Healthy | 91.95 | 94.76 | 93.33 | |||
| Overall | 93.26 | 93.22 | 93.21 | |||
| XG Boost | Disease | 92.17 | 92.98 | 92.58 | 92.56 | 89.43 |
| Healthy | 92.95 | 92.14 | 92.54 | |||
| Overall | 92.56 | 92.56 | 92.56 | |||
| Ada Boost | Disease | 82.46 | 82.46 | 82.46 | 82.06 | 83.52 |
| Healthy | 82.53 | 82.53 | 82.53 | |||
| Overall | 82.49 | 82.49 | 82.49 |
| Metric | Image | Model | ||
|---|---|---|---|---|
| RF | XG Boost | Ada Boost | ||
| Learned Perceptual Image Patch Similarity (LPIPS) | 1 | 0.046 | 0.054 | 0.086 |
| 2 | 0.010 | 0.014 | 0.027 | |
| 3 | 0.010 | 0.014 | 0.042 | |
| 4 | 0.006 | 0.029 | 0.038 | |
| Average | 0.026 | 0.02775 | 0.04825 | |
| Structural Similarity Index (SSIM) | 1 | 0.9840 | 0.9797 | 0.9629 |
| 2 | 0.9940 | 0.9921 | 0.9863 | |
| 3 | 0.9948 | 0.9921 | 0.9819 | |
| 4 | 0.9973 | 0.9885 | 0.9785 | |
| Average | 0.992525 | 0.9881 | 0.9774 | |
| Mean Square Error (MSE) | 1 | 1.3898 | 1.7793 | 3.3205 |
| 2 | 0.6257 | 0.819 | 1.3649 | |
| 3 | 0.5735 | 0.8241 | 1.7484 | |
| 4 | 0.2368 | 1.1017 | 1.9488 | |
| Average | 0.70645 | 1.131025 | 2.09565 | |
| Metric | Method | ||
|---|---|---|---|
| Method of This Study | K-Means | Watershed Segmentation | |
| Learned Perceptual Image Patch Similarity (LPIPS) | 0.046 | 0.076 | 0.082 |
| Structural Similarity Index (SSIM) | 0.9840 | 0.9558 | 0.9255 |
| Mean Square Error (MSE) | 1.3898 | 2.6713 | 3.4319 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, R.; Jin, F.; Ji, M.; Zuo, Y. Research on the Method of Identifying the Severity of Wheat Stripe Rust Based on Machine Vision. Agriculture 2023, 13, 2187. https://doi.org/10.3390/agriculture13122187
Gao R, Jin F, Ji M, Zuo Y. Research on the Method of Identifying the Severity of Wheat Stripe Rust Based on Machine Vision. Agriculture. 2023; 13(12):2187. https://doi.org/10.3390/agriculture13122187
Chicago/Turabian StyleGao, Ruonan, Fengxiang Jin, Min Ji, and Yanan Zuo. 2023. "Research on the Method of Identifying the Severity of Wheat Stripe Rust Based on Machine Vision" Agriculture 13, no. 12: 2187. https://doi.org/10.3390/agriculture13122187
APA StyleGao, R., Jin, F., Ji, M., & Zuo, Y. (2023). Research on the Method of Identifying the Severity of Wheat Stripe Rust Based on Machine Vision. Agriculture, 13(12), 2187. https://doi.org/10.3390/agriculture13122187








